A Single-Domain VNAR Nanobody Binds with High-Affinity and Selectivity to the Heparin Pentasaccharide Fondaparinux
Abstract
:1. Introduction
2. Results
2.1. Characterization of Biotinylated Fondaparinux (b-FP)Subsection
2.2. Anti-Fondaparinux VNARs
2.3. Structural Analysis and Molecular Modelling
2.4. Fc Fusions and Fondaparinux Binding
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Antigen Preparation
4.2.1. Biotinylation of Fondaparinux
4.2.2. MS Analysis
4.2.3. Oligosaccharide PAGE
4.2.4. Isothermal Fluorescence Titration (IFT)
4.2.5. Phage Display Selection Campaign
4.2.6. Monoclonal Phage ELISA
4.2.7. Generation of VNAR-Fc Fusions
4.2.8. Surface Plasmon Resonance
4.2.9. Fermentation and Purification of FP-Binding VNAR Fonda054-D09
4.2.10. NMR Spectroscopy
4.2.11. Molecular Modelling
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AT-III | Antithrombin-III |
| b-FP | Biotinylated fondaparinux |
| BSA | Bovine Serum Albumin |
| CDR | Complementarity-determining region |
| CFU | Colony-forming unit |
| CS | Chondroitin sulfate |
| D2O | Deuterium oxide |
| DNA | Deoxyribonucleic acid |
| DOAJ | Directory of open access journals |
| DS | Dermatan sulfate |
| EDTA | Ethylenediaminetetraacetic acid |
| ELISA | Enzyme-linked Immunosorbent Assay |
| FDA | Food and Drug Administration |
| FP | Fondaparinux |
| GAG | Glycosaminoglycans |
| GD2 | Tumor-associated ganglioside |
| H2O | Water |
| HP | Heparin |
| HPLC | High Performance Liquid Chromatography |
| HRP | Horseradish peroxidase |
| HS | Heparan Sulfate |
| HV | Hypervariable regions |
| IFT | Isothermal Fluorescence Titration |
| IgG | Immunglobulin G |
| IgNAR | Immunoglobulin new antigen receptor |
| Kd | Dissociation Constant |
| kDa | Kilodalton |
| LB medium | Luria Broth medium |
| LD | Linear dichroism |
| LMWH | Low-Molecular-Weight Heparin |
| M | Molar |
| m/z | Mass to Charge Ratio |
| MDPI | Multidisciplinary Digital Publishing Institute |
| MES | 2-(N-Morpholino)-ethansulfon-acid |
| min | Minutes |
| mM | Milimolar |
| MS | Mass Spectrometry |
| NaCl | Sodium chloride |
| NaOH | Sodium Hydroxide |
| NaPi | Sodium Phosphate |
| nM | Nanomolar |
| NMR | Nuclear magnetic resonance |
| OD600 | Optical Density at 600 nm |
| PAGE | Oligosaccharide-Polyacrylamide Gel Electrophoresis |
| PBS | Phosphate-buffered saline |
| PEG | Polyethylenglycol |
| pH | Potential of hydrogen |
| rpm | Revolutions per minute |
| RT | Room temperature |
| sdAbs | Single-domain antibodies |
| SDS-PAGE | Sodium dodecyl-sulfate polyacrylamide gel electrophoresis |
| SEC | Size Exclusion Chromatography |
| SN | Supernatant |
| SPR | Surface plasmon resonance |
| TLA | Three letter acronym |
| Tris-HCl | Trizma Hydrochlorid -hydrochlorid |
| Trp | Tryptophan |
| VHHs | Heavy-chain variable domains |
| VHs | Heavy chains |
| VNARs | Variable domain of new antigen receptor |
| µg/mL | Microgram per milliliter |
References
- Gesslbauer, B.; Derler, R.; Handwerker, C.; Seles, E.; Kungl, A.J. Exploring the glycosaminoglycan-protein interaction network by glycan-mediated pull-down proteomics. Electrophoresis 2016, 37, 1437–1447. [Google Scholar] [CrossRef] [PubMed]
- Sampaio, L.O.; Tersariol, I.L.; Lopes, C.C.; Bouças, R.I.; Nascimento, F.D.; Rocha, H.A.; Nader, H.B. Heparins and heparan sulfates. Structure, distribution and protein interactions. In Insights into Carbohydrate Structure and Biological Function; Verli, H., Ed.; Transportation Research Network: Kerala, India, 2006; pp. 1–17. ISBN 81-7895-243-2. [Google Scholar]
