Probing Peptide Assembly and Interaction via High-Resolution Imaging Techniques: A Mini Review
Abstract
:1. Introduction
2. Probing Peptide Assembly via High-Resolution Imaging Techniques
2.1. Micelles
2.2. Tubes
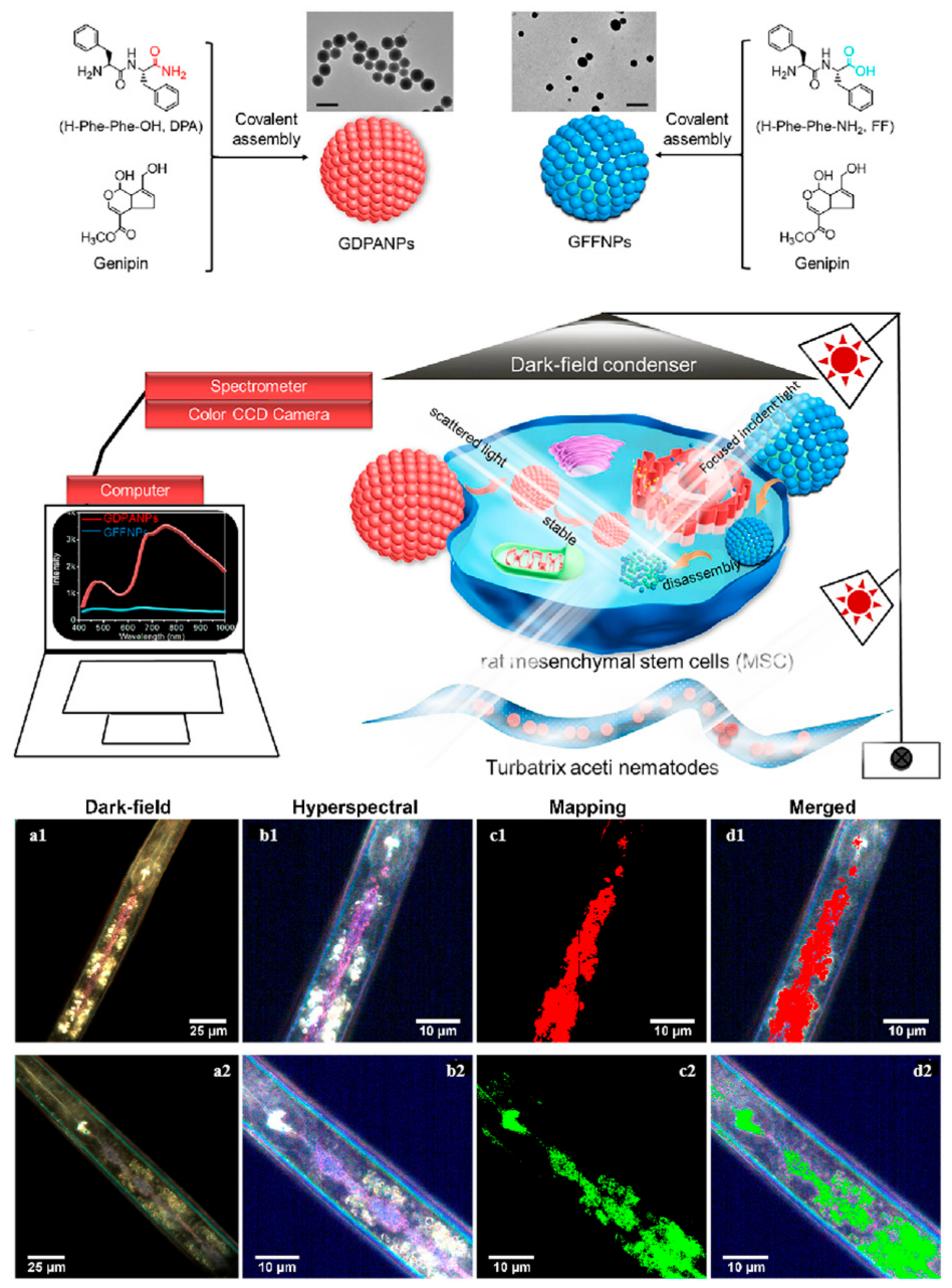
2.3. Particles
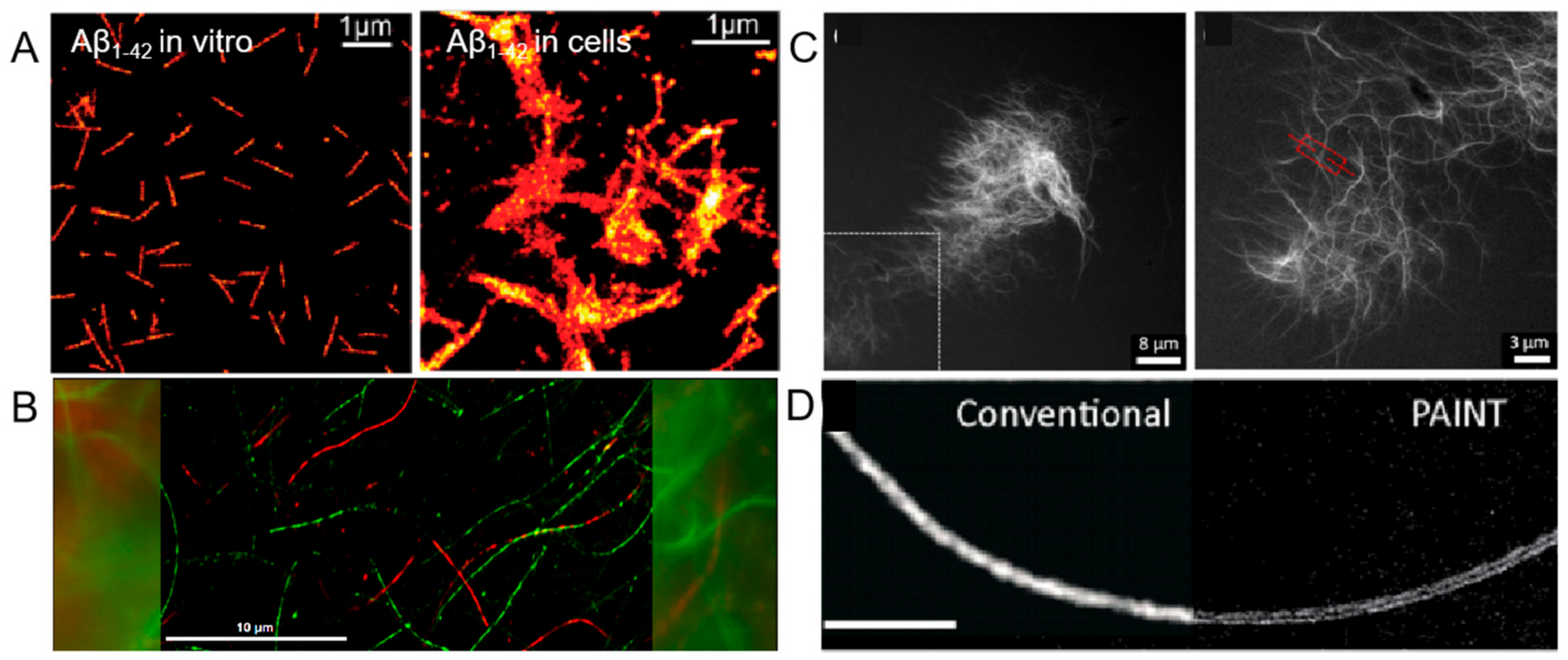
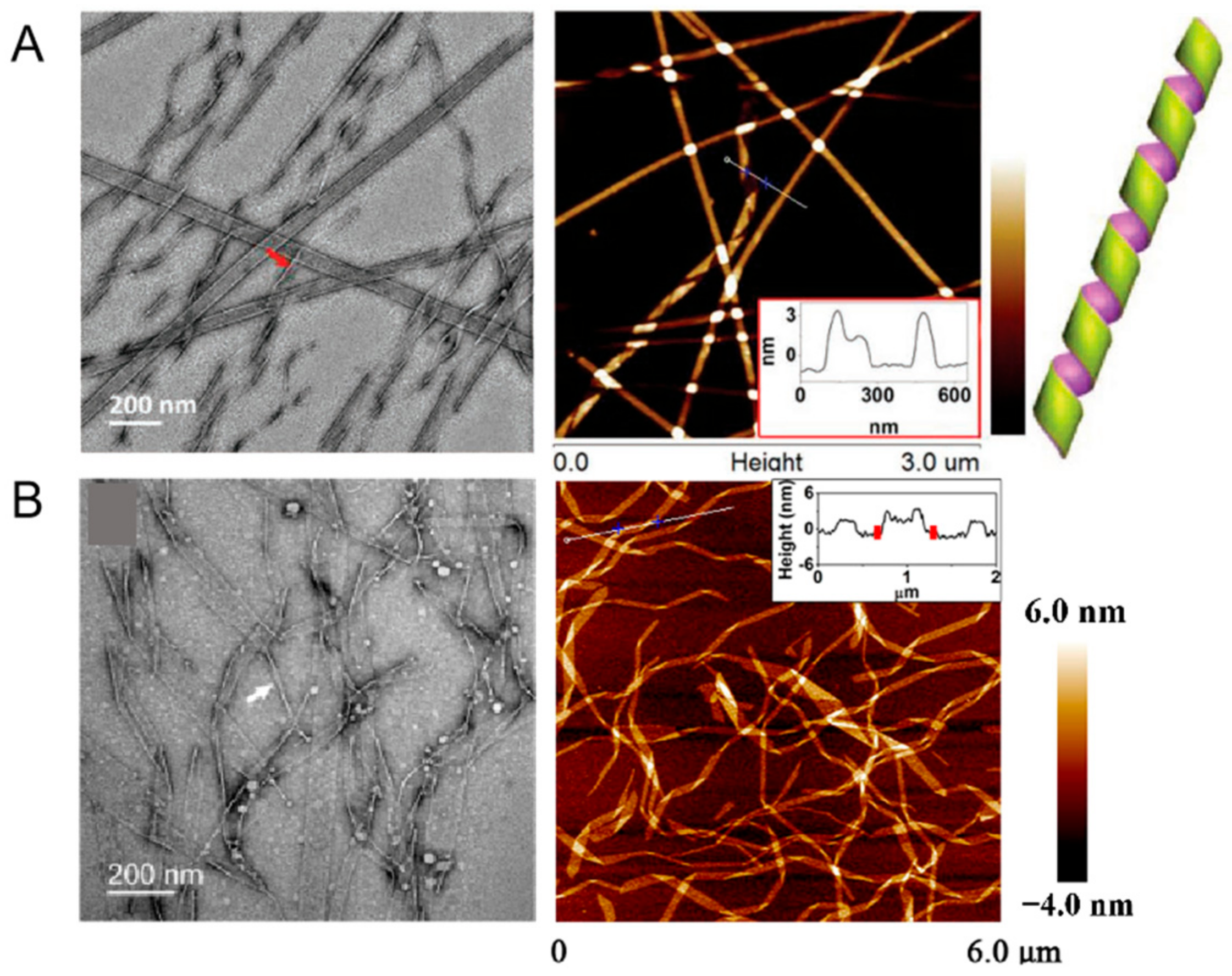
2.4. Fibers
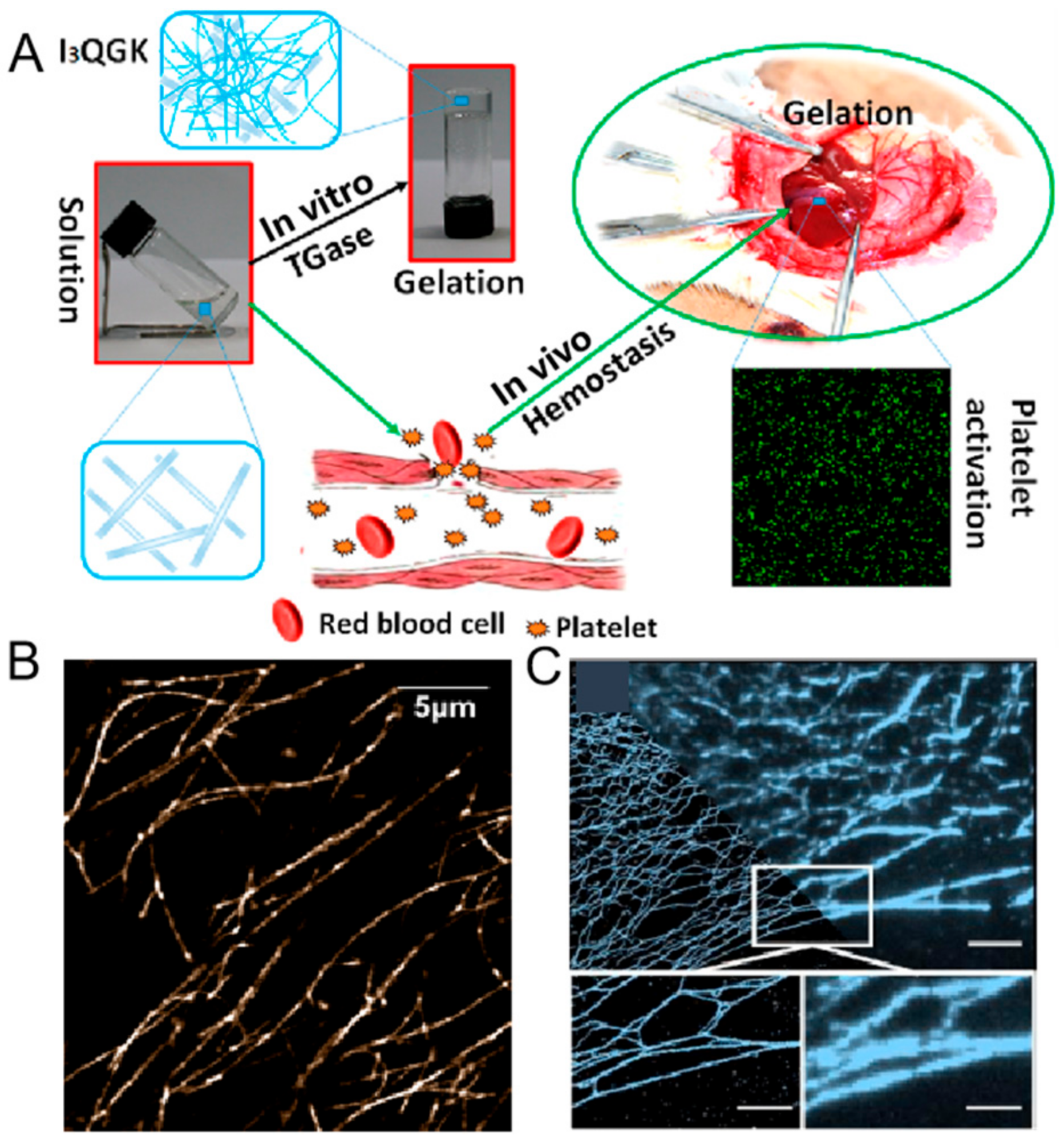
2.5. Hydrogel
3. Probing Peptide Interaction via High-Resolution Imaging Techniques
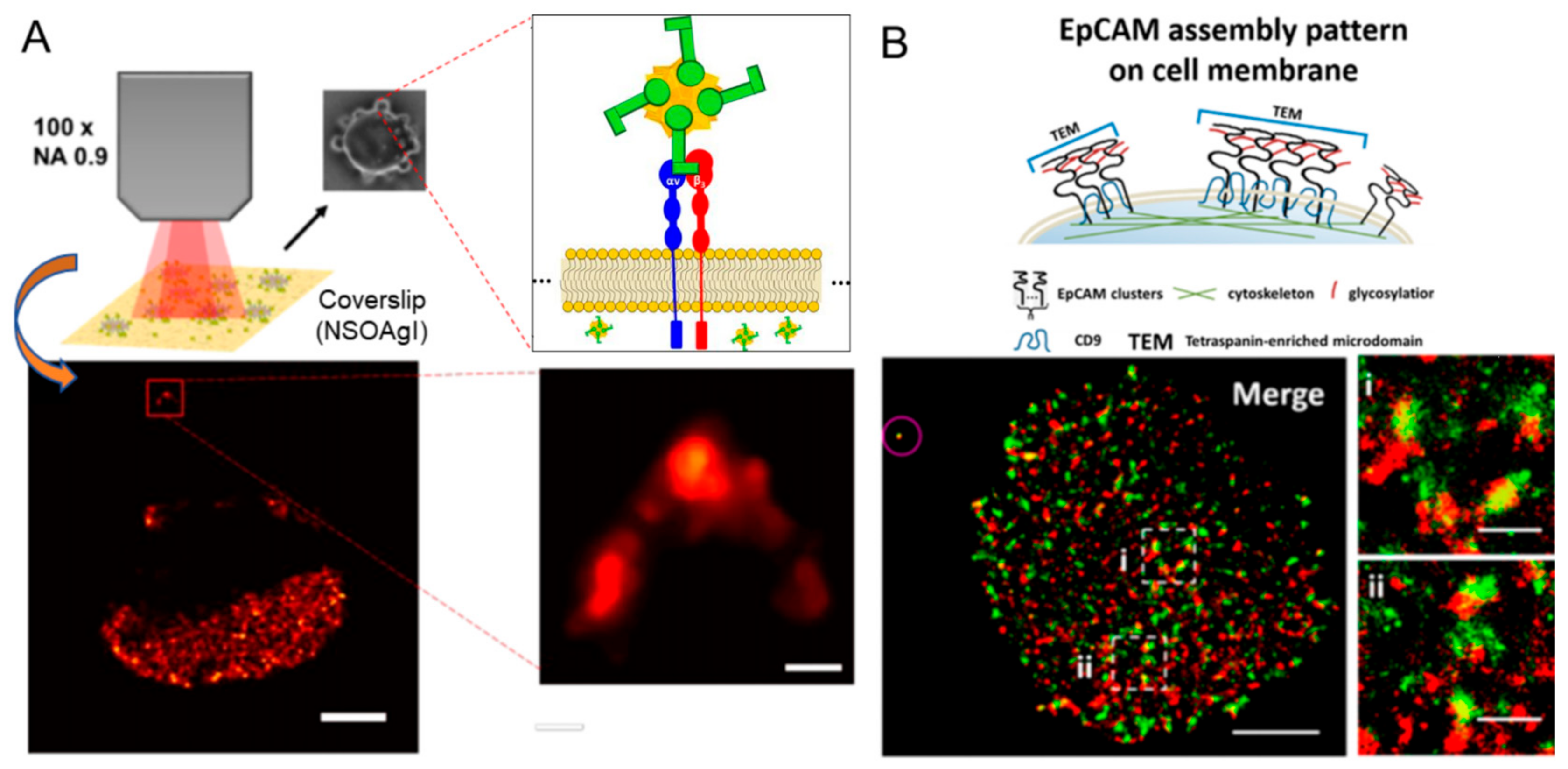
3.1. Peptide–Protein Interaction
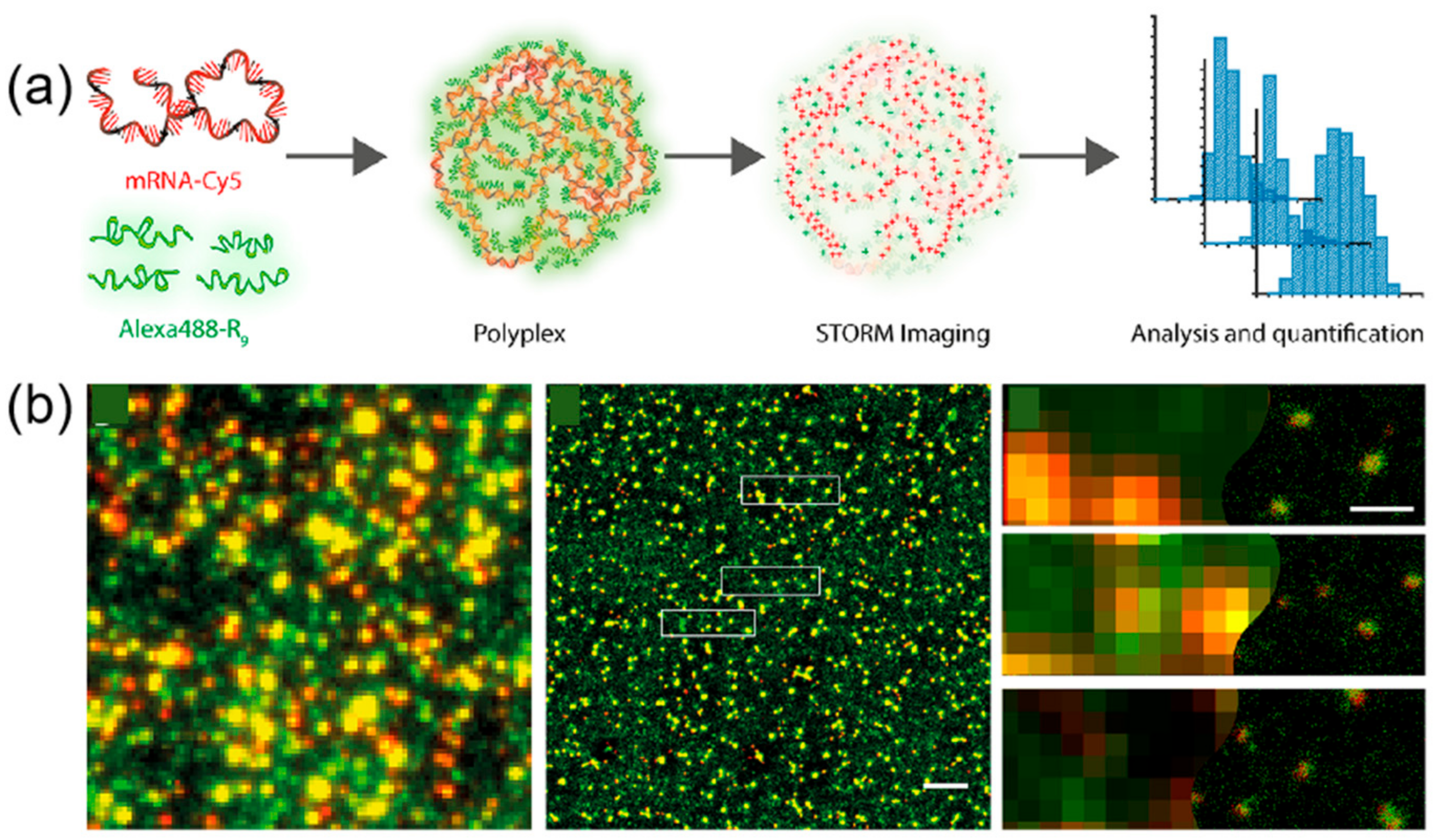
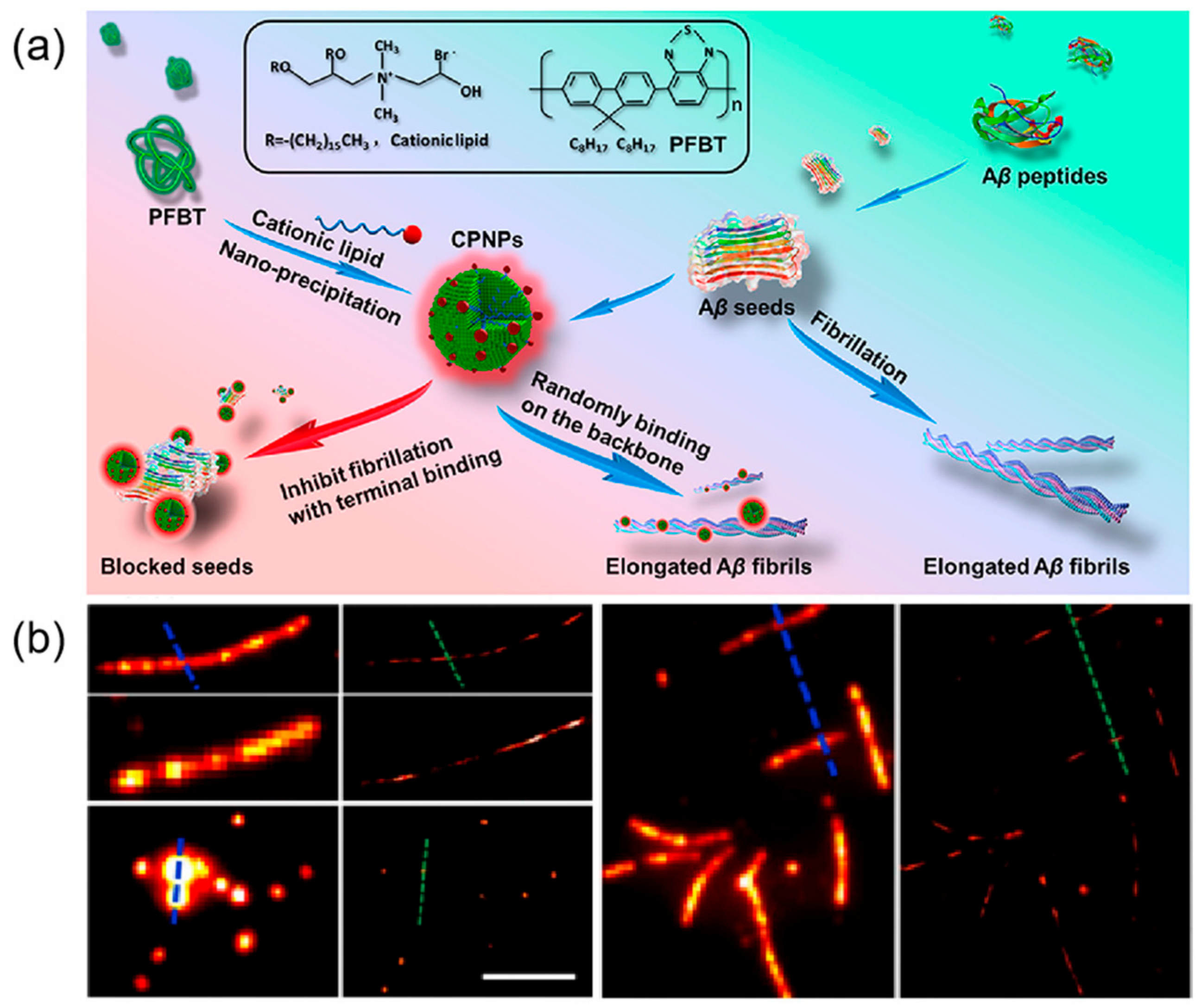
3.2. Peptide–Polymer Interaction
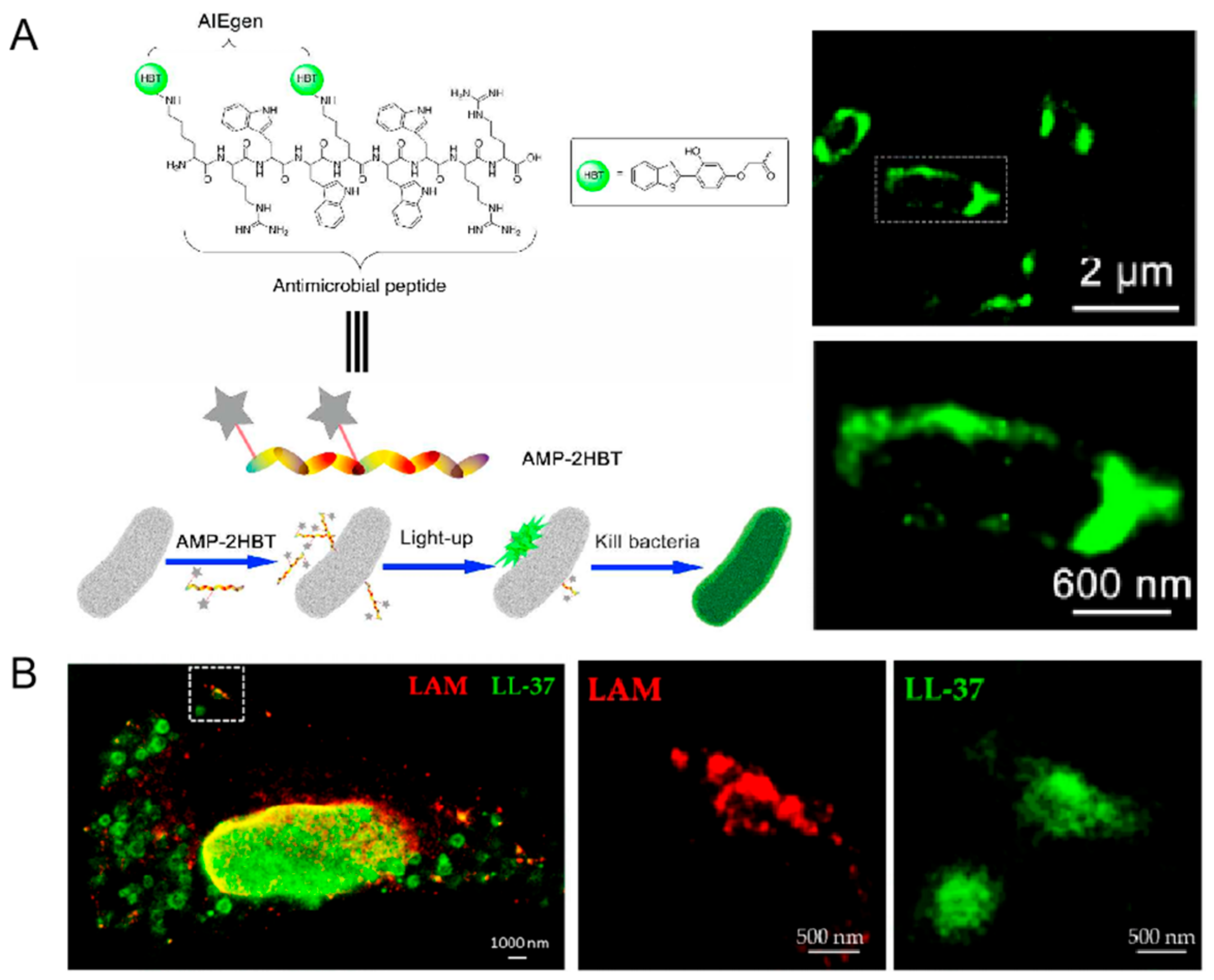
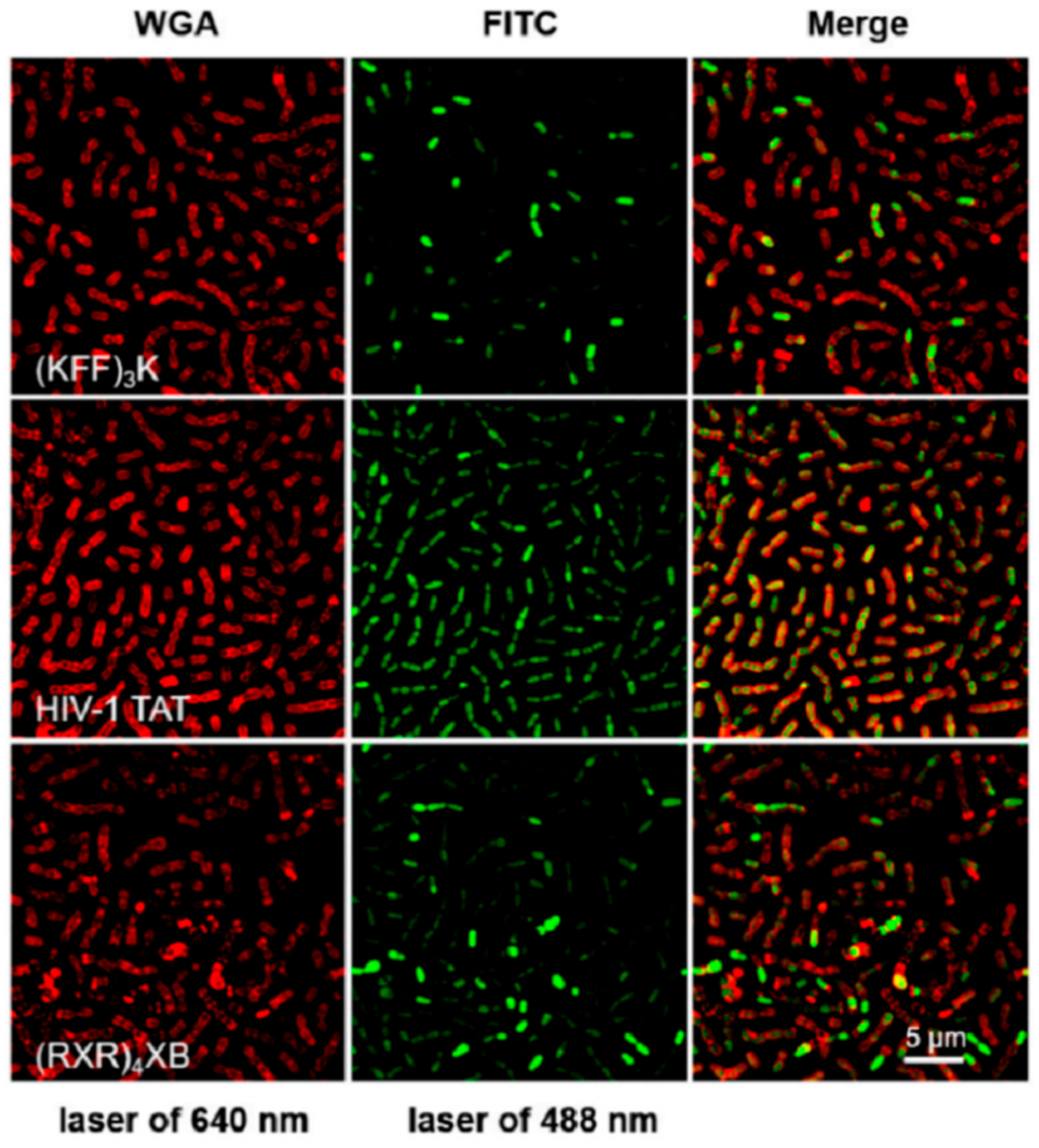
3.3. Peptide–Microbe Interaction
4. Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Chang, R.; Yuan, C.; Zhou, P.; Xing, R.; Yan, X. Peptide self-assembly: From ordered to disordered. Acc. Chem. Res. 2024, 57, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Levin, A.; Hakala, T.A.; Schnaider, L.; Bernardes, G.J.L.; Gazit, E.; Knowles, T.P.J. Biomimetic peptide self-assembly for functional materials. Nat. Rev. Chem. 2020, 4, 615–634. [Google Scholar] [CrossRef] [PubMed]
- Lou, S.; Wang, X.; Yu, Z.; Shi, L. Peptide tectonics: Encoded structural complementarity dictates programmable Self-Assembly. Adv. Sci. 2019, 6, 1802043. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Fu, L.; Hu, Z.; Zhong, Y. A mini-review on peptide-based self-assemblies and their biological applications. Nanotechnology 2021, 33, 062004. [Google Scholar] [CrossRef]
- Hamley, I.W. Small bioactive peptides for biomaterials design and therapeutics. Chem. Rev. 2017, 117, 14015–14041. [Google Scholar] [CrossRef]
- Wang, J.; Liu, K.; Xing, R.; Yan, X. Peptide self-assembly: Thermodynamics and kinetics. Chem. Soc. Rev. 2016, 45, 5589–5604. [Google Scholar] [CrossRef]
- Mason, T.O.; Buell, A.K. The Kinetics, Thermodynamics and Mechanisms of Short Aromatic Peptide Self-Assembly. Adv. Exp. Med. Biol. 2019, 1174, 61–112. [Google Scholar] [CrossRef]
- Zaguri, D.; Zimmermann, M.R.; Meisl, G.; Levin, A.; Rencus-Lazar, S.; Knowles, T.P.J.; Gazit, E. Kinetic and thermodynamic driving factors in the assembly of Phenylalanine-Based modules. ACS Nano 2021, 15, 18305–18311. [Google Scholar] [CrossRef]
- Thurston, B.A.; Tovar, J.D.; Ferguson, A.L. Thermodynamics, morphology, and kinetics of early-stage self-assembly of π-conjugated oligopeptides. Mol. Simul. 2016, 42, 955–975. [Google Scholar] [CrossRef]
