Abstract
Although volcanoes represent extreme environments for life, they harbour bacterial communities. Vulcano Island (Aeolian Islands, Sicily) presents an intense fumarolic activity and widespread soil degassing, fed by variable amounts of magmatic gases (dominant at La Fossa Crater) and hydrothermal fluids (dominant at Levante Bay). The aim of this study is to analyse the microbial communities from the different environments of Vulcano Island and to evaluate their possible correlation with the composition of the gas emissions. Microbial analyses were carried out on soils and pioneer plants from both La Fossa Crater and Levante Bay. Total DNA has been extracted from all the samples and sequenced through Illumina MiSeq platform. The analysis of microbiome composition and the gases sampled in the same sites could suggest a possible correlation between the two parameters. We can suggest that the ability of different bacterial genera/species to survive in the same area might be due to the selection of particular genetic traits allowing the survival of these microorganisms. On the other side, the finding that microbial communities inhabiting different sites exhibiting different emission profiles are similar might be explained on the basis of a possible sharing of metabolic abilities related to the gas composition.
1. Introduction
In ecology, the term “extreme” commonly refers to unfavourable environmental factors that depress the ability of organisms to function [1]. Understanding the mechanisms underlying the adaptation of microbes to extreme environments is of fundamental importance to deeply investigate processes that led to the evolution of Earth [2]; for example, it has been shown that microbial communities are mainly shaped by environmental conditions and the microbes inhabiting extreme habitats evolve faster than those populating benign environments [1]. Bacterial communities from natural extreme environments represent not only a gene reservoir for potential biotechnological applications (e.g., the discovery of the Taq polymerase, isolated from the thermophilic bacteria Thermus aquaticus), but they can be used as a model system to explore relationships between diversity and environmental factors [3]. Adaptative features of these extremophiles permit them to survive under such extreme and hostile environmental conditions [4].
In this context, volcanic environments arouse interest in the scientific community, as they are considered analogous to some of the earliest environments on Earth [5]. In the last decades, several studies focused on understanding the distribution of microorganisms in volcanic environments [6], such as Antarctic and Icelandic volcanoes. These studies were motivated by the desire to elucidate how the geochemically extreme conditions of such environments can influence microbial diversity both on the surface and in the subsurface of the Earth. Interest in the microbial diversity of geothermal and volcanic soils has been growing, and high diversity, including novel and rare bacteria, has been detected so far [7].
The Aeolian volcanic archipelago is situated north of Sicily (Italy), along the southern margin of the Tyrrhenian Sea and is underlain by a 15 to 20 km thick continental crust. Lipari, Vulcano, and Salina are three summits of a volcanic pile located along a regional crustal lineament crossing the north-eastern coast of Sicily and continuing as far as the Aeolian Islands. Each island in the Aeolian arc has had a different tectonic and magmatic history and their activity is not always correlated in time [8]. As usually occurs, shallow hydrothermal systems are associated with the Aeolian volcanoes [9]. Vulcano and the other Aeolian Islands have been investigated for the microbial populations [10], leading to the isolation of novel halotolerant and thermophilic species [11,12,13,14]. The thermal springs in the bay Baia di Levante of Vulcano Island (Aeolian Islands) host dozens of aerobic and anaerobic, thermophilic, and hyperthermophilic microorganisms belonging to Bacteria and Archaea domains [9].
Despite the abundance of researches about the microbial communities from these environments, from a scientific viewpoint, very little is known about the possible correlation between interstitial soil gases and soil bacteria.
Due to their detectability in almost every environment and the high information content, volatile (organic) compounds (VOCs) have been widely studied in relation to different organisms, functions, and interactions [15]. VOCs produced by bacteria have been correlated with inhibition of laccase activity [16], alteration of nitrification processes [17], antagonistic interactions with other species [18], and plant growth promotion [19]. Asensio et al. found microorganism, roots, and physio-chemical factors to be involved in VOCs emissions [20].
It is known that geothermal fluxes shape microbial soil communities [21] and that bacteria produce volatile compounds [22], but there is an almost complete lack of information about the possibility that bacteria may actually participate in the production of these interstitial gases.
Volcanoes are extreme environments for plants too. The role of bacteria associated with plants represents, at the moment, a very interesting topic. Symbiotic bacteria can establish a relationship with the host plant that gives benefits to both parts [23]. In an extreme environment, it is reliable to suppose that the role of bacteria for the survival of the plant in harsh conditions (lack of organic matter, high or low values of pH, presence of high concentration of metals and heavy metals) might be of major importance [24]. Microbial communities associated with plants in diverse conditions, including extreme environments, have been characterized for their potential biotechnological applications in industry, medicine, and agriculture [25]. Plants belonging to the Poa genus are pioneer plants widely found in volcanic environments [26,27,28,29], thus they result in interesting models to study the endophytic communities in extreme environments.
In this study, we aim to investigate the diversity of bacterial communities of Vulcano Island, Aeolian Islands (Italy). Physicochemical characteristics of fluids from the different sampling areas analysed in this study were investigated, in order to establish a possible correlation of these data with the composition of bacterial communities, and to evaluate the possibility that bacteria may be involved in the production of these interstitial soil gases.
2. Materials and Methods
2.1. Study Area: Vulcano Island
Vulcano is the southernmost island of the Aeolian archipelago (Sicily, southern Italy), a volcanic arc consisting of seven islands and nine sea mountains with an extension of 22 km2. The base of the volcanic cone leans on the Calabro-Peloritano block at an average depth of 1000 m below sea level, for a diameter of about 15 km [30]. The evolution of the island is linked to a trans-tensional tectonics, related to the subduction process [31].
Sampling Areas
Samples for microbiological and geochemical analyses were mostly collected from two areas marked by intense exhalative activity: La Fossa Crater and the Levante Bay, along the eastern side of the isthmus connecting Vulcano Island to the Vulcanello peninsula.
The north-western sector of the crater rim of La Fossa Crater is currently affected by intense fumarolic degassing, with outlet temperatures ranging from the boiling point of water up to 450 °C. The fumarolic fluids have a typical magmatic composition, with dry gas fraction dominated by CO2 and with relevant concentrations of HCl, SO2, H2S, HF, and CO [32,33]. The mineralogy of the crater fumaroles is dominated by sulfides and sulfosalts, deposited due to a sublimate formation under high-temperature reducing conditions [34,35,36]. Notable quantities of ammonium minerals were also recognized [37]. Rapidly changing temperatures, magmatic fluids input, soil permeability, etc., offer a multitude of environment in a very narrow area. On the volcanic edifice, pioneer plants belonging to the Poa genus start to grow at the altitude of nearly 360 m above the sea level.
The Levante Bay area is characterized by the presence of low-temperature fumaroles (T < 100 °C), both subaerial and submerged, that originated by boiling of a shallow geothermal aquifer heated by the uprising hot magmatic fluids. The discharged fluids have a typical hydrothermal composition, being characterized by higher contents of CH4 and H2S relative to the crater fumaroles. Pioneer plants are present in Levante Bay, in high CO2 fluxes areas, and in high salinity environments. This site and, in particular, the submarine gas vents were selected as natural laboratories for research studies regarding, e.g., (i) ocean acidification [38] and (ii) thermophilic bacteria [11].
2.2. Interstitial Soil Gas Sampling and Analyses
Sampling was performed on May 2017 in different sites of Vulcano Island, Aeolian Islands (Sicily, Italy). In this work, analyses were concentrated at La Fossa Crater (38°24′15″ N 14°57′42″ E) and Levante Bay (38°25′02″ N 14°57′34″ E).
Interstitial soil gases were collected from 8 sampling sites, as follows: (i) 3 sites on the northern flank of La Fossa Crater at different distances from fumarolic emissions (CSCS, CSF56, CSF94), (ii) 1 site on the western flank of the volcanic edifice characterized by the presence of a pioneer plant (CPBS), and (iii) 4 sites in the Levante Bay (LSV11, LSV13, LSV14, LPBS), selected according to in situ soil CO2 flux measurements. Soil CO2 fluxes were measured according to the accumulation chamber (AC) method [39], using a cylindrical chamber (basal area 200 cm2 and inner volume 3060 cm3) and a Licor® (NE, USA) Li-820 Infra-Red (IR) spectrophotometer, as described in Venturi et al. [40].
Gas samples were collected by inserting a stainless-steel probe within the soil down to the sampling depth (5, 10, or 30 cm) connected, through silicon tubing and a PTFE three-way valve, to a 60 mL plastic syringe in order to pump the gas into (i) a 12 mL glass vial equipped with a silicon rubber membrane and (ii) a 1 L Supelco’s (Darmstadt, Germany) Tedlar® gas sampling bag. The detailed description of the sampling equipment and procedure is reported in previously published literature [40,41,42,43]. After sampling, soil temperature was measured with a thermocouple inserted within the soil down to the sampling depth.