- Yip, G.W.; Smollich, M.; Götte, M. Therapeutic value of glycosaminoglycans in cancer. Mol. Cancer Ther. 2006, 5, 2139–2148. [Google Scholar] [CrossRef] [PubMed]
- Mihov, D.; Spiess, M. Glycosaminoglycans: Sorting determinants in intracellular protein traffic. Int. J. Biochem. Cell Biol. 2015, 68, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Kjellén, L.; Lindahl, U. Specificity of glycosaminoglycan-protein interactions. Curr. Opin. Struct. Biol. 2018, 50, 101–108. [Google Scholar] [CrossRef]
- Shi, D.; Sheng, A.; Chi, L. Glycosaminoglycan-Protein Interactions and Their Roles in Human Disease. Front. Mol. Biosci. 2021, 8, 639666. [Google Scholar] [CrossRef] [PubMed]
- Handel, T.M.; Johnson, Z.; Crown, S.E.; Lau, E.K.; Proudfoot, A.E. Regulation of protein function by glycosaminoglycans—As exemplified by chemokines. Annu. Rev. Biochem. 2005, 74, 385–410. [Google Scholar] [CrossRef]
- Hjelm, R.; Schedin-Weiss, S. High affinity interaction between a synthetic, highly negatively charged pentasaccharide and alpha- or beta-antithrombin is predominantly due to nonionic interactions. Biochemistry 2007, 46, 3378–3384. [Google Scholar] [CrossRef] [PubMed]
- Sinaÿ, P.; Jacquinet, J.-C.; Petitou, M.; Duchaussoy, P.; Lederman, I.; Choay, J.; Torri, G. Total synthesis of a heparin pentasaccharide fragment having high affinity for antithrombin III. Carbohydr. Res. 1984, 132, C5–C9. [Google Scholar] [CrossRef]
- Asada, M.; Shinomiya, M.; Suzuki, M.; Honda, E.; Sugimoto, R.; Ikekita, M.; Imamura, T. Glycosaminoglycan affinity of the complete fibroblast growth factor family. Biochim. Biophys. Acta 2009, 1790, 40–48. [Google Scholar] [CrossRef] [PubMed]
- de Paz, J.L.; Seeberger, P.H. Deciphering the glycosaminoglycan code with the help of microarrays. Mol. Biosyst. 2008, 4, 707–711. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Kett, W.C.; Severin, I.C.; Agyekum, I.; Duan, J.; Amster, I.J.; Proudfoot, A.E.I.; Coombe, D.R.; Woods, R.J. The Interaction of Heparin Tetrasaccharides with Chemokine CCL5 Is Modulated by Sulfation Pattern and pH. J. Biol. Chem. 2015, 290, 15421–15436. [Google Scholar] [CrossRef] [PubMed]
- Townley, R.A.; Bülow, H.E. Deciphering functional glycosaminoglycan motifs in development. Curr. Opin. Struct. Biol. 2018, 50, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Dimitrov, A.S. Therapeutic Antibodies: Methods and Protocols; Humana: New York, NY, USA, 2009; ISBN 9781934115923. [Google Scholar]
- Muyldermans, S. Nanobodies: Natural single-domain antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef] [PubMed]
- Saerens, D.; Ghassabeh, G.H.; Muyldermans, S. Single-domain antibodies as building blocks for novel therapeutics. Curr. Opin. Pharmacol. 2008, 8, 600–608. [Google Scholar] [CrossRef]
- Tasumi, S.; Velikovsky, C.A.; Xu, G.; Gai, S.A.; Wittrup, K.D.; Flajnik, M.F.; Mariuzza, R.A.; Pancer, Z. High-affinity lamprey VLRA and VLRB monoclonal antibodies. Proc. Natl. Acad. Sci. USA 2009, 106, 12891–12896. [Google Scholar] [CrossRef]
- Holt, L.J.; Basran, A.; Jones, K.; Chorlton, J.; Jespers, L.S.; Brewis, N.D.; Tomlinson, I.M. Anti-serum albumin domain antibodies for extending the half-lives of short lived drugs. Protein Eng. Des. Sel. 2008, 21, 283–288. [Google Scholar] [CrossRef]
- Simmons, D.P.; Abregu, F.A.; Krishnan, U.V.; Proll, D.F.; Streltsov, V.A.; Doughty, L.; Hattarki, M.K.; Nuttall, S.D. Dimerisation strategies for shark IgNAR single domain antibody fragments. J. Immunol. Methods 2006, 315, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Els Conrath, K.; Lauwereys, M.; Wyns, L.; Muyldermans, S. Camel single-domain antibodies as modular building units in bispecific and bivalent antibody constructs. J. Biol. Chem. 2001, 276, 7346–7350. [Google Scholar] [CrossRef]