- Panda, J.J.; Chauhan, V.S. Short peptide based self-assembled nanostructures: Implications in drug delivery and tissue engineering. Polym. Chem. 2014, 5, 4418–4436. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, W.; Chen, C.; Wang, J.; Zhang, L.; Xu, H. Rational design and self-assembly of short amphiphilic peptides and applications. Curr. Opin. Colloid Interface Sci. 2018, 35, 112–123. [Google Scholar] [CrossRef]
- Sivagnanam, S.; Arul, A.; Ghosh, S.; Dey, A.; Ghorai, S.; Das, P. Concentration-dependent fabrication of short-peptide-based different self-assembled nanostructures with various morphologies and intracellular delivery property. Mater. Chem. Front. 2019, 3, 2110–2119. [Google Scholar] [CrossRef]
- Guo, C.; Luo, Y.; Zhou, R.; Wei, G. Triphenylalanine peptides self-assemble into nanospheres and nanorods that are different from the nanovesicles and nanotubes formed by diphenylalanine peptides. Nanoscale 2014, 6, 2800–2811. [Google Scholar] [CrossRef]
- Nambiar, M.; Nepal, M.; Chmielewski, J. Self-Assembling Coiled-Coil Peptide Nanotubes with Biomolecular Cargo Encapsulation. ACS Biomater. Sci. Eng. 2019, 5, 5082–5087. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.; Jin, S.-M.; Han, E.H.; Ko, E.; Ahn, M.; Bang, W.-Y.; Bang, J.-K.; Lee, E. Structure-dependent antimicrobial theranostic functions of self-assembled short peptide nanoagents. Biomacromolecules 2017, 18, 3600–3610. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, T.; Du, Y.; Zhao, B.; Patel, H.P.; Boldt, R.; Auernhammer, G.K.; Fery, A.; Li, J.; Thiele, J. Titanium dioxide nanoparticles embedded in assembled dipeptide hydrogels for microfluidic photodegradation. J. Colloid Interface Sci. 2023, 654, 405–412. [Google Scholar] [CrossRef]
- Wu, A.; Guo, Y.; Li, M.; Li, Q.; Zang, H.; Li, J. Tunable chirality of self-assembled dipeptides mediated by bipyridine derivative. Angew. Chem. Int. Ed. 2023, 62, e202314368. [Google Scholar] [CrossRef]
- Das, R.; Gayakvad, B.; Shinde, S.D.; Rani, J.; Jain, A.; Sahu, B. Ultrashort peptides—A glimpse into the structural modifications and their applications as biomaterials. ACS Appl. Bio Mater. 2020, 3, 5474–5499. [Google Scholar] [CrossRef]
- Hosoyama, K.; Lazurko, C.; Muñoz, M.; McTiernan, C.D.; Alarcon, E.I. Peptide-based functional biomaterials for soft-tissue repair. Front. Bioeng. Biotechnol. 2019, 7, 205. [Google Scholar] [CrossRef]