The chemical analyses of major gas constituents were performed on gas samples stored in the glass vials using a Shimadzu (Duisburg, Germany) 15A gas chromatrograph (GC), equipped with a thermal conductivity detector (TCD) and either a 10 m long 5A Molecular Sieve column (for N2, O2, Ar and H2) or a 3 m long column filled with 80/100 mesh Porapak Q (for CO2 and H2S). The analyses of VOCs were performed using (i) a Shimadzu 14A GC equipped with a flame ionization detector (FID) and a 10 m long stainless-steel column filled with 23% SP 1700 on Chromosorb PAW (80/100 mesh) for the analysis of light hydrocarbons (C1–C3), and (ii) a Thermo Trace Ultra GC coupled with a Thermo DSQ Quadrupole Mass Spectrometer (MS) for the analysis of C4+ VOCs. The analytical procedure and instrumental specifications are described in Venturi et al. [43].
The analysis of carbon isotopic composition of CO2 and CH4 (expressed as δ13CO2 and δ13CH4, respectively, in ‰ vs. V-PDB) was performed on gas samples stored in the Tedlar® bags by using a Cavity Ring-Down Picarro G2201-i spectrometer after proper gas sample treatments, as described in Venturi et al. [43].
2.3. Biological Sample Collection
Samples were collected into 50 mL sterile falcon tubes. Then, 20 g of soil samples were taken in the same sites where interstitial soil gases were measured, immediately before the gas sampling.
A pioneer plant, belonging to the Poa genus, present both on the western flank of the volcanic crater and in Levante Bay, was included in the study. Each plant sample analysed was composed of a pool of three plants collected in the same site. Plants were divided into two main compartments: Aerial part and roots. Moreover, rhizospheric and bulk soils were collected from each site, resulting in four different samples for each plant (Levante Bay: LPA, LPR, LPRS, LPBS; La Fossa Crater: CPA, CPR, CPRS, CPBS).
Table 1 reports the list of samples collected from each site.

Table 1.
List of samples collected on Vulcano Island.
2.4. Extraction of Genomic DNA and Next Generation Sequencing
DNA extraction was performed from each sample by using the PowerLyzer® PowerSoil® DNA Isolation Kit (MO BIO laboratories, Inc., Carlsbad, CA, USA) following the manufacturer’s instruction. Concentration and purity of extracted DNA were checked by 0.8% agarose gel electrophoresis. Bacterial 16S rRNA gene contains conserved sequences and nine hypervariable regions, named V1–V9 [44,45]. Hypervariable regions are used as molecular markers for bacterial identification in HTS analysis [46]. In particular, in this study, we sequenced the V3–V4 regions using the primer 341F and 805R [47] according to the protocol reported in the 16S Metagenomic Sequencing Library Preparation protocol from Illumina [48]. Library preparation and demultiplexing were performed following Illumina’s standard pipeline [49]. Libraries were sequenced in a single run using Illumina MiSeq technology with pair-end sequencing strategy with MiSeq Reagent Kit v3. PCR amplification, library construction, and sequencing were performed by an external company (IGA Technology Services, Udine, Italy). Sequence files were deposited in the NCBI sequence read archive (SRA) under the accession PRJNA508599.
2.5. Sequence Analysis
Primers were removed from all the sequences using Cutadapt [50], version 2.4. Sequences were clustered following the DADA2 pipeline (version 1.8) described at https://benjjneb.github.io/dada2/tutorial.html [51], using the R software version 3.4.3 [52]. Sequences were trimmed, quality checked (Figure S1), denoised, and then forward and reverse reads were merged. An amplicon sequence variant (ASV) table was obtained, a higher-resolution analogue of the traditional OTU table [53]. Chimeras were removed; then the taxonomy was assigned to the output sequences through the Silva database (version 128) [54]. Tables produced by the DADA2 pipeline were imported into phyloseq (through the phyloseq R package version 1.22.3), for further analyses of microbiome data. Representative sequences that were not assigned to domain Bacteria were removed from the dataset (Tables S1 and S2).
2.6. Statistical Testing
All statistical analyses were performed into the R environment, version 3.4.3 [52,55]. Good’s coverage estimator was calculated using the formula: 1-(n/N)∙100, where n is the number of sequences found only one time in a specimen (singletons) and N is the total number of sequences assigned to an ASV in that specimen [56].
Bacterial diversity was estimated using the vegan package (version 2.5–4, Dixon 2003). The function ‘diversity’ was used to compute the Shannon index (H), whereas species evenness (J) was estimated as a function of the Shannon diversity and the number of ASVs detected in the sample (S), according to the Pielou’s formula J = H/log(S) [57]. Differences in bacterial diversity between the different sampling sites were tested using the Student’s t-test (‘t.test’ R function), and the possible correlation between samples’ Shannon diversity and environmental parameters was calculated with the ‘cor.test’ function through the Pearson method.
Beta-diversity was analysed with the Bray–Curtis dissimilarity, using the ‘vegdist’ function of the vegan package. Correspondence analysis (CA) and canonical correspondence analysis (CCA) were performed using the ‘cca’ function of the vegan package.
Different community structures were tested using permutational multivariate analysis of variance (‘adonis2’ function of the vegan R package) with 1000 permutations.
The possible correlation between taxa distribution and gas composition of soil samples was evaluated with a Mantel test of correlation (‘mantel’ function of vegan package with Bray–Curtis distance).
The composition of all interstitial gases detected was fitted onto ordination analyses previously produced using the ‘envfit’ function of the vegan package.
2.7. Isolation of Culturable Bacterial Strains
Isolation of bacteria was performed on Tryptic Soy Agar (TSA) medium using as starting material 1 g of soil (for samples CSCS, CSF56, CSF94, CPBS, CPRS, LSV11, LSV13, LSV14, LPBS, LPRS), and 0.5 g of surface sterilized plant tissue (samples CPA, CPR, LPA, LPR) [58].
Samples were serial diluted and plated on TSA medium, in triplicate. Bacterial plate counts were carried out after 48 h incubation at 30 °C. For each sample, a variable number of colonies (based on the different number of isolates obtained from the samples) was isolated on TSA medium and stored at −80 °C in 20% glycerol for further analysis.
Cell lysates of bacterial isolates were prepared by processing with thermal lysis (95 °C for 10 min, 0 °C for 5 min) an isolated colony dissolved in 20 µL of distilled sterile water.
2.8. Random Amplified Polymorphic DNA (RAPD) Analysis
Random amplification of DNA fragments [59] was performed on DNA samples obtained from thermal lysis. Reactions were performed in a 25 μL total volume, as described in Chiellini et al. [58] using primer 1253 (5′-GTT TCCGCCC-3′) [58,60]. Amplicons were analysed by 2% agarose gel electrophoresis. For each biological sample, isolates showing the same RAPD profile were grouped together into the same haplotype.
2.9. Taxonomical Characterization of Culturable Bacterial Strains
One strain for each RAPD haplotype was chosen for the amplification of the 16S rRNA gene, following the protocol described in Chiellini et al. [58]. Amplification of 16S rRNA genes was performed in a final volume of 20 μL, with DreamTaq DNA Polymerase reagents (Thermofisher Scientific, Waltham, MA, USA), 0.5 μM of P0 and P6 primers (P0, 5′-GAGAGTTTGATCCTGGCTCAG; P6 5′-CTACGGCTACCTTGTTACGA) [61] and 1 μL cell lysate was used as template [61]. Amplification conditions were the following: 90″ denaturation at 95 °C, 30 cycles of 30″ at 95 °C, 30″ at 50 °C, and 1′ at 72 °C, followed by a final extension of 10′ at 72 °C. Sequencing of the 16S rRNA amplicon was performed by and external company (IGA Technology Services) with primer P0. Taxonomic affiliation of the 16S rRNA gene sequences was performed using the Classifier tool of Ribosomal Database Project (RDP) [62]. Each sequence obtained was submitted to GenBank with the accession numbers from MK249884 to MK249979.
3. Results
3.1. Geochemical Features of Interstitial Soil Gases
3.1.1. La Fossa Crater
Sampling sites at La Fossa Crater were characterized by widely varying soil temperatures at 5 cm depth, sharply decreasing as the distance from the fumarolic emission increased, i.e., 94 °C at less than 1 m (CSF94), 56 °C at about 10 m (CSF56), and 31 °C at about 50 m (CSCS). Similarly, the chemical composition of soil gases changed as a function of the distance from the fumarolic vent. Although N2 was the dominant gas constituent (from 507 to 877 mmol/mol), CO2, H2S, and H2 progressively increased approaching the fumarolic vent, while N2, Ar, and O2 showed an opposite behaviour (Table S3). The isotopic composition of CO2 was slightly heavier than that of the fumarolic gas (−0.86‰ vs. V-PDB; data not reported), with δ13CO2 values progressively increasing from 1.11‰ to 2.39‰ vs. V-PDB moving away from the fumarolic vent. Considering that the isotopic signature of CO2 in the fumarolic gases implies an origin related to mixing of magmatic and carbonate sources [32], the δ13CO2 values of the interstitial gases, which are enriched in 13C, were likely produced by partial consumption of volcanogenic CO2 occurring in the soil due to microbial activity [43]. CH4 showed similar concentrations in CSF94 and CSF56 (6.2 and 7.1 μmol/mol, respectively) and markedly decreased in CSCS (2.3 μmol/mol), while the δ13CH4 values markedly increased from −47.2‰ and –44.8‰ vs. V-PDB in CSF94 and CSF56 to −5.5‰ vs. V-PDB in CSCS. Such a wide range of δ13CH4 values was likely caused by oxidation, at different degrees, of volcanogenic CH4 [43], the latter likely being produced by thermogenic processes acting on pre-existing organic matter at relatively high temperatures typical of a volcanic environment [63].