- Greenberg, A.S.; Avila, D.; Hughes, M.; Hughes, A.; McKinney, E.C.; Flajnik, M.F. A new antigen receptor gene family that under-goes rearrangement and extensive somatic diversification in sharks. Nature 1995, 374, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Nuttall, S.D. Overview and discovery of IgNARs and generation of VNARs. In Single Domain Antibodies; Humana Press: Totowa, NJ, USA, 2012; pp. 27–36. [Google Scholar]
- Roux, K.H.; Greenberg, A.S.; Greene, L.; Strelets, L.; Avila, D.; McKinney, E.C.; Flajnik, M.F. Structural analysis of the nurse shark (new) antigen receptor (NAR): Molecular convergence of NAR and unusual mammalian immunoglobulins. Proc. Natl. Acad. Sci. USA 1998, 95, 11804–11809. [Google Scholar] [CrossRef] [PubMed]
- Stanfield, R.L.; Dooley, H.; Flajnik, M.F.; Wilson, I.A. Crystal structure of a shark single-domain antibody V region in complex with lysozyme. Science 2004, 305, 1770–1773. [Google Scholar] [CrossRef] [PubMed]
- Streltsov, V.A.; Carmichael, J.A.; Nuttall, S.D. Structure of a shark IgNAR antibody variable domain and modeling of an early-developmental isotype. Protein Sci. 2005, 14, 2901–2909. [Google Scholar] [CrossRef] [PubMed]
- Fennell, B.J.; Darmanin-Sheehan, A.; Hufton, S.E.; Calabro, V.; Wu, L.; Müller, M.R.; Cao, W.; Gill, D.; Cunningham, O.; Finlay, W.J.J. Dissection of the IgNAR V domain: Molecular scanning and orthologue database mining define novel IgNAR hallmarks and affinity maturation mechanisms. J. Mol. Biol. 2010, 400, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Zielonka, S.; Weber, N.; Becker, S.; Doerner, A.; Christmann, A.; Christmann, C.; Uth, C.; Fritz, J.; Schäfer, E.; Steinmann, B.; et al. Shark Attack: High affinity binding proteins derived from shark vNAR domains by stepwise in vitro affinity maturation. J. Biotechnol. 2014, 191, 236–245. [Google Scholar] [CrossRef]
- Damen, L.A.A.; van de Westerlo, E.M.A.; Versteeg, E.M.M.; van Wessel, T.; Daamen, W.F.; van Kuppevelt, T.H. Construction and evaluation of an antibody phage display library targeting heparan sulfate. Glycoconj. J. 2020, 37, 445–455. [Google Scholar] [CrossRef]
- Smits, N.C.; Lensen, J.F.M.; Wijnhoven, T.J.M.; ten Dam, G.B.; Jenniskens, G.J.; van Kuppevelt, T.H. Phage display-derived human antibodies against specific glycosaminoglycan epitopes. Methods Enzymol. 2006, 416, 61–87. [Google Scholar] [CrossRef]
- Uchimura, K.; Lemjabbar-Alaoui, H.; van Kuppevelt, T.H.; Rosen, S.D. Use of a phage display antibody to measure the enzymatic activity of the Sulfs. Methods Enzymol. 2010, 480, 51–64. [Google Scholar] [CrossRef]
- Bruinsma, I.B.; te Riet, L.; Gevers, T.; ten Dam, G.B.; van Kuppevelt, T.H.; David, G.; Küsters, B.; de Waal, R.M.W.; Verbeek, M.M. Sulfation of heparan sulfate associated with amyloid-beta plaques in patients with Alzheimer’s disease. Acta Neuropathol. 2010, 119, 211–220. [Google Scholar] [CrossRef]
- Kurup, S.; Wijnhoven, T.J.M.; Jenniskens, G.J.; Kimata, K.; Habuchi, H.; Li, J.-P.; Lindahl, U.; van Kuppevelt, T.H.; Spillmann, D. Characterization of anti-heparan sulfate phage display antibodies AO4B08 and HS4E4. J. Biol. Chem. 2007, 282, 21032–21042. [Google Scholar] [CrossRef]