- Qi, G.; Gao, Y.; Wang, L.; Wang, H. Self-Assembled Peptide-Based nanomaterials for biomedical imaging and therapy. Adv. Mater. 2018, 30, 1703444. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, Y.; Fei, R.; Cao, C.; Wang, M.; Wang, J.; Bai, J.; Cox, H.; Waigh, T.; Lu, J.R.; et al. Hydrogelation of the short self-assembling peptide I3QGK regulated by transglutaminase and use for rapid hemostasis. ACS Appl. Mater. Interfaces 2016, 8, 17833–17841. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Wang, Z.; Wang, Q.; Yang, C.; Liu, J. Self-assembled peptide-based nanoprobes for disease theranostics and disease-related molecular imaging. Small Methods 2019, 4, 1900403. [Google Scholar] [CrossRef]
- Rauf, S.; Susapto, H.H.; Kahin, K.; Alshehri, S.; Abdelrahman, S.; Lam, J.H.; Asad, S.; Jadhav, S.; Sundaramurthi, D.; Gao, X.; et al. Self-assembling tetrameric peptides allow in situ 3D bioprinting under physiological conditions. J. Mater. Chem. B 2021, 9, 1069–1081. [Google Scholar] [CrossRef]
- Kornmueller, K.; Lehofer, B.; Leitinger, G.; Amenitsch, H.; Prassl, R. Peptide self-assembly into lamellar phases and the formation of lipid-peptide nanostructures. Nano Res. 2017, 11, 913–928. [Google Scholar] [CrossRef] [PubMed]
- Pignataro, M.F.; Herrera, M.G.; Dodero, V.I. Evaluation of peptide/protein self-assembly and aggregation by spectroscopic methods. Molecules 2020, 25, 4854. [Google Scholar] [CrossRef]
- Banerji, B.; Chatterjee, M.; Pal, U.; Maiti, N.C. Formation of annular protofibrillar assembly by cysteine tripeptide: Unraveling the interactions with NMR, FTIR, and molecular dynamics. J. Phys. Chem. B 2017, 121, 6367–6379. [Google Scholar] [CrossRef]
- Abelein, A. Metal binding of Alzheimer’s amyloid-Β and its effect on peptide Self-Assembly. Acc. Chem. Res. 2023, 56, 2653–2663. [Google Scholar] [CrossRef]
- Pashuck, E.T.; Duchet, B.J.R.; Hansel, C.S.; Maynard, S.A.; Chow, L.W.; Stevens, M.M. Controlled Sub-Nanometer epitope spacing in a Three-Dimensional Self-Assembled peptide hydrogel. ACS Nano 2016, 10, 11096–11104. [Google Scholar] [CrossRef]
- Min, K.; Kim, D.; Lee, H.; Lin, L.; Kim, D. Direct synthesis of a covalently self-assembled peptide nanogel from a tyrosine-rich peptide monomer and its biomineralized hybrids. Angew. Chem. Int. Ed. 2018, 57, 5630–5634. [Google Scholar] [CrossRef]
- Hu, X.; Liao, M.; Gong, H.; Zhang, L.; Cox, H.; Waigh, T.A.; Lu, J.R. Recent advances in short peptide self-assembly: From rational design to novel applications. Curr. Opin. Colloid Interface Sci. 2019, 45, 1–13. [Google Scholar] [CrossRef]