The organic fraction of interstitial soil gases was largely dominated by alkanes (C2 to C11 compounds, mainly represented by ethane), with relative abundances ranging from 61% to 79% of ΣVOCs (Table S3), followed by aromatics, which were characterized by relative abundances varying from 12% to 26% of ΣVOCs and decreasing as the distance from the fumarolic emission increased. A similar trend was observed for cycloalkanes and S-bearing compounds (≤2.96% and ≤8.75% of ΣVOCs), whilst the relative abundance of O-bearing species sharply increased as the distance from the fumarolic vent increased (from 0.26% to 7.40% of ΣVOCs). Alkenes (only represented by iso-butene) were present at relative abundances varying in a narrow range, from 1.06% to 1.99% of ΣVOCs. Overall, ΣVOCs concentrations ranged from 0.15 to 0.53 μmol/mol and decreased moving away from the fumarolic emission.
3.1.2. Levante Bay
Sampling sites at Levante Bay were characterized by soil temperatures ranging from 30.9 to 37.5 °C and strongly different soil CO2 fluxes (hereafter, ΦCO2), with values of 6.5, 53, and 346 g m−2 day−1 in LSV13, LSV11, and LSV14, respectively (Table S3). Similarly, the chemical composition of interstitial soil gases varied widely among the selected sites. N2 was the dominant gas component (from 537 to 963 mmol/mol), but the CO2/N2 ratio progressively increased with increasing ΦCO2 values (from 0.02 to 0.77), the CO2 concentrations varying from 15 to 411 mmol/mol. H2S and H2 were only detected in LSV11 and LSV14, where O2 and Ar concentrations were from 36 to 44 mmol/mol and around 7 mmol/mol, respectively, and CH4 ranged from 1.7 to 1.9 μmol/mol. Differently, LSV13 was characterized by lower O2 (8.7 mmol/mol) and higher Ar (13 mmol/mol) and CH4 (5.1 μmol/mol) contents (Table S3). The isotopic compositions of CO2 and CH4 (Table S3) were largely different among the selected sites, with heavier values in sites characterized by higher ΦCO2 (from −3.33‰ to −1.15‰ vs. V-PDB and from 4.30‰ to 6.47‰ vs. V-PDB, respectively) and lighter values in LSV13 (−8.13‰ and −46.2‰ vs. V-PDB, respectively). Methane concentration was 840 μmol/mol, i.e., two orders of magnitude higher than those of the interstitial soil gases. The isotopic compositions of CO2 and CH4 in this gas sample were −2.21‰ and −16.2‰ vs. PDB, respectively.
Alkanes dominated the organic fraction of interstitial soil gases (Table S3), being represented by C2 to C11 compounds with overall relative abundances ≥74% of ΣVOCs in high ΦCO2 sampling sites and by C2 to C6 compounds with overall relative abundance of 51% of ΣVOCs in LSV13. Ethane was the most abundant alkane, followed by propane. Aromatics (dominated by benzene) represented the second most abundant organic functional group, with relative abundances ≤19% of ΣVOCs in LSV11 and LSV14 and of 43% in LSV13. Iso-butene was constantly present at ca. 2% of ΣVOCs, whereas cycloalkanes and S-bearing species were only detected in LSV11 and LSV14 (up to 1.37 and 3.5% of ΣVOCs). On the other hand, O-bearing compounds were present at relative abundances ≤0.37% of ΣVOCs in LSV11 and LSV14 and of 3.39% in V13. Overall, ΣVOCs concentrations ranged from 0.12 (in LSV13) to 1.48 (in LSV11) μmol/mol.
3.1.3. Pioneer Plant Sites
The chemical composition of interstitial soil gases from the two pioneer plants bulk soil (temperatures around 30 °C) was largely dominated by N2 (up to 965 mmol/mol). No relevant difference was observed in terms of inorganic constituents between CPBS and LPBS, being characterized by CO2 concentrations of 16 and 21 mmol/mol, respectively, and Ar and O2 of 14 and 15 mmol/mol and 5.2 and 3.1 mmol/mol, respectively. H2S and H2 were not detected (Table S3). LPBS, characterized by a low ΦCO2 value (2.6 g m−2 day−1), was slightly enriched in CH4 (5.7 μmol/mol) with respect to CPBS (2.8 μmol/mol). Although the δ13CH4 values were similar in the two sites (−45.4‰ and −47.7‰ vs. V-PDB in CPBS and LPBS, respectively), the δ13CO2 values strongly differed, the interstitial soil gas from CPBS being much heavier (8.60‰ vs. V-PDB) with respect to that from LPBS (−15.5‰ vs. V-PDB) (Table S3).
The chemical composition of the organic fraction of the two interstitial soil gases was different in terms of relative abundances of alkanes and aromatics. The former (C2 to C6 compounds) represented 63% and 58% of ΣVOCs in CPBS and LPBS, respectively, whereas the latter accounted for 24% and 37% of ΣVOCs in the two sites, respectively (Table S3). Cycloalkanes and S-bearing species were not detected, whereas iso-butene was present at ca. 1.6% ΣVOCs in both sites. A sharp difference was observed in the relative abundances of O-bearing compounds, accounting for 12% and 3.8% of ΣVOCs in CPBS and LPBS, respectively. Overall, ΣVOCs concentrations were 0.10 and 0.14 μmol/mol in CPBS and LPBS, respectively (Table S3).
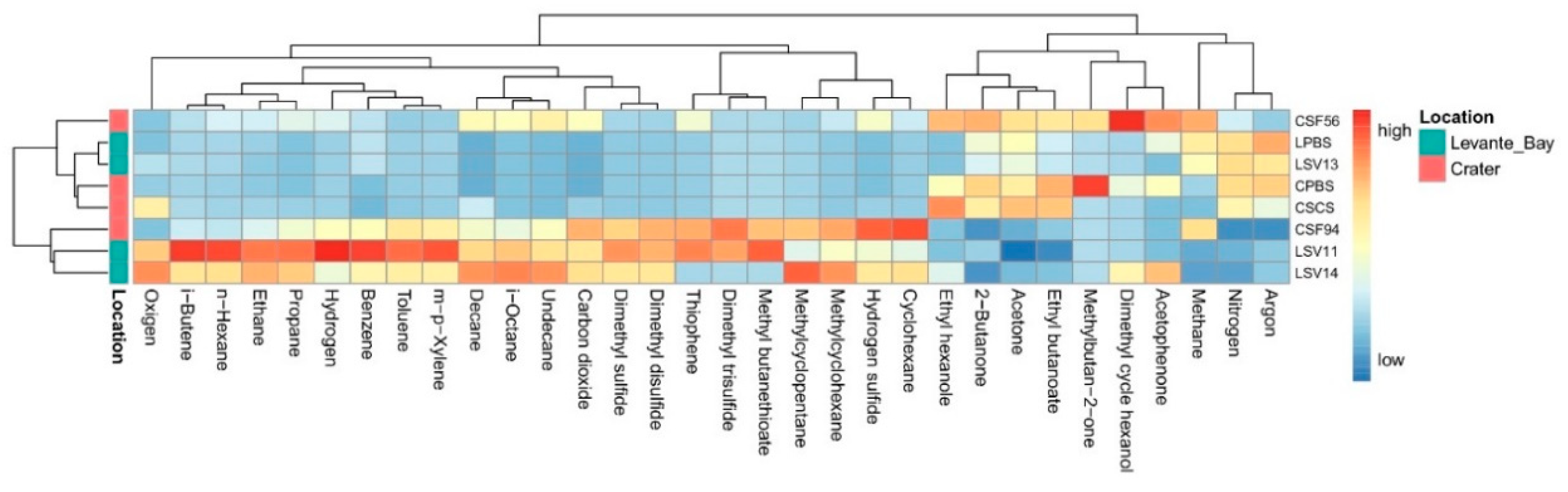
An overall view of the interstitial gases composition of every sample site is shown in Figure 1.
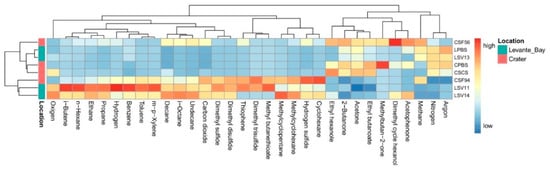
Figure 1.
Heatmap showing the scaled measures of detected interstitial soil gases.
3.2. Molecular Characterization of Bacterial Communities
Bacterial communities of soil and plant tissues samples collected in both emissive and non-emissive areas of Vulcano Island were examined through NGS analysis. Sequencing yielded 2,360,693 paired sequences (2 × 300 bp). More than 60% of the initial pairs were correctly merged (1,446,377 sequences) with a mean of 103,313 sequences per sample. Quality filtering steps produced 975,139 high-quality sequences that were correctly mapped into 5073 ASVs with an average of 69,653 sequences per sample. Samples reported a Good’s coverage estimator ranging from 99% to 100% indicating that roughly 1% of the reads in a given sample came from ASVs that appear only once in that sample [64].