- ten Dam, G.B.; Kurup, S.; van de Westerlo, E.M.A.; Versteeg, E.M.M.; Lindahl, U.; Spillmann, D.; van Kuppevelt, T.H. 3-O-sulfated oligosaccharide structures are recognized by anti-heparan sulfate antibody HS4C3. J. Biol. Chem. 2006, 281, 4654–4662. [Google Scholar] [CrossRef] [PubMed]
- van Kuppevelt, T.H.; Dennissen, M.A.; van Venrooij, W.J.; Hoet, R.M.; Veerkamp, J.H. Generation and application of type-specific anti-heparan sulfate antibodies using phage display technology. Further evidence for heparan sulfate heterogeneity in the kidney. J. Biol. Chem. 1998, 273, 12960–12966. [Google Scholar] [CrossRef] [PubMed]
- Nissim, A.; Hoogenboom, H.R.; Tomlinson, I.; Flynn, G.; Midgley, C.; Lane, D.; Winter, G. Antibody fragments from a ‘single pot’phage display library as immunochemical reagents. EMBO J. 1994, 13, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Stocki, P.; Szary, J.; Rasmussen, C.L.M.; Demydchuk, M.; Northall, L.; Logan, D.B.; Gauhar, A.; Thei, L.; Moos, T.; Walsh, F.S.; et al. Blood-brain barrier transport using a high affinity, brain-selective VNAR antibody targeting transferrin receptor 1. FASEB J. 2021, 35, e21172. [Google Scholar] [CrossRef]
- Feng, M.; Bian, H.; Wu, X.; Fu, T.; Fu, Y.; Hong, J.; Fleming, B.D.; Flajnik, M.F.; Ho, M. Construction and next-generation sequencing analysis of a large phage-displayed VNAR single-domain antibody library from six naïve nurse sharks. Antib. Ther. 2019, 2, 1–11. [Google Scholar] [CrossRef]
- Wu, T.T.; Johnson, G.; Kabat, E.A. Length distribution of CDRH3 in antibodies. Proteins 1993, 16, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.T.; Kok, B.H.; Leow, C.Y.; Leow, C.H. Exploring shark VNAR antibody against infectious diseases using phage display technology. Fish Shellfish Immunol. 2023, 140, 108986. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.; Velez, J.; Singh, M.; Cerny, J.; Flajnik, M.F. Mutational pattern of the nurse shark antigen receptor gene (NAR) is similar to that of mammalian Ig genes and to spontaneous mutations in evolution: The translesion synthesis model of somatic hypermutation. Int. Immunol. 1999, 11, 825–833. [Google Scholar] [CrossRef]
- Liu, J.L.; Anderson, G.P.; Goldman, E.R. Isolation of anti-toxin single domain antibodies from a semi-synthetic spiny dogfish shark display library. BMC Biotechnol. 2007, 7, 78. [Google Scholar] [CrossRef] [PubMed]
- Stanfield, R.L.; Dooley, H.; Verdino, P.; Flajnik, M.F.; Wilson, I.A. Maturation of shark single-domain (IgNAR) antibodies: Evidence for induced-fit binding. J. Mol. Biol. 2007, 367, 358–372. [Google Scholar] [CrossRef] [PubMed]
- Cardin, A.D.; Weintraub, H.J. Molecular modeling of protein-glycosaminoglycan interactions. Arterioscler. Thromb. Vasc. Biol. 1989, 9, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Proudfoot, A.E.I.; Johnson, Z.; Bonvin, P.; Handel, T.M. Glycosaminoglycan Interactions with Chemokines Add Complexity to a Complex System. Pharmaceuticals 2017, 10, 70. [Google Scholar] [CrossRef]
- Solari, V.; Rudd, T.R.; Guimond, S.E.; Powell, A.K.; Turnbull, J.E.; Yates, E.A. Heparan sulfate phage display antibodies recognise epitopes defined by a combination of sugar sequence and cation binding. Org. Biomol. Chem. 2015, 13, 6066–6072. [Google Scholar] [CrossRef] [PubMed]
- Thompson, S.M.; Fernig, D.G.; Jesudason, E.C.; Losty, P.D.; van de Westerlo, E.M.A.; van Kuppevelt, T.H.; Turnbull, J.E. Heparan sulfate phage display antibodies identify distinct epitopes with complex binding characteristics: Insights into protein binding specificities. J. Biol. Chem. 2009, 284, 35621–35631. [Google Scholar] [CrossRef]