- Shao, Q.; Wong, K.M.; Seroski, D.T.; Wang, Y.; Liu, R.; Paravastu, A.K.; Hudalla, G.A.; Hall, C.K. Anatomy of a selectively coassembled β-sheet peptide nanofiber. Proc. Nat. Acad. Sci. USA 2020, 117, 4710–4717. [Google Scholar] [CrossRef] [PubMed]
- Touve, M.A.; Carlini, A.S.; Gianneschi, N.C. Self-assembling peptides imaged by correlated liquid cell transmission electron microscopy and MALDI-imaging mass spectrometry. Nat. Commun. 2019, 10, 4837. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.; Zhang, X.; Hu, J.; Zhang, Y. Self-Assembly of Amyloid-Like peptides at interfaces investigated by atomic force microscopy. Sci. Adv. Mater. 2017, 9, 65–76. [Google Scholar] [CrossRef]
- Li, M.; Xi, N.; Wang, Y.; Liu, L. Nanoscale multiparametric imaging of Peptide-Assembled nanofibrillar hydrogels by atomic force microscopy. IEEE Trans. Nanotechnol. 2019, 18, 315–328. [Google Scholar] [CrossRef]
- Danino, D.; Egelman, E.H. Direct imaging and computational cryo-electron microscopy of ribbons and nanotubes. Curr. Opin. Colloid Interface Sci. 2018, 34, 100–113. [Google Scholar] [CrossRef]
- Pujals, S.; Feiner-Gracia, N.; Delcanale, P.; Voets, I.; Albertazzi, L. Super-resolution microscopy as a powerful tool to study complex synthetic materials. Nat. Rev. Chem. 2019, 3, 68–84. [Google Scholar] [CrossRef]
- Dhiman, S.; Andrian, T.; Gonzalez, B.S.; Tholen, M.M.E.; Wang, Y.; Albertazzi, L. Can super-resolution microscopy become a standard characterization technique for materials chemistry? Chem. Sci. 2022, 13, 2152–2166. [Google Scholar] [CrossRef]
- Delcanale, P.; Miret-Ontiveros, B.; Arista-Romero, M.; Pujals, S.; Albertazzi, L. Nanoscale mapping functional sites on nanoparticles by points accumulation for imaging in nanoscale topography (PAINT). ACS Nano 2018, 12, 7629–7637. [Google Scholar] [CrossRef]
- Liu, Y.; Cui, S.; Ma, W.; Wu, Y.; Xin, R.; Bai, Y.; Chen, Z.; Xu, J.; Ge, J. Direct imaging of protein clusters in metal–organic frameworks. J. Am. Chem. Soc. 2024, 146, 12565–12576. [Google Scholar] [CrossRef]
- Wang, Y.; Rodriguez, P.E.D.S.; Woythe, L.; Sánchez, S.; Samitier, J.; Zijlstra, P.; Albertazzi, L. Multicolor super-resolution microscopy of protein corona on single nanoparticles. ACS Appl. Mater. Interfaces 2022, 14, 37345–37355. [Google Scholar] [CrossRef]
- Bonilla, J.C.; Clausen, M.P. Super-resolution microscopy to visualize and quantify protein microstructural organization in food materials and its relation to rheology: Egg white proteins. Food Hydrocolloids 2022, 124, 107281. [Google Scholar] [CrossRef]