Representative sequences for each ASV were correctly classified into 520 bacterial genera belonging to 23 phyla according to the Silva database (see Materials and Methods), with a different distribution across samples. The analysis of the taxonomic composition revealed that more than 83% of the ASVs were classified into five phyla: Proteobacteria (32.22%), Planctomycetes (16.44%), Actinobacteria (13.56%), Firmicutes (11.93%), and Bacteroidetes (8.95%).
Comparison between La Fossa Crater and Levante Bay Soil and Plant Samples
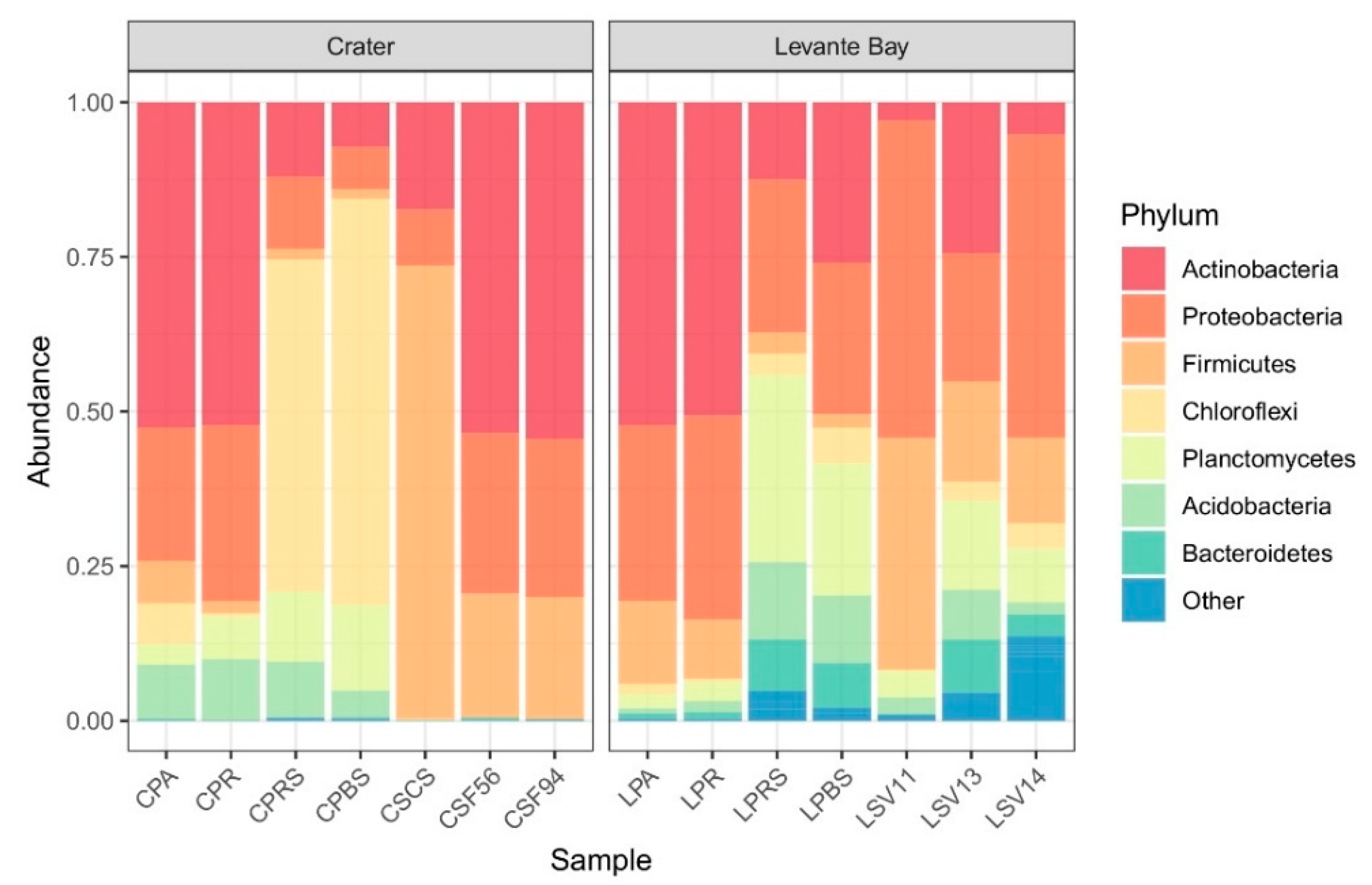
Bacterial communities of soil samples from La Fossa Crater (at different soil temperatures) and Levante Bay (at different soil CO2 fluxes) were analysed, and data obtained are shown in Figure 2.
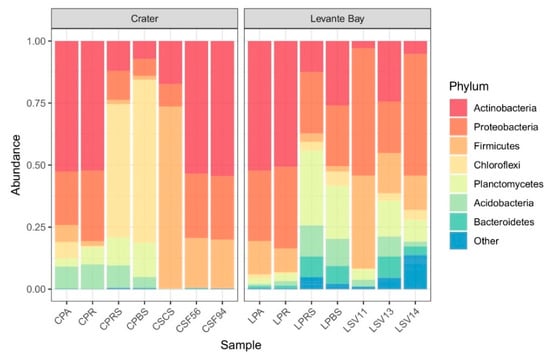
Figure 2.
Bar plot showing the relative abundances of bacterial phyla in each sample (from Crater, on the left, and Levante Bay, on the right). Amplicon sequence variants (ASVs) representing <3% of the whole community were reported as “Other”.
Exploring the different communities at the phylum level, the Crater soil samples showed the presence of three main phyla (Actinobacteria, Proteobacteria, and Firmicutes) that, in the samples CSCS, CSF56, and CSF94, comprise more of the 99% of the detected ASVs. Both CSF94 and CSF56 showed a frequency of over 50% in Actinobacteria (54.5% and 53.5%, respectively), of 25.6% and 25.9% in Proteobacteria, and of 19.7% and 20% in Firmicutes. The sample CSCS showed frequencies of 73.3% in Firmicutes, 17.3% in Actinobacteria, and 9.1% in Proteobacteria. Conversely, the sample CPBS presented a frequency in Chloroflexi equal to 65.6%, and in Planctomycetes of 13.9%, followed by Actinobacteria (7.3%), Proteobacteria (6.8%), Acidobacteria (4.3%), and Firmicutes (1.6%).
In Levante Bay soil samples, the main phyla detected were Proteobacteria (51.3%, 20.7%, 49%, and 24.4%), Firmicutes (37.5%, 16.2%, 13.9%, and 2.2%), Planctomycetes (4.4%, 14.4%, 8.9%, and 21.4%), and Actinobacteria (2.9%, 24.4%, 5.2%, and 26%) in LSV11, LSV13, LSV14, and LPBS, respectively.
Bacterial communities of Poa plant aerial part, roots, rhizosphere, and bulk soil from both Levante Bay and La Fossa Crater were analysed too (Figure 2). All the samples presented a high frequency in Actinobacteria and Proteobacteria, except for the samples CPBS and CPRS in which Chloroflexi are predominant (65.6% and 53.7%, respectively).