- Krieger, E.; Geretti, E.; Brandner, B.; Goger, B.; Wells, T.N.; Kungl, A.J. A structural and dynamic model for the interaction of interleukin-8 and glycosaminoglycans: Support from isothermal fluorescence titrations. Proteins 2004, 54, 768–775. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Abrahams, J.P.; Skinner, R.; Petitou, M.; Pike, R.N.; Carrell, R.W. The anticoagulant activation of antithrombin by heparin. Proc. Natl. Acad. Sci. USA 1997, 94, 14683–14688. [Google Scholar] [CrossRef] [PubMed]
- Manoutcharian, K.; Gevorkian, G. Shark VNAR phage display libraries: An alternative source for therapeutic and diagnostic recombinant antibody fragments. Fish Shellfish Immunol. 2023, 138, 108808. [Google Scholar] [CrossRef] [PubMed]
- Dooley, H.; Flajnik, M.F.; Porter, A.J. Selection and characterization of naturally occurring single-domain (IgNAR) antibody fragments from immunized sharks by phage display. Mol. Immunol. 2003, 40, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Zielonka, S.; Empting, M.; Grzeschik, J.; Könning, D.; Barelle, C.J.; Kolmar, H. Structural insights and biomedical potential of IgNAR scaffolds from sharks. MAbs 2015, 7, 15–25. [Google Scholar] [CrossRef]
- Dooley, H.; Stanfield, R.L.; Brady, R.A.; Flajnik, M.F. First molecular and biochemical analysis of in vivo affinity maturation in an ectothermic vertebrate. Proc. Natl. Acad. Sci. USA 2006, 103, 1846–1851. [Google Scholar] [CrossRef]
- Kovaleva, M.; Ferguson, L.; Steven, J.; Porter, A.; Barelle, C. Shark variable new antigen receptor biologics—A novel technology platform for therapeutic drug development. Expert Opin. Biol. Ther. 2014, 14, 1527–1539. [Google Scholar] [CrossRef]
- Sterner, E.; Flanagan, N.; Gildersleeve, J.C. Perspectives on Anti-Glycan Antibodies Gleaned from Development of a Community Resource Database. ACS Chem. Biol. 2016, 11, 1773–1783. [Google Scholar] [CrossRef] [PubMed]
- Temme, J.S.; Butler, D.L.; Gildersleeve, J.C. Anti-glycan antibodies: Roles in human disease. Biochem. J. 2021, 478, 1485–1509. [Google Scholar] [CrossRef]
- Kappler, K.; Hennet, T. Emergence and significance of carbohydrate-specific antibodies. Genes Immun. 2020, 21, 224–239. [Google Scholar] [CrossRef] [PubMed]
- Keam, S.J. GKL Fondaparinux sodium. Drugs 2002, 62, 1673–1685. [Google Scholar] [CrossRef] [PubMed]
- Walenga, J.M.; Jeske, W.P.; Frapaise, F.X.; Bick, R.L.; Fareed, J.; Samama, M.M. Fondaparinux: A synthetic heparin pentasaccharide as a new antithrombotic agent. Expert Opin. Investig. Drugs 2002, 11, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, M.; Tan, L.; Pan, N.; Zhang, L. The clinical use of Fondaparinux: A synthetic heparin pentasaccharide. Prog. Mol. Biol. Transl. Sci. 2019, 163, 41–53. [Google Scholar]
- Cole, C.L.; Hansen, S.U.; Baráth, M.; Rushton, G.; Gardiner, J.M.; Avizienyte, E.; Jayson, G.C. Synthetic heparan sulfate oligosaccharides inhibit endothelial cell functions essential for angiogenesis. PLoS ONE 2010, 5, e11644. [Google Scholar] [CrossRef]
- Kawashima, H.; Atarashi, K.; Hirose, M.; Hirose, J.; Yamada, S.; Sugahara, K.; Miyasaka, M. Oversulfated chondroitin/dermatan sulfates containing GlcAbeta1/IdoAalpha1-3GalNAc(4,6-O-disulfate) interact with L- and P-selectin and chemokines. J. Biol. Chem. 2002, 277, 12921–12930. [Google Scholar] [CrossRef]
- Schenauer, M.R.; Yu, Y.; Sweeney, M.D.; Leary, J.A. CCR2 chemokines bind selectively to acetylated heparan sulfate octasaccharides. J. Biol. Chem. 2007, 282, 25182–25188. [Google Scholar] [CrossRef] [PubMed]