- Sun, N.; Jia, Y.; Bai, S.; Yang, Y.; Dai, L.; Li, J. Spatial mapping and quantitative evaluation of protein corona on PEGylated mesoporous silica particles by super-resolution fluorescence microscopy. J. Colloid Interface Sci. 2024, 653, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Betzig, E.; Patterson, G.H.; Sougrat, R.; Lindwasser, O.W.; Olenych, S.; Bonifacino, J.S.; Davidson, M.W.; Lippincott-Schwartz, J.; Hess, H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science 2006, 313, 1642–1645. [Google Scholar] [CrossRef]
- Klar, T.A.; Jakobs, S.; Dyba, M.; Egner, A.; Hell, S.W. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Nat. Acad. Sci. USA 2000, 97, 8206–8210. [Google Scholar] [CrossRef]
- Ma, Y.; Wen, K.; Liu, M.; Zheng, J.; Chu, K.; Smith, Z.J.; Liu, L.; Gao, P. Recent advances in structured illumination microscopy. J. Phys. Photonics 2021, 3, 024009. [Google Scholar] [CrossRef]
- Saxena, M.; Eluru, G.; Gorthi, S.S. Structured illumination microscopy. Adv. Opt. Photonics 2015, 7, 241. [Google Scholar] [CrossRef]
- Blom, H.; Widengren, J. Stimulated emission depletion microscopy. Chem. Rev. 2017, 117, 7377–7427. [Google Scholar] [CrossRef]
- Fürstenberg, A.; Heilemann, M. Single-molecule localization microscopy—Near-molecular spatial resolution in light microscopy with photoswitchable fluorophores. Phys. Chem. Chem. Phys. 2013, 15, 14919. [Google Scholar] [CrossRef]
- Carsten, A.; Wolters, M.; Aepfelbacher, M. Super-resolution fluorescence microscopy for investigating bacterial cell biology. Mol. Microbiol. 2023, 121, 646–658. [Google Scholar] [CrossRef]
- Yang, Z.; Sharma, A.; Qi, J.; Peng, X.; Lee, D.Y.; Hu, R.; Lin, D.; Qu, J.; Kim, J.S. Super-resolution fluorescent materials: An insight into design and bioimaging applications. Chem. Soc. Rev. 2016, 45, 4651–4667. [Google Scholar] [CrossRef]
- Sun, N.; Bai, S.; Dai, L.; Jia, Y. Super-resolution microscopy as a versatile tool in probing molecular assembly. Int. J. Mol. Sci. 2024, 25, 11497. [Google Scholar] [CrossRef] [PubMed]
- Onogi, S.; Shigemitsu, H.; Yoshii, T.; Tanida, T.; Ikeda, M.; Kubota, R.; Hamachi, I. In situ real-time imaging of self-sorted supramolecular nanofibres. Nat. Chem. 2016, 8, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Yamamoto, Y.; Inaba, H.; Matsuura, K. Strategy toward in-cell self-assembly of an artificial viral capsid from a fluorescent protein-modified β-annulus peptide. ACS Synth. Biol. 2024, 13, 1842–1850. [Google Scholar] [CrossRef]