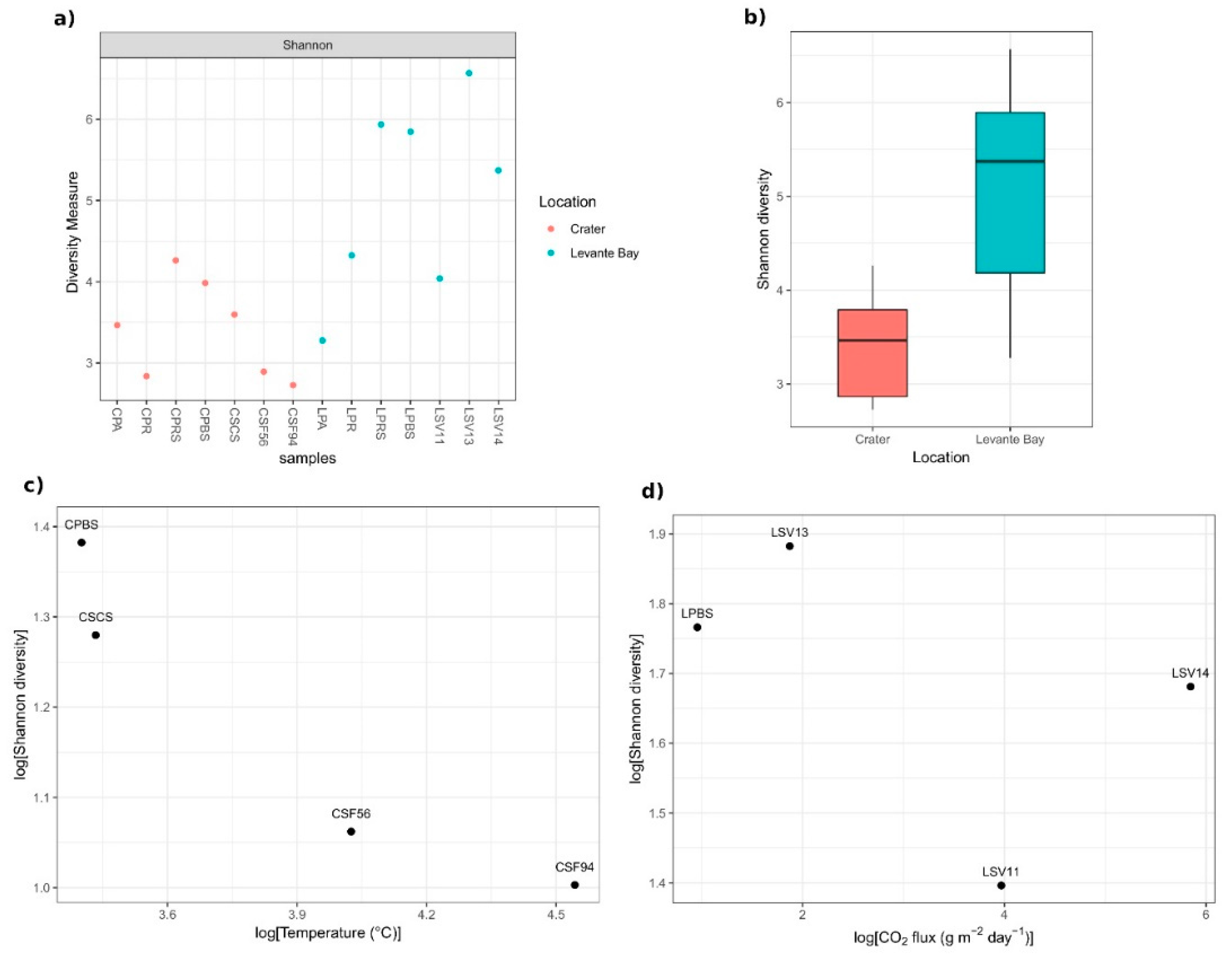
Levante Bay samples showed a higher biodiversity than Crater samples (Figure 3a,b), and the Student’s t-test between the Shannon indices and evenness of the two sampling sites resulted in statistically significant p-values (p-value = 0.0033 and 0.0028, respectively).
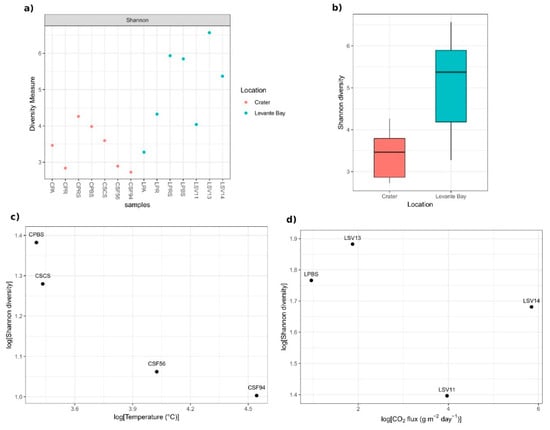
Figure 3.
Shannon diversity of samples bacterial communities. (a) Samples’ Shannon diversity indices, calculated on raw data; (b) box-whiskers plot of the indices based on the different sampling area: Ends of the whiskers represent the lowest and the highest point still within 1.5 of the inter quartile range (IQR); (c) relation between Crater samples soil temperatures and Shannon diversity indices (log transformation of the data has been performed); (d) absence of correlation between Levante Bay samples CO2 fluxes and Shannon diversity indices (log transformation of the data has been performed).
A reduction in the Shannon diversity of the Crater soil samples was observed as the distance of the sampling site to the fumarolic emission decreases (so, as the soil temperature enhancing) (Pearson’s r = −0.94, p-value = 0.06) (Figure 3c). The analysis of the Levante Bay soil samples did not reveal a linear correlation between the diversity and the soil CO2 fluxes (Pearson’s r = −0.50, p-value = 0.50) (Figure 3d).
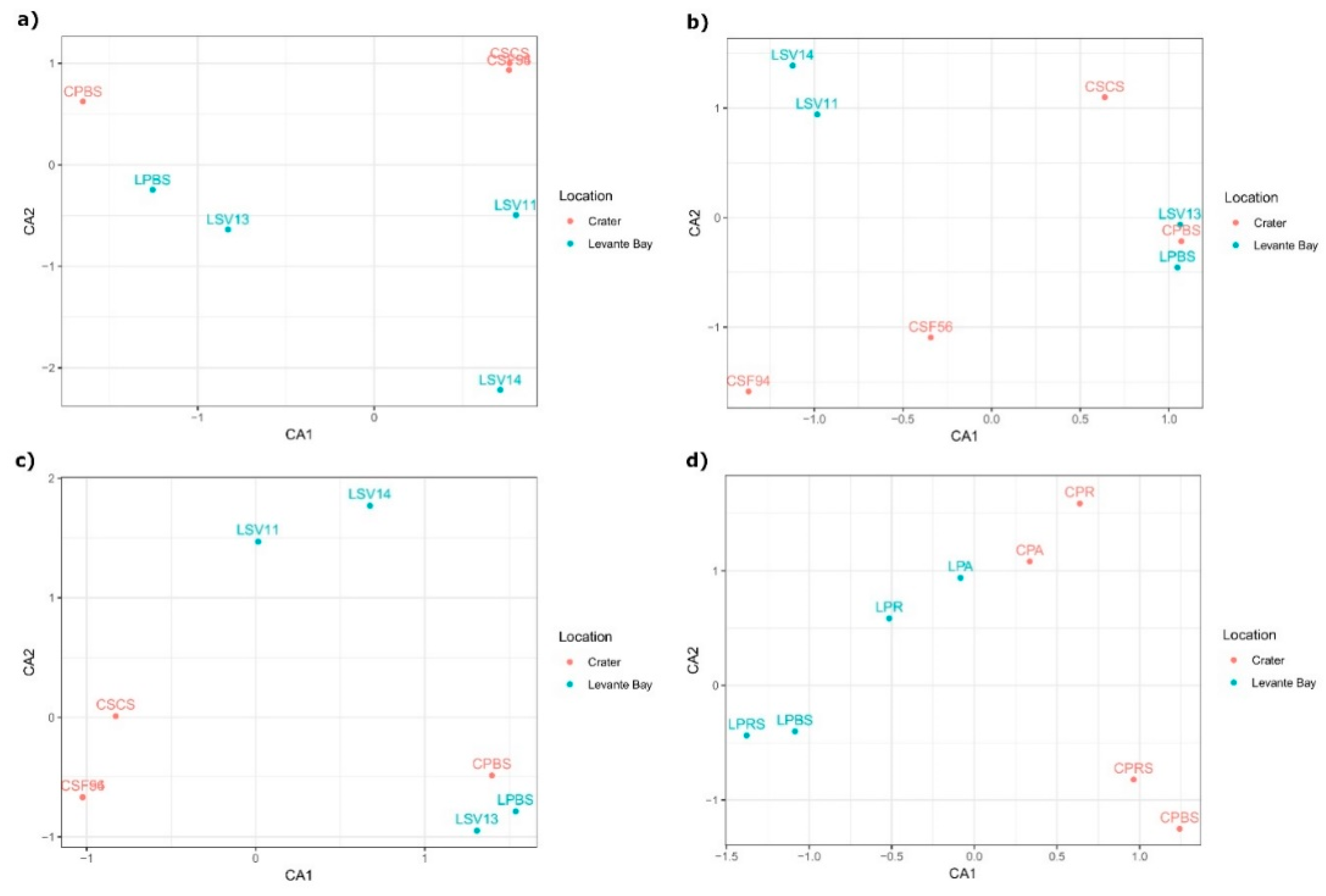
Correspondence analysis (CA) showed a high similarity between the Crater soil samples CSF94, CSF56, and CSCS, while Levante Bay soil bacterial communities of the different samples resulted in being diverse from each other (Figure 4a). Different community structures were tested using permutational multivariate analysis of variance, resulting in a coefficient of determination R2 = 0.24 and in a non-statistically significant p-value (p-value = 0.09).
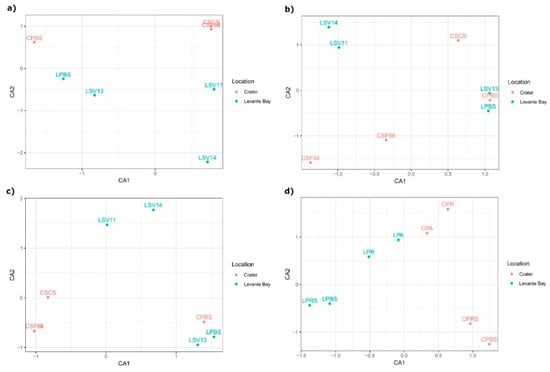
Figure 4.
Correspondence analysis (CA) ordination analysis of samples based on (a) soil microbiome taxonomic composition; (b) soil gas emissions; (c) soil microbiome taxonomic composition collapsed at the genus level; (d) plant microbiome taxonomic composition.
To evaluate whether the differences between the two sampling sites, in terms of composition of the bacterial community, can be possibly correlated to the gas emissions present in these areas, a CA analysis was performed on the samples on the basis of the gas emissions (Figure 4b). In this case, the samples LSV11 and LSV14 appeared to be similar to each other, as did LPBS, CPBS, and LSV13, whereas the other samples were separated. Permutational multivariate analysis of variance resulted in a p-value = 0.97, showing no interstitial gases differences between the sampling sites.