- Spillmann, D.; Witt, D.; Lindahl, U. Defining the interleukin-8-binding domain of heparan sulfate. J. Biol. Chem. 1998, 273, 15487–15493. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.-H.; Karnaukhova, E.; Rajabi, M.; Reeder, K.; Chen, T.; Dhawan, S.; Kozlowski, S. Oversulfated chondroitin sulfate binds to chemokines and inhibits stromal cell-derived factor-1 mediated signaling in activated T cells. PLoS ONE 2014, 9, e94402. [Google Scholar] [CrossRef] [PubMed]
- Kieber-Emmons, T.; Monzavi-Karbassi, B.; Hutchins, L.F.; Pennisi, A.; Makhoul, I. Harnessing benefit from targeting tumor associated carbohydrate antigens. Hum. Vaccin. Immunother. 2017, 13, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, Y.; Ju, T. Aberrant Glycosylation as Immune Therapeutic Targets for Solid Tumors. Cancers 2023, 15, 3536. [Google Scholar] [CrossRef]
- Sterner, E.; Peach, M.L.; Nicklaus, M.C.; Gildersleeve, J.C. Therapeutic Antibodies to Ganglioside GD2 Evolved from Highly Selective Germline Antibodies. Cell Rep. 2017, 20, 1681–1691. [Google Scholar] [CrossRef] [PubMed]
- Kailemia, M.J.; Park, D.; Lebrilla, C.B. Glycans and glycoproteins as specific biomarkers for cancer. Anal. Bioanal. Chem. 2017, 409, 395–410. [Google Scholar] [CrossRef] [PubMed]
- Puiu, M.; Nativi, C.; Bala, C. Early detection of tumour-associated antigens: Assessment of point-of-care electrochemical im-munoassays. TrAC Trends Anal. Chem. 2023, 160, 116981. [Google Scholar] [CrossRef]
- Gerlza, T.; Trojacher, C.; Kitic, N.; Adage, T.; Kungl, A.J. Development of Molecules Antagonizing Heparan Sulfate Proteoglycans. In Seminars in Thrombosis and Hemostasis; Thieme Medical Publishers, Inc.: New York, NY, USA, 2021; pp. 316–332. [Google Scholar]
- Gerlza, T.; Hecher, B.; Jeremic, D.; Fuchs, T.; Gschwandtner, M.; Falsone, A.; Gesslbauer, B.; Kungl, A.J. A combinatorial approach to biophysically characterise chemokine-glycan binding affinities for drug development. Molecules 2014, 19, 10618–10634. [Google Scholar] [CrossRef]
- Clackson, T.; Lowman, H.B. Phage Display; Oxford University Press Inc.: New York, NY, USA, 2004. [Google Scholar]
- Häsler, J.; Rutkowski, J.L. Semi-Synthetic Nurse Shark VNAR Libraries for Making and Using Selective Binding Compounds. U.S. Patent US10479990B2, 19 November 2019. [Google Scholar]
- Braunagel, M.; Little, M. Construction of a semisynthetic antibody library using trinucleotide oligos. Nucleic Acids Res. 1997, 25, 4690–4691. [Google Scholar] [CrossRef]
- Häsler, J.; Flajnik, M.F.; Williams, G.; Walsh, F.S.; Rutkowski, J.L. VNAR single-domain antibodies specific for BAFF inhibit B cell development by molecular mimicry. Mol. Immunol. 2016, 75, 28–37. [Google Scholar] [CrossRef]
- Hoogenboom, H.R.; de Bruïne, A.P.; Hufton, S.E.; Hoet, R.M.; Arends, J.-W.; Roovers, R.C. Antibody phage display technology and its applications. Immunotechnology 1998, 4, 1–20. [Google Scholar] [CrossRef]
- Kretzschmar, T.; Von Rüden, T. Antibody discovery: Phage display. Curr. Opin. Biotechnol. 2002, 13, 598–602. [Google Scholar] [CrossRef] [PubMed]
- Grzesiek, S.; Bax, A. Improved 3D triple-resonance NMR techniques applied to a 31 kDa protein. J. Magn. Reson. 1992, 96, 432–440. [Google Scholar] [CrossRef]
- Kay, L.E.; Xu, G.Y.; Yamazaki, T. Enhanced-sensitivity triple-resonance spectroscopy with minimal H2O saturation. J. Magn. Reson. 1994, 109, 129–133. [Google Scholar] [CrossRef]
- Clubb, R.T.; Thanabal, V.; Wagner, G. A constant-time three-dimensional triple-resonance pulse scheme to correlate intraresidue 1HN, 15N, and 13C’chemical shifts in 15N—13C-labelled proteins. J. Magn. Reson. 1992, 97, 213–217. [Google Scholar]