- Kumar, M.; Sementa, D.; Narang, V.; Riedo, E.; Ulijn, R.V. Self-assembly propensity dictates lifetimes in transient naphthalimide–dipeptide nanofibers. Chem. Eur. J. 2020, 26, 8372–8376. [Google Scholar] [CrossRef]
- Trent, A.; Marullo, R.; Lin, B.; Black, M.; Tirrell, M. Structural properties of soluble peptide amphiphile micelles. Soft Matter 2011, 7, 9572–9582. [Google Scholar] [CrossRef]
- Castelletto, V.; De Mello, L.R.; Seitsonen, J.; Hamley, I.W. Micellization of lipopeptides containing toll-like receptor agonist and integrin binding sequences. ACS Appl. Mater. Interfaces 2024, 16, 68713–68723. [Google Scholar] [CrossRef]
- Paramonov, S.E.; Jun, H.-W.; Hartgerink, J.D. Self-assembly of peptide−amphiphile nanofibers: The roles of hydrogen bonding and amphiphilic packing. J. Am. Chem. Soc. 2006, 128, 7291–7298. [Google Scholar] [CrossRef]
- Muraoka, T.; Koh, C.; Cui, H.; Stupp, S.I. Light-triggered bioactivity in three dimensions. Angew. Chem. Int. Ed. 2009, 48, 5946–5949. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, W.; Wang, D.; Wang, J.; Li, Z.; Hu, X.; King, S.; Rogers, S.; Lu, J.R.; Xu, H. Controlling the diameters of nanotubes self-assembled from designed peptide bolaphiles. Small 2018, 14, 1703216. [Google Scholar] [CrossRef]
- Zhao, Y.; Deng, L.; Wang, J.; Xu, H.; Lu, J.R. Solvent controlled structural transition of KI4K self-assemblies: From nanotubes to nanofibrils. Langmuir 2015, 31, 12975–12983. [Google Scholar] [CrossRef]
- Liu, Y.; Naumenko, E.; Akhatova, F.; Zou, Q.; Fakhrullin, R.; Yan, X. Self-assembled peptide nanoparticles for enhanced dark-field hyperspectral imaging at the cellular and invertebrate level. Chem. Eng. J. 2021, 424, 130348. [Google Scholar] [CrossRef]
- Schierle, G.S.K.; Van De Linde, S.; Erdelyi, M.; Esbjörner, E.K.; Klein, T.; Rees, E.; Bertoncini, C.W.; Dobson, C.M.; Sauer, M.; Kaminski, C.F. In situ measurements of the formation and morphology of intracellular β-amyloid fibrils by super-resolution fluorescence imaging. J. Am. Chem. Soc. 2011, 133, 12902–12905. [Google Scholar] [CrossRef] [PubMed]
- Cox, H.; Georgiades, P.; Xu, H.; Waigh, T.A.; Lu, J.R. Self-assembly of mesoscopic peptide surfactant fibrils investigated by STORM super-resolution fluorescence microscopy. Biomacromolecules 2017, 18, 3481–3491. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, R.M.P.; Van Der Zwaag, D.; Albertazzi, L.; Lee, S.S.; Meijer, E.W.; Stupp, S.I. Super-resolution microscopy reveals structural diversity in molecular exchange among peptide amphiphile nanofibres. Nat. Commun. 2016, 7, 11561. [Google Scholar] [CrossRef]