Subsequently, a CA analysis was performed on the microbiome taxa composition collapsed at the genus level (Figure 4c): Both the CA analysis and the CCA analysis constrained on the interstitial gases composition (Figure S2) resulted in the same spatial distribution of the samples, which appeared to be comparable to the distribution of the samples based on the gases composition: In particular, samples LSV13, LPBS, and CPBS, and LV11 and LSV14 formed two clusters that are conserved in both the distributions. However, a contrasting pattern was observed for the samples CSF94, CSF56, and CSCS, when comparing microbiome and gases ordinations. Indeed, a Mantel test showed an overall non-significant correlation between taxa and gases composition (p-value > 0.1).
To assess the possibility that one (or more) gases could affect the taxonomic distribution, the fitting of the interstitial gases composition into taxa ordination analysis was performed, resulting in significant p-values for decane (p = 0.009), argon (p = 0.018), i-octane (p = 0.029), and undecane (p = 0.044) (Figure S3).
Beta diversity between plant samples was analysed through CA analysis (Figure 4d) and revealed a marked separation between Levante Bay and La Fossa Crater samples, on the CA1. Permutational multivariate analysis of variance between the two sites gave back a R2 = 0.23 and a p-value = 0.047, resulting in a statistically significant difference between the plants bacterial communities of La Fossa Crater and Levante Bay.
3.3. Culturable Bacterial Community Analysis
Due to the very scarce amount of microbiological studies concerning the cultivable microbiome of Vulcano Island, we performed the isolation of cultivable bacteria from the samples collected in this study. The analysis of the titer of cultivable bacteria in the samples revealed that they contained from 5 × 102 to 1.5 × 105 CFU/g (Table 2).

Table 2.
Table counts of cultivable bacteria in the collected samples.
All the isolates were screened through RAPD analysis in order to split them into groups, on the basis of their RAPD profile (hereinafter haplotype), assuming that isolates exhibiting the same haplotype correspond to the same strain. For one strain for each haplotype, a 16S rRNA gene sequencing was performed, in order to taxonomically affiliate the strain. Table S4 reports the list of strains isolated from the samples collected in this study.
4. Discussion
The comprehension of the mechanisms that may shape bacterial communities living in extreme environments is crucial from both evolutionary and ecological viewpoint. In this context, Vulcano Island (Aeolian Islands, Sicily, Italy) represents an interesting field of study for analysing bacterial communities of extreme environments, since it offers a multitude of different situations in a very narrow area. Even though previous studies have been carried out on microbial communities of Vulcano Island, very little is known about the possible correlation between interstitial soil gases and soil bacteria. Thus, in this work, we aimed to analyse bacterial communities from different substrates of Vulcano Island and explore the possibility that these bacteria may participate in the production of interstitial gases.
4.1. Bacterial Communities Study
A wide number of studies on extreme environments microbial communities has been carried out in the last decades. Chan et al. performed a similar bacterial communities’ study on Malaysian circumneutral hot springs, finding that Firmicutes and Proteobacteria dominated this environment. Medrano-Santillana et al. found that the bacterial communities in volcanic soils were characterized by the prevalence of Firmicutes, Proteobacteria, and Actinobacteria [65,66]. The same phyla were disclosed in this study, along with others, such as Planctomycetes and Bacteroidetes, in agreement with other studies showing that members of these phyla have been found in a wide variety of environments, confirming their strong versatility.
4.1.1. Soil Samples Analysis
Analysing the composition of the bacterial communities of La Fossa Crater and Levante Bay samples, it was shown that Crater samples exhibited a lower biodiversity with respect to Levante Bay ones (Figure 3a,b). Considering the Shannon diversity of the bacterial communities of La Fossa Crater and Levante Bay soil samples, it was shown that Crater samples exhibited a negative trend between the Shannon diversity index and the proximity to the fumarolic emission, in agreement with previous studies showing that extreme environments are less diverse than other ones [67]. Moreover, Crater samples, with the exception of CPBS sample, showed a major taxonomic uniformity compared to Levante Bay ones, confirming that more emissive environments select only few taxa able to survive under these conditions (Figure 4a).
The correlation between the soil bacterial composition and interstitial soil gases composition was analysed. The ordination analyses resulted in different spatial distribution of samples between the taxa composition CA plot and the gases CA plot (Figure 4a,b). These data did not highlight the existence of a direct influence of bacteria on the interstitial gases’ composition, or vice versa. However, bacterial communities of Levante Bay samples differentiate from each other on the basis of the sampling sites (they present differences in the relative abundances of the main phyla detected, that are Proteobacteria, Firmicutes, Planctomycetes, and Actinobacteria), but belong to the same clusters in the interstitial gases ordination analysis; therefore, we can hypothesize that, in spite of their taxonomical diversity, they could share metabolic abilities related to the gas composition. On the other hand, we can also speculate that sites with similar bacterial communities’ composition and different interstitial gases (i.e., Crater samples) could host different genetic repertoire responsible for different functions within the same taxonomic unit.
Performing a CA analysis on the taxa composition collapsed at the genus level (Figure 4c); the distribution of samples obtained appeared to have some similarities to the distribution based on interstitial gases composition. This could suggest the possibility of a direct relation between some soil bacterial genera and interstitial soil gases. However, a Mantel test did not support statistical correlation, possibly in relation to the relatively low number of samples analysed. Anyway, we can hypothesize that some genetic traits, possibly related to the presence of some interstitial gases, could be shared among the same genus.
Further, the fitting of the interstitial gases’ composition into the taxa ordination analysis was performed to evaluate the possibility that one (or more) gases could affect the taxonomic distribution. The analysis suggested that decane, argon, i-octane, and undecane could be related with the different composition of the microbial communities; however, data obtained from this study do not allow to understand whether the gases affect the taxa distribution, or if bacteria are involved in the production of such gases. Moreover, since these results do not represent a cause-effect relation, it is not possible to know which group of bacteria is responsible for such evidence.
4.1.2. Plant Samples Analysis
It is known that microbial communities associated with plants can establish relationships with the host, giving benefits to both parts [23]. Accordingly, in an extreme environment, it is plausible that the role of bacteria could be of major importance for the survival of the plant [24].
In this study, we choose a pioneer plant belonging to the Poa genus, since they are widely distributed in volcanic environments, as a model to study the bacterial communities associated with it.
The bacterial communities of plants tissues and the surrounding soil from both Levante Bay and La Fossa Crater presented the predominance of Actinobacteria and Proteobacteria, except for two Crater soil samples (bulk soil and rhizospheric soil) in which Chloroflexi are the most abundant phyla detected (Figure 2). In a previous study [68], Chloroflexi phylum has been found as dominant group in the Atacama Desert, the oldest and driest desert located in South America. So, it is possible that the aridity might represent a driving force for the development of such communities. At the same time, the absence of this phylum in the Crater soil samples CSCS, CSF56, and CSF94 could be due to the presence of higher fumarolic emissions in these sites.
Evaluating the distances between the samples, ordination analysis resulted in a marked separation between Levante Bay and Crater samples showing that, despite the differences between the plant compartments analysed, the composition of bacterial communities between the two sampling areas is more different than between the plant compartments of the same area (Figure 4d).
4.2. Cultivable Bacteria
Due to the very scarce amount of microbiological studies concerning the cultivable microbiome of Vulcano Island, we also isolated cultivable bacteria from the samples for further analyses and future perspectives. About the soil samples, the vital titer resulted in a difference between sampling areas: In particular, samples collected in La Fossa Crater showed a lower amount of cultivable bacteria, with a total absence in the samples CSF94 and CSF56, collected at the temperatures of 94 °C and 56 °C, respectively, and in proximity of the fumarolic emission. Concerning the plants samples, there are no obvious differences between the sampling sites.
Analysing the 16S rRNA gene of the isolates (Table S4), a prevalence of the Bacillus genus in Levante Bay soil and plant tissue samples was observed, while in Crater samples, a higher diversity of cultivable bacteria was detected, with isolates mainly belonging to Arthrobacter and Staphylococcus genera, and the almost total absence of Bacillus genus.
5. Conclusions
Vulcano Island is an interesting place harbouring different geochemical situations able to shape bacterial communities. In this context, our work represents one of the first geo-microbiological approaches for the study of volcanic environments. Data obtained on the composition of the bacterial communities from different sites collapsed at the genus level with different emission profiles suggested the possibility of a correlation between the two parameters. It is plausible to imagine that the ability of different bacterial genera/species to survive in the same area might be due to the selection of particular genetic traits, allowing the survival of these microorganisms. On the other side, the finding that microbial communities inhabiting different sites exhibiting different emission profiles are similar might be explained on the basis of a possible sharing of metabolic abilities related to the gas composition.
Moreover, analysing the gases individually, a possible correlation between decane, argon, i-octane, and undecane and the different composition of the microbial communities emerged, even though it is not possible to know which group of bacteria is responsible for such evidence.