- Wittekind, M.; Mueller, L. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha-and beta-carbon resonances in proteins. J. Magn. Reson. 1993, 101, 201–205. [Google Scholar] [CrossRef]
- Muhandiram, D.; Kay, L.E. Gradient-enhanced triple-resonance three-dimensional NMR experiments with improved sensitivity. J. Magn. Reson. 1994, 103, 203–216. [Google Scholar] [CrossRef]
- Schleucher, J.; Sattler, M.; Griesinger, C. Coherence selection by gradients without signal attenuation: Application to the three-dimensional HNCO experiment. Angew. Chem. Int. Ed. Engl. 1993, 32, 1489–1491. [Google Scholar] [CrossRef]
- Olejniczak, E.T.; Xu, R.X.; Fesik, S.W. A 4D HCCH-TOCSY experiment for assigning the side chain 1 H and 13 C resonances of proteins. J. Biomol. NMR 1992, 2, 655–659. [Google Scholar] [CrossRef]
- Bax, A.; Clore, G.M.; Gronenborn, A.M. 1H-1H correlation via isotropic mixing of 13C magnetization, a new three-dimensional approach for assigning 1H and 13C spectra of 13C-enriched proteins. J. Magn. Reson. 1990, 88, 425–431. [Google Scholar] [CrossRef]
- Kay, L.E.; Xu, G.-Y.; Singer, A.U.; Muhandiram, D.R.; Forman-Kay, J.D. A gradient-enhanced HCCH-TOCSY experiment for recording side-chain 1H and 13C correlations in H2O samples for proteins. J. Magn. Reson. 1993, 101, 333–337. [Google Scholar] [CrossRef]
- Grzesiek, S.; Anglister, J.; Bax, A. Correlation of backbone amide and aliphatic side-chain resonances in 13C/15N-enriched proteins by isotropic mixing of 13C magnetization. J. Magn. Reson. 1993, 101, 114–119. [Google Scholar] [CrossRef]
- Montelione, G.T.; Lyons, B.A.; Emerson, S.D.; Tashiro, M. An efficient triple resonance experiment using carbon-13 isotropic mixing for determining sequence-specific resonance assignments of isotopically-enriched proteins. J. Am. Chem. Soc. 1992, 114, 10974–10975. [Google Scholar] [CrossRef]
- Davis, A.L.; Keeler, J.; Laue, E.D.; Moskau, D. Experiments for recording pure-absorption heteronuclear correlation spectra using pulsed field gradients. J. Magn. Reson. 1992, 98, 207–216. [Google Scholar] [CrossRef]
- Skinner, S.P.; Fogh, R.H.; Boucher, W.; Ragan, T.J.; Mureddu, L.G.; Vuister, G.W. CcpNmr AnalysisAssign: A flexible platform for integrated NMR analysis. J. Biomol. NMR 2016, 66, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, T.; Güntert, P.; Wüthrich, K. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J. Biomol. NMR 2002, 24, 171–189. [Google Scholar] [CrossRef] [PubMed]
- Güntert, P.; Mumenthaler, C.; Wüthrich, K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997, 273, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Nederveen, A.J.; Doreleijers, J.F.; Vranken, W.; Miller, Z.; Spronk, C.A.E.M.; Nabuurs, S.B.; Güntert, P.; Livny, M.; Markley, J.L.; Nilges, M.; et al. RECOORD: A recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. PROTEINS Struct. Funct. Bioinform. 2005, 59, 662–672. [Google Scholar] [CrossRef]
- Brünger, A.T.; Adams, P.D.; Clore, G.M.; DeLano, W.L.; Gros, P.; Grosse-Kunstleve, R.W.; Jiang, J.S.; Kuszewski, J.; Nilges, M.; Pannu, N.S.; et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998, 54, 905–921. [Google Scholar]
- Shen, Y.; Delaglio, F.; Cornilescu, G.; Bax, A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 2009, 44, 213–223. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Tejero, R.; Montelione, G.T. Evaluating protein structures determined by structural genomics consortia. PROTEINS Struct. Funct. Bioinform. 2007, 66, 778–795. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger, LLC. The PyMOL Molecular Graphics Systemed; Schrödinger, LLC: New York, NY, USA, 2015. [Google Scholar]