- Beun, L.H.; Albertazzi, L.; Van Der Zwaag, D.; De Vries, R.; Stuart Ma, C. Unidirectional living growth of self-assembled protein nanofibrils revealed by super-resolution microscopy. ACS Nano 2016, 10, 4973–4980. [Google Scholar] [CrossRef]
- Kumar, M.; Son, J.; Huang, R.H.; Sementa, D.; Lee, M.; O’Brien, S.; Ulijn, R.V. In situ, noncovalent labeling and stimulated emission depletion-based super-resolution imaging of supramolecular peptide nanostructures. ACS Nano 2020, 14, 15056–15063. [Google Scholar] [CrossRef]
- Pujals, S.; Tao, K.; Terradellas, A.; Gazit, E.; Albertazzi, L. Studying structure and dynamics of self-assembled peptide nanostructures using fluorescence and super resolution microscopy. Chem. Commun. 2017, 53, 7294–7297. [Google Scholar] [CrossRef]
- Cox, H.; Xu, H.; Waigh, T.A.; Lu, J.R. Single-molecule study of peptide gel dynamics reveals states of prestress. Langmuir 2018, 34, 14678–14689. [Google Scholar] [CrossRef]
- Fuentes, E.; Boháčová, K.; Fuentes-Caparrós, A.M.; Schweins, R.; Draper, E.R.; Adams, D.J.; Pujals, S.; Albertazzi, L. PAINT-ing fluorenylmethoxycarbonyl (FMOC)-diphenylalanine hydrogels. Chem. Eur. J. 2020, 26, 9869–9873. [Google Scholar] [CrossRef]
- De Albuquerque, C.D.L.; Schultz, Z.D. Super-resolution surface-enhanced RAMAN scattering imaging of single particles in cells. Anal. Chem. 2020, 92, 9389–9398. [Google Scholar] [CrossRef]
- Jing, Y.; Zhou, L.; Chen, J.; Xu, H.; Sun, J.; Cai, M.; Jiang, J.; Gao, J.; Wang, H. Quantitatively mapping the assembly pattern of EpCAM on cell membranes with peptide probes. Anal. Chem. 2020, 92, 1865–1873. [Google Scholar] [CrossRef] [PubMed]
- Tyler, J.; Ralston, C.Y.; Rad, B. Sneaking in SpyCatcher using cell penetrating peptides for in vivo imaging. Nanotechnology 2023, 34, 425101. [Google Scholar] [CrossRef] [PubMed]
- Tas, R.P.; Albertazzi, L.; Voets, I.K. Small peptide–protein interaction pair for genetically encoded, fixation compatible peptide-PAINT. Nano Lett. 2021, 21, 9509–9516. [Google Scholar] [CrossRef] [PubMed]
- Arnal-Pastor, M.; González-Mora, D.; García-Torres, F.; Pradas, M.M.; Vallés-Lluch, A. Interaction between acrylic substrates and RAD16-I peptide in its self-assembling. J. Polym. Res. 2016, 23, 173. [Google Scholar] [CrossRef]
- Feiner-Gracia, N.; Olea, R.A.; Fitzner, R.; Boujnouni, N.E.; Van Asbeck, A.H.; Brock, R.; Albertazzi, L. Super-resolution imaging of structure, molecular composition, and stability of single oligonucleotide polyplexes. Nano Lett. 2019, 19, 2784–2792. [Google Scholar] [CrossRef]