Additional analyses are still required, involving a major number of samples and a targeted analysis on archaea communities, combined with gas composition profiling.
Supplementary Materials
The following are available online at https://www.mdpi.com/1424-2818/11/8/140/s1, Figure S1: Plots of the estimated error rates of the amplicon dataset, Table S1: Number of reads throughout the filtering steps, Table S2: Relative abundances of reads through the filtering steps, Table S3: Geochemical data of the sampling sites, Table S4: List of the isolated strains and their taxonomic characterization, Figure S2: CCA ordination analysis of soil microbiome taxonomic composition collapsed at the genus level, constrained on the interstitial gases composition, Figure S3: Plot of the gases fitted on the taxa distribution.
Author Contributions
Conceptualization, C.F., S.D.D., S.V., C.C., R.F. and F.T.; data curation, C.F. and S.D.D.; formal analysis, S.D.D. and G.B.; investigation, C.F., S.V. and C.C.; project administration, R.F. and F.T.; supervision, G.B., R.F. and F.T.; writing—original draft, C.F., S.D.D. and S.V.; writing—review and editing, C.F., S.D.D., S.V., C.C., G.B., R.F. and F.T.
Funding
This research received no external funding.
Conflicts of Interest
Funders had no role in the design of the study, in the collection, analyses, or interpretation of data, and in the writing of the manuscript, or in the decision to publish the results.
References
- Li, S.J.; Hua, Z.S.; Huang, L.N.; Li, J.; Shi, S.H.; Chen, L.X.; Kuang, J.L.; Liu, J.; Hu, M.; Shu, W.S. Microbial communities evolve faster in extreme environments. Sci. Rep. 2014, 4, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chiellini, C.; Miceli, E.; Bacci, G.; Fagorzi, C.; Coppini, E.; Fibbi, D.; Bianconi, G.; Mengoni, A.; Canganella, F.; Fani, R. Spatial structuring of bacterial communities in epilithic biofilms in the Acquarossa river (Italy). FEMS Microbiol. Ecol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Canfora, L.; Lo Papa, G.; Vittori Antisari, L.; Bazan, G.; Dazzi, C.; Benedetti, A. Spatial microbial community structure and biodiversity analysis in “extreme” hypersaline soils of a semiarid Mediterranean area. Appl. Soil Ecol. 2015. [Google Scholar] [CrossRef]
- Yadav, A.N.; Verma, P.; Kumar, M.; Pal, K.K.; Dey, R.; Gupta, A.; Padaria, J.C.; Gujar, G.T.; Kumar, S.; Suman, A.; et al. Diversity and phylogenetic profiling of niche-specific Bacilli from extreme environments of India. Ann. Microbiol. 2015, 65, 611–629. [Google Scholar] [CrossRef]
- Van Kranendonk, M.J.; Pirajno, F. Geochemistry of metabasalts and hydrothermal alteration zones associated with c. 3.45 Ga chert and barite deposits: Implications for the geological setting of the Warrawoona Group, Pilbara Craton, Australia. Geochem. Explor. Environ. Anal. 2005, 4, 253–278. [Google Scholar] [CrossRef]
- Herrera, A.; Cockell, C.S. Exploring microbial diversity in volcanic environments: A review of methods in DNA extraction. J. Microbiol. Methods 2007, 70, 1–12. [Google Scholar] [CrossRef]
- Stott, M.B.; Crowe, M.A.; Mountain, B.W.; Smirnova, A.V.; Hou, S.; Alam, M.; Dunfield, P.F. Isolation of novel bacteria, including a candidate division, from geothermal soils in New Zealand. Environ. Microbiol. 2008, 10, 2030–2041. [Google Scholar] [CrossRef]
- Crisci, G.M.; De Rosa, R.; Esperan, S.; Mazzuoli, R.; Sonnino, M.; Scienze, D.; Calabria, U. Temporal evolution of a three component system: The island of Lipari (Aeolian Arc, southern Italy) GM. Bull. Volcanol. 1991, 53, 207–221. [Google Scholar] [CrossRef]
- Maugeri, T.L.; Lentini, V.; Gugliandolo, C.; Italiano, F.; Cousin, S.; Stackebrandt, E. Bacterial and archaeal populations at two shallow hydrothermal vents off Panarea Island (Eolian Islands, Italy). Extremophiles 2009, 13, 199–212. [Google Scholar] [CrossRef]
- Amend, J.P.; Rogers, K.L.; Shock, E.L.; Gurrieri, S.; Inguaggiato, S. Energetics of chemolithoautotrophy in the hydrothermal system of Vulcano Island, southern Italy. Geobiology 2003, 1, 37–58. [Google Scholar] [CrossRef]
- Caccamo, D.; Gugliandolo, C.; Stackebrandt, E.; Maugeri, T.L. Bacillus vulcani sp. nov., a novel thermophilic species isolated from a shallow marine hydrothermal vent. Int. J. Syst. Evol. Microbiol. 2000, 50, 2009–2012. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, T.L.; Gugliandolo, C.; Caccamo, D.; Stackebrandt, E. A polyphasic taxonomic study of thermophilic bacilli from shallow, marine vents. Syst. Appl. Microbiol. 2001, 24, 572–587. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, T.L.; Gugliandolo, C.; Caccamo, D.; Stackebrandt, E. Three novel halotolerant and thermophilic Geobacillus strains from shallow marine vents. Syst. Appl. Microbiol. 2002, 25, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Gugliandolo, C.; Maugeri, T.L.; Caccamo, D.; Stackebrandt, E. Bacillus aeolius sp. nov. a novel thermophilic, halophilic marine Bacillus species from Eolian Islands (Italy). Syst. Appl. Microbiol. 2003, 26, 172–176. [Google Scholar] [CrossRef]
- Insam, H.; Seewald, M.S.A. Volatile organic compounds (VOCs) in soils. Biol. Fertil. Soils 2010, 46, 199–213. [Google Scholar] [CrossRef]
- Mackie, A.E.; Wheatley, R.E. Effects and incidence of volatile organic compound interactions between soil bacterial and fungal isolates. Soil Biol. Biochem. 1999. [Google Scholar] [CrossRef]
- Bending, G.D.; Lincoln, S.D. Inhibition of soil nitrifying bacteria communities and their activities by glucosinolate hydrolysis products. Soil Biol. Biochem. 2000. [Google Scholar] [CrossRef]
- Orlandini, V.; Maida, I.; Fondi, M.; Perrin, E.; Papaleo, M.C.; Bosi, E.; de Pascale, D.; Tutino, M.L.; Michaud, L.; Lo Giudice, A.; et al. Genomic analysis of three sponge-associated Arthrobacter Antarctic strains, inhibiting the growth of Burkholderia cepacia complex bacteria by synthesizing volatile organic compounds. Microbiol. Res. 2014, 169, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Fincheira, P.; Quiroz, A. Microbial volatiles as plant growth inducers. Microbiol. Res. 2018, 208, 63–75. [Google Scholar] [CrossRef]
- Asensio, D.; Peñuelas, J.; Filella, I.; Llusià, J. On-line screening of soil VOCs exchange responses to moisture, temperature and root presence. Plant Soil 2007, 291, 249–261. [Google Scholar] [CrossRef]
- Gagliano, A.L.; Tagliavia, M.; D’Alessandro, W.; Franzetti, A.; Parello, F.; Quatrini, P. So close, so different: Geothermal flux shapes divergent soil microbial communities at neighbouring sites. Geobiology 2016. [Google Scholar] [CrossRef] [PubMed]
- Kai, M.; Haustein, M.; Molina, F.; Petri, A.; Scholz, B.; Piechulla, B. Bacterial volatiles and their action potential. Appl. Microbiol. Biotechnol. 2009, 81, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Rosenblueth, M.; Martínez-Romero, E. Bacterial Endophytes and Their Interactions with Hosts. Mol. Plant-Microbe Interact. 2007, 19, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Kong, Z.; Glick, B.R. The Role of Plant Growth-Promoting Bacteria in Metal Phytoremediation, 1st ed.; Elsevier Ltd.: Amsterdam, The Netherlands, 2017; Volume 71, ISBN 0065-2911. [Google Scholar]
- Verma, P.; Yadav, A.N.; Kumar, V.; Singh, D.P.; Saxena, A.K. Beneficial plant-microbes interactions: Biodiversity of microbes from diverse extreme environments and its impact for crop improvement. In Plant-Microbe Interactions in Agro-Ecological Perspectives; Springer: Singapore, 2017; ISBN 9789811065934. [Google Scholar]
- Fridriksson, S. Plant Colonization of a Volcanic Island, Surtsey, Iceland. Arct. Alp. Res. 2006, 19, 425. [Google Scholar] [CrossRef]
- Korablev, A.P.; Smirnov, V.E.; Neshataeva, V.Y.; Khanina, L.G. Plant Life-Forms and Environmental Filtering during Primary Succession on Loose Volcanic Substrata (Kamchatka, Russia). Biol. Bull. 2018, 45, 255–264. [Google Scholar] [CrossRef]