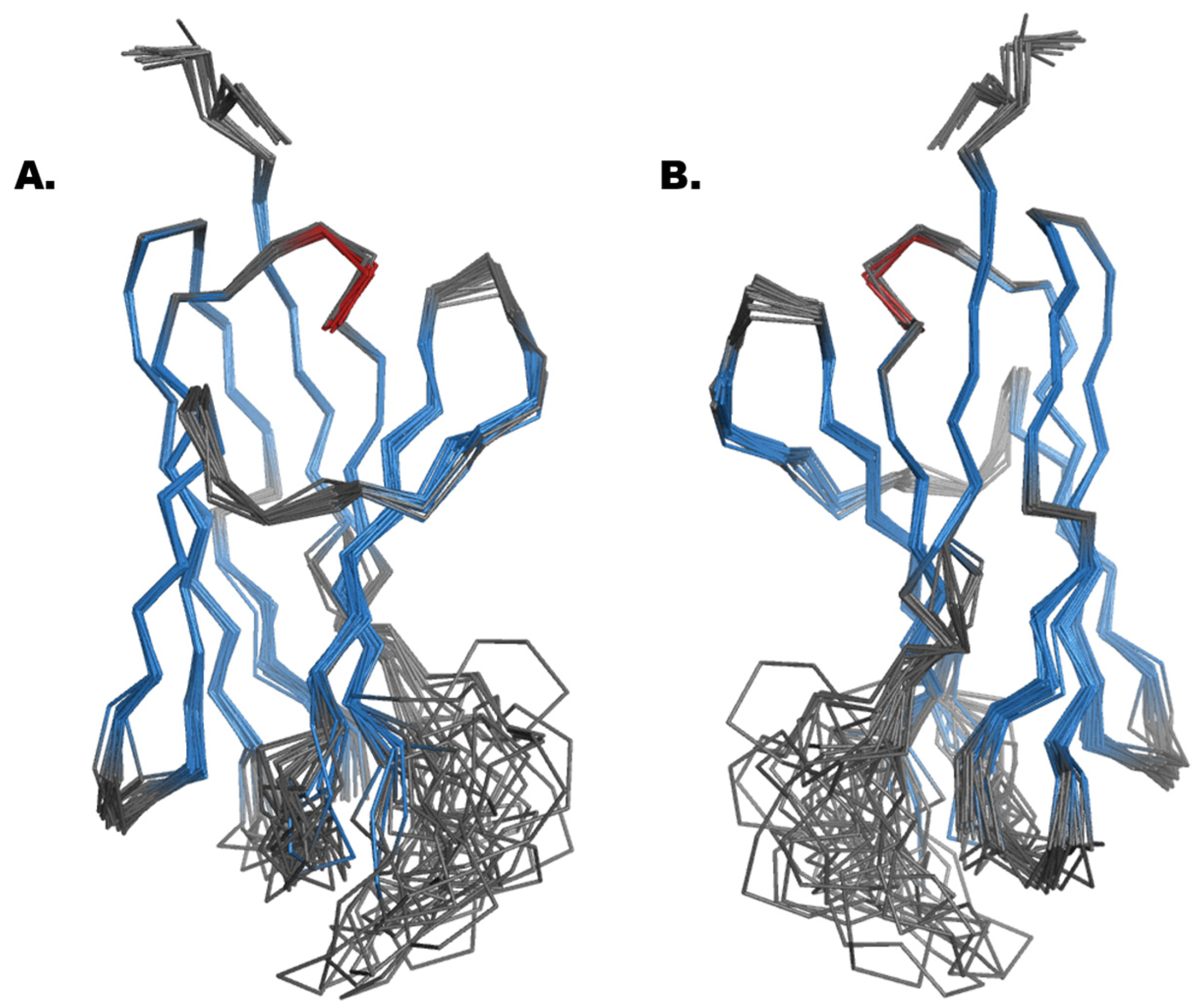
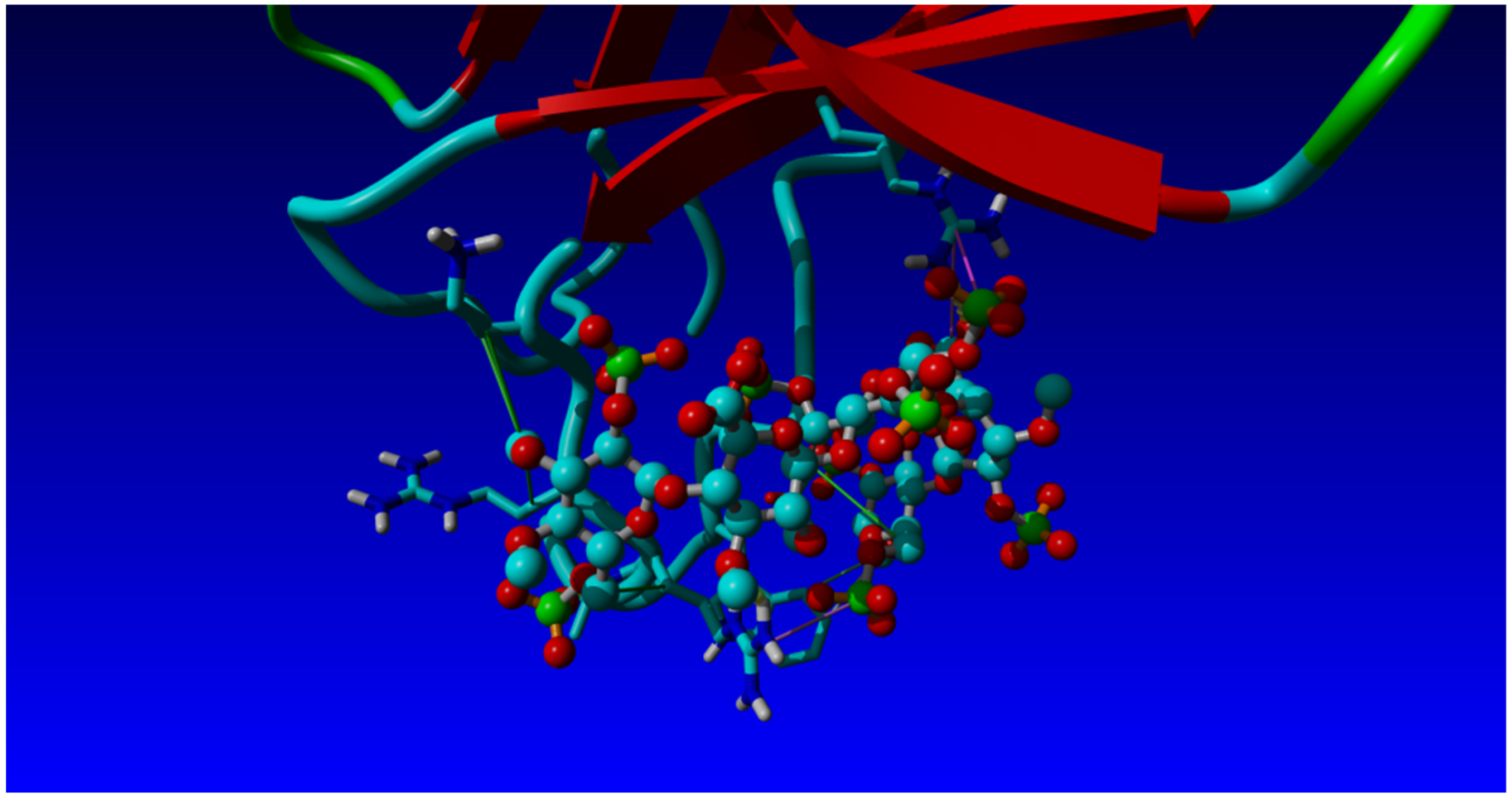

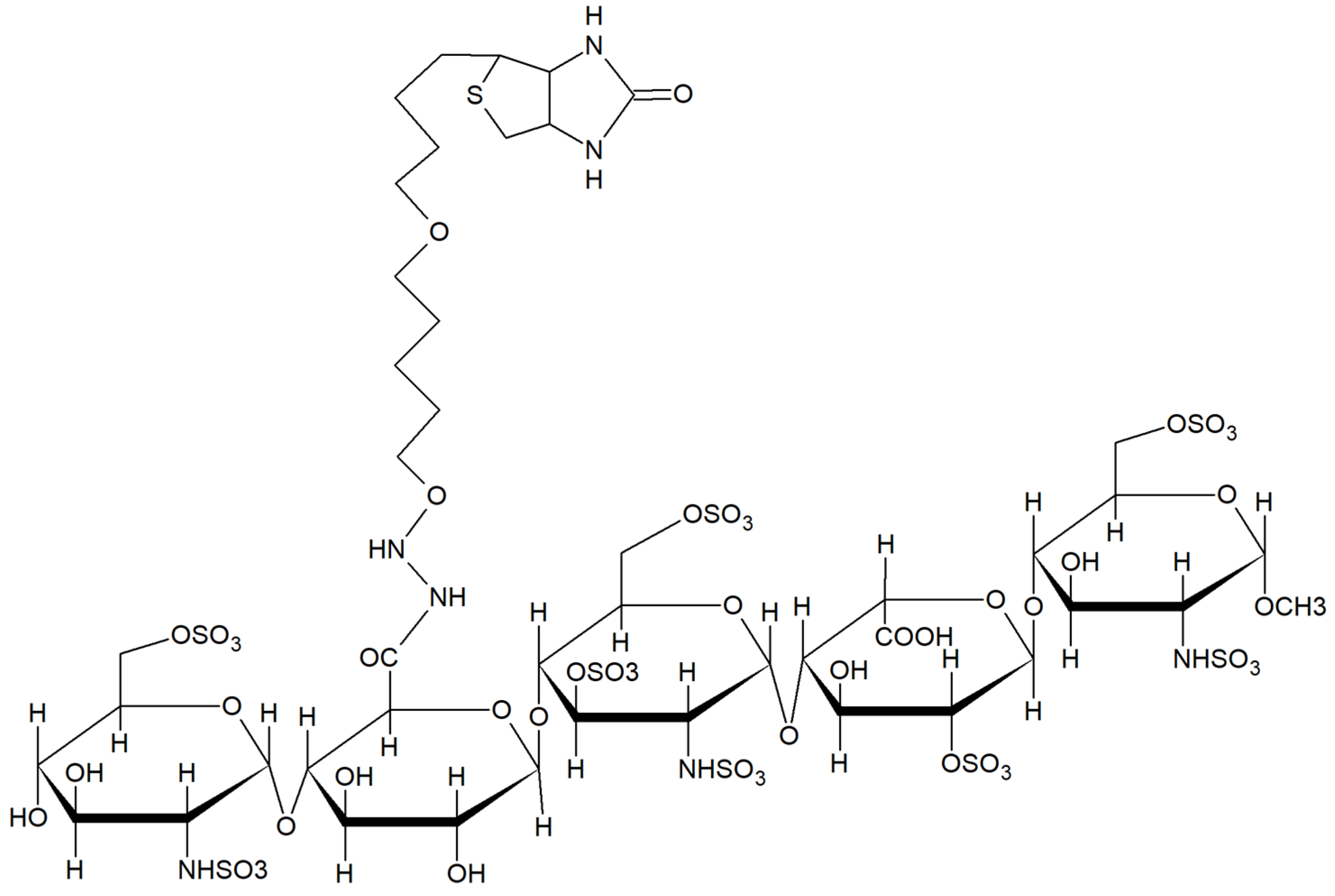
| Clone | CDR3 | CDR1 | VNAR Type (Isoform Family) |
|---|---|---|---|
| Fonda051-A01 | NVYSVTHSIQGKLRAI | PYYALA | Type 4 |
| Fonda051-A02 | NVYVHRRKTPYLTKQ | VDVARA | Type 4 |
| Fonda051-A03 | NVYCVTHSLQGKLRAM | PYYAPA | Type 4 |
| Fonda051-A05 | NVYGqCCNRRRL | SNCALP | Type 2 |
| Fonda051-A06 | NVYVYDHPQYRGGFGH | RQFAPA | Type 4 |
| Fonda051-A07 | NVYSVTHSIQGKLRAI | PYYALA | Type 4 |
| Fonda051-A08 | NVYVYDHPQYRGGVGH | RQFALA | Type 4 |
| Clone | CDR3 | CDR1 | VNAR Type |
|---|---|---|---|
| Fonda054-A01 | PRLYRSSCQGSSRR | SNCALP | Type 2 |
| Fonda054-A06 | PRLYRSSCQGSSRR | SNCALP | Type 2 |
| Fonda054-F01 | PRLFRSSCqGSSRR | SNCALP | Type 2 |
| Fonda054-G10 | PRLYSSSCqGSSRR | SNCALP | Type 2 |
| Fonda054-H05 | PRVYRSSCqGSSRR | SNCALP | Type 2 |
| Fonda054-D09 | HCRRRCGDVWC | SICALS | Type 2 |
| Fonda054-A02 | NRSYVESYDIWSKLL | SNCALS | Type 2 |
| Distance constraints | |
|---|---|
| Total | 1912 (100%) |
| Intraresidue, |i-j| = 0 | 343 (17.9%) |
| Sequential, |i-j| = 1 | 456 (23.8%) |
| medium-range, 1 < |i-j| < 5 | 191 (10.0%) |
| long-range, |i-j| >= 5 | 922 (48.2%) |
| Dihedral angle constraints | |
| Total | 150 |
| Violations | |
| RMS of distance violation/constraint | 0.02 Å |
| Maximum distance violation | 0.28 Å |
| RMS of dihedral angle violation/constraint | 0.07° |
| Maximum dihedral angle violation | 1.20° |
| Deviations from Ideal Geometry | |
| RMS deviation for bond angles | 1.6° |
| RMS deviation for bond lengths | 0.019 Å |
| RMSD Values | |
| Backbone | All: 1.5 Å Ordered: 0.3 Å |
| Heavy atoms | All: 2.1 Å Ordered: 0.7 Å |
| Ramachandran Plot | |
| Most favored regions | 90.5% |
| Additionally allowed regions | 9.0% |
| Generously allowed regions | 0.5% |
| Disallowed regions | 0.0% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gschwandtner, M.; Derler, R.; Talker, E.; Trojacher, C.; Gubensäk, N.; Becker, W.; Gerlza, T.; Klaus, Z.; Stocki, P.; Walsh, F.S.; et al. A Single-Domain VNAR Nanobody Binds with High-Affinity and Selectivity to the Heparin Pentasaccharide Fondaparinux. Int. J. Mol. Sci. 2025, 26, 4045. https://doi.org/10.3390/ijms26094045
Gschwandtner M, Derler R, Talker E, Trojacher C, Gubensäk N, Becker W, Gerlza T, Klaus Z, Stocki P, Walsh FS, et al. A Single-Domain VNAR Nanobody Binds with High-Affinity and Selectivity to the Heparin Pentasaccharide Fondaparinux. International Journal of Molecular Sciences. 2025; 26(9):4045. https://doi.org/10.3390/ijms26094045
Chicago/Turabian StyleGschwandtner, Martha, Rupert Derler, Elisa Talker, Christina Trojacher, Nina Gubensäk, Walter Becker, Tanja Gerlza, Zangger Klaus, Pawel Stocki, Frank S. Walsh, and et al. 2025. "A Single-Domain VNAR Nanobody Binds with High-Affinity and Selectivity to the Heparin Pentasaccharide Fondaparinux" International Journal of Molecular Sciences 26, no. 9: 4045. https://doi.org/10.3390/ijms26094045
APA StyleGschwandtner, M., Derler, R., Talker, E., Trojacher, C., Gubensäk, N., Becker, W., Gerlza, T., Klaus, Z., Stocki, P., Walsh, F. S., Rutkowski, J. L., & Kungl, A. (2025). A Single-Domain VNAR Nanobody Binds with High-Affinity and Selectivity to the Heparin Pentasaccharide Fondaparinux. International Journal of Molecular Sciences, 26(9), 4045. https://doi.org/10.3390/ijms26094045