- Ye, Z.; Wei, L.; Li, Y.; Xiao, L. Efficient modulation of β-amyloid peptide fibrillation with polymer nanoparticles revealed by super-resolution optical microscopy. Anal. Chem. 2019, 91, 8582–8590. [Google Scholar] [CrossRef]
- Chen, J.; Gao, M.; Wang, L.; Li, S.; He, J.; Qin, A.; Ren, L.; Wang, Y.; Tang, B.Z. Aggregation-induced emission probe for study of the bactericidal mechanism of antimicrobial peptides. ACS Appl. Mater. Interfaces 2018, 10, 11436–11442. [Google Scholar] [CrossRef]
- Deshpande, D.; Grieshober, M.; Wondany, F.; Gerbl, F.; Noschka, R.; Michaelis, J.; Stenger, S. Super-resolution microscopy reveals a direct interaction of intracellular mycobacterium tuberculosis with the antimicrobial peptide LL-37. Int. J. Mol. Sci. 2020, 21, 6741. [Google Scholar] [CrossRef]
- Zhu, J.; Liang, Z.; Yao, H.; Wu, Z. Identifying cell-penetrating peptides for effectively delivering antimicrobial molecules into streptococcus suis. Antibiotics 2024, 13, 725. [Google Scholar] [CrossRef]
- Zhang, M.; Deng, Y.; Zhou, Q.; Gao, J.; Zhang, D.; Pan, X. Advancing micro-nano supramolecular assembly mechanisms of natural organic matter by machine learning for unveiling environmental geochemical processes. Environ. Sci.-Process Impacts 2025, 27, 24–45. [Google Scholar] [CrossRef]
- Jing, Y.; Zhang, C.; Yu, B.; Lin, D.; Qu, J. Super-resolution microscopy: Shedding new light on in vivo imaging. Front. Chem. 2021, 9, 746900. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Yang, Z.; Lin, J.; Zhou, W.; Sun, N.; Jia, Y. Probing Peptide Assembly and Interaction via High-Resolution Imaging Techniques: A Mini Review. Int. J. Mol. Sci. 2025, 26, 3998. https://doi.org/10.3390/ijms26093998
Zhang X, Yang Z, Lin J, Zhou W, Sun N, Jia Y. Probing Peptide Assembly and Interaction via High-Resolution Imaging Techniques: A Mini Review. International Journal of Molecular Sciences. 2025; 26(9):3998. https://doi.org/10.3390/ijms26093998
Chicago/Turabian StyleZhang, Xiaoming, Zhanshu Yang, Jiaxuan Lin, Wei Zhou, Nan Sun, and Yi Jia. 2025. "Probing Peptide Assembly and Interaction via High-Resolution Imaging Techniques: A Mini Review" International Journal of Molecular Sciences 26, no. 9: 3998. https://doi.org/10.3390/ijms26093998
APA StyleZhang, X., Yang, Z., Lin, J., Zhou, W., Sun, N., & Jia, Y. (2025). Probing Peptide Assembly and Interaction via High-Resolution Imaging Techniques: A Mini Review. International Journal of Molecular Sciences, 26(9), 3998. https://doi.org/10.3390/ijms26093998