- Moral, R.; Wood, D.M. Early primary succession on the volcano Mount St. Helens. J. Veg. Sci. 2006. [Google Scholar] [CrossRef]
- Clarkson, B.D.; Clarkson, B.R.; Juvik, J.O. Pattern and process of vegetation change (succession) on two northern New Zealand island volcanoes. Surtsey Res. 2015, 45–48. [Google Scholar]
- Gabbianelli, G. Submarine morphology and tectonics of Vulcano (Aeolian Islands, Southern Tyrrhenian Sea). Acta Vulcanol. 1991, 1, 135–141. [Google Scholar]
- Calanchi, N.; Peccerillo, A.; Tranne, C.A.; Lucchini, F.; Rossi, P.L.; Kempton, P.; Barbieri, M.; Wu, T.W. Petrology and geochemistry of volcanic rocks from the island of Panarea: Implications for mantle evolution beneath the Aeolian island arc (Southern Tyrrhenian sea). J. Volcanol. Geotherm. Res. 2002, 115, 367–395. [Google Scholar] [CrossRef]
- Capasso, G.; Favara, R.; Inguaggiato, S. Chemical features and isotopic composition of gaseous manifestations on Vulcano Island, Aeolian Islands, Italy: An interpretative model of fluid circulation. Geochim. Cosmochim. Acta 1997. [Google Scholar] [CrossRef]
- Inguaggiato, S.; Mazot, A.; Diliberto, I.S.; Inguaggiato, C.; Madonia, P.; Rouwet, D.; Vita, F. Total CO2 output from Vulcano island (Aeolian Islands, Italy). Geochem. Geophys. Geosyst. 2012, 13, 1–19. [Google Scholar] [CrossRef]
- Garavelli, A.; LAviano, R.; Vurro, F. Sublimate deposition from hydrothermal fluids at the Fossa. Eur. J. Mineral. 1997, 9, 423–432. [Google Scholar] [CrossRef]
- Cheynet, B.; Dall’Aglio, M.; Garavelli, A.; Grasso, M.F.; Vurro, F. Trace elements from fumaroles at Vulcano Island (Italy): Rates of transport and a thermochemical model. J. Volcanol. Geotherm. Res. 2000. [Google Scholar] [CrossRef]
- Pinto, D.; Garavelli, A.; Mitolo, D. Baliczunicite, Bi2O(SO4)(2), a new fumarole mineral from La Fossa crater, Vulcano, Aeolian Islands, Italy. Mineral. Mag. 2014. [Google Scholar] [CrossRef]
- Demartin, F.; Campostrini, I.; Castellano, C.; Gramaccioli, C.M.; Russo, M. D’ansite-(Mn), Na21Mn2+(SO4)10Cl3 and d’ansite-(Fe), Na21Fe2+(SO4)10Cl3, two new minerals from volcanic fumaroles. Mineral. Mag. 2012. [Google Scholar] [CrossRef]
- Boatta, F.; D’Alessandro, W.; Gagliano, A.L.; Liotta, M.; Milazzo, M.; Rodolfo-Metalpa, R.; Hall-Spencer, J.M.; Parello, F. Geochemical survey of Levante Bay, Vulcano Island (Italy), a natural laboratory for the study of ocean acidification. Mar. Pollut. Bull. 2013, 73, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Chiodini, G.; Cioni, R.; Guidi, M.; Raco, B.; Marini, L.; Chiodini, G.; Cioni, R.; Guidi, M.; Raco, B.; Marini, L. Soil CO2 flux measurements in volcanic and geothermal areas. Appl. Geochem. 1998, 13, 543–552. [Google Scholar] [CrossRef]
- Venturi, S.; Tassi, F.; Cabassi, J.; Vaselli, O.; Minardi, I.; Neri, S.; Caponi, C.; Capasso, G.; Di Martino, R.M.R.; Ricci, A.; et al. A multi-instrumental geochemical approach to assess the environmental impact of CO2-rich gas emissions in a densely populated area: The case of Cava dei Selci (Latium, Italy). Appl. Geochem. 2019, 101, 109–126. [Google Scholar] [CrossRef]
- Tassi, F.; Venturi, S.; Cabassi, J.; Capecchiacci, F.; Nisi, B.; Vaselli, O. Volatile organic compounds (VOCs) in soil gases from Solfatara crater (Campi Flegrei, Southern Italy): Geogenic source(s) vs. biogeochemical processes. Appl. Geochem. 2015, 56, 37–49. [Google Scholar] [CrossRef]
- Tassi, F.; Venturi, S.; Cabassi, J.; Vaselli, O.; Gelli, I.; Cinti, D.; Capecchiacci, F. Organic Geochemistry Biodegradation of CO2, CH4 and volatile organic compounds (VOCs) in soil gas from the Vicano—Cimino hydrothermal system (central Italy). Org. Geochem. 2015, 86, 81–93. [Google Scholar] [CrossRef]
- Venturi, S.; Tassi, F.; Magi, F.; Cabassi, J.; Ricci, A.; Capecchiacci, F.; Caponi, C.; Nisi, B.; Vaselli, O. Carbon isotopic signature of interstitial soil gases reveals the potential role of ecosystems in mitigating geogenic greenhouse gas emissions: Case studies from hydrothermal systems in Italy. Sci. Total Environ. 2019, 655, 887–898. [Google Scholar] [CrossRef]
- Chakravorty, S.; Helb, D.; Burday, M.; Connell, N.; Alland, D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 2007. [Google Scholar] [CrossRef]
- Petrosino, J.F.; Highlander, S.; Luna, R.A.; Gibbs, R.A.; Versalovic, J. Metagenomic pyrosequencing and microbial identification. Clin. Chem. 2009, 55, 856–866. [Google Scholar] [CrossRef]
- Huse, S.M.; Dethlefsen, L.; Huber, J.A.; Welch, D.M.; Relman, D.A.; Sogin, M.L. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008. [Google Scholar] [CrossRef]
- Herlemann, D.P.R.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl. Environ. Microbiol. 2013. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 1994, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Dada, S.P.H. High resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. 2014. Available online: http//www.R-project.org/ (accessed on 20 August 2019).
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011. [Google Scholar] [CrossRef]
- Good, I.J. The Population Frequencies of Species and the Estimation of Population Parafeters. Biometrika 1953, 40, 237–264. [Google Scholar] [CrossRef]
- Hill, M.O. Diversity and Evenness: A Unifying Notation and Its Consequences. Ecology 1973. [Google Scholar] [CrossRef]
- Chiellini, C.; Maida, I.; Emiliani, G.; Mengoni, A.; Mocali, S.; Fabiani, A.; Biffi, S.; Maggini, V.; Gori, L.; Vannacci, A.; et al. Endophytic and rhizospheric bacterial communities isolated from the medicinal plants echinacea purpurea and echinacea angustifolia. Int. Microbiol. 2015, 17, 165–174. [Google Scholar] [CrossRef]
- Williams, J.G.K.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990. [Google Scholar] [CrossRef]
- Mori, E.; Liò, P.; Daly, S.; Damiani, G.; Perito, B.; Fani, R. Molecular nature of RAPD markers from Haemophilus influenzae Rd genome. Res. Microbiol. 1999. [Google Scholar] [CrossRef]
- Di Cello, F.; Fani, R. A molecular strategy for the study of natural bacterial communities by PCR-based techniques. Minerva Biotecnol. 1996, 8, 126–134. [Google Scholar]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014. [Google Scholar] [CrossRef]
- Capaccioni, B.; Tassi, F.; Vaselli, O. Organic and inorganic geochemistry of low temperature gas discharges at the Baia di Levante beach, Vulcano Island, Italy. J. Volcanol. Geotherm. Res. 2001, 108, 173–185. [Google Scholar] [CrossRef]
- Bacci, G.; Cerri, M.; Lastrucci, L.; Ferranti, F.; Ferri, V.; Foggi, B.; Gigante, D.; Venanzoni, R.; Viciani, D.; Mengoni, A.; et al. Applying predictive models to decipher rhizobacterial modifications in common reed die-back affected populations. Sci. Total Environ. 2018. [Google Scholar] [CrossRef]
- Chan, C.S.; Chan, K.G.; Ee, R.; Hong, K.W.; Urbieta, M.S.; Donati, E.R.; Shamsir, M.S.; Goh, K.M. Effects of physiochemical factors on prokaryotic Biodiversity in Malaysian circumneutral hot springs. Front. Microbiol. 2017. [Google Scholar] [CrossRef]
- Medrano-Santillana, M.; Souza-Brito, E.M.; Duran, R.; Gutierrez-Corona, F.; Reyna-López, G.E. Bacterial diversity in fumarole environments of the Paricutín volcano, Michoacán (Mexico). Extremophiles 2017. [Google Scholar] [CrossRef]
- Lanzén, A. Analysis of Sequencing Data in Environmental Genomics Exploring the Diversity of the Microbial Biosphere. Ph.D. Thesis, University of Bergen, Bergen, Norway, 2013. [Google Scholar]
- Neilson, J.W.; Quade, J.; Ortiz, M.; Nelson, W.M.; Legatzki, A.; Tian, F.; LaComb, M.; Betancourt, J.L.; Wing, R.A.; Soderlund, C.A.; et al. Life at the hyperarid margin: Novel bacterial diversity in arid soils of the Atacama Desert, Chile. Extremophiles 2012, 16, 553–566. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).