Fish Assemblage Structure in the Northwestern Hawaiian Islands Is Associated with the Architectural Complexity of Coral-Reef Habitats
Abstract
1. Introduction
2. Materials and Methods
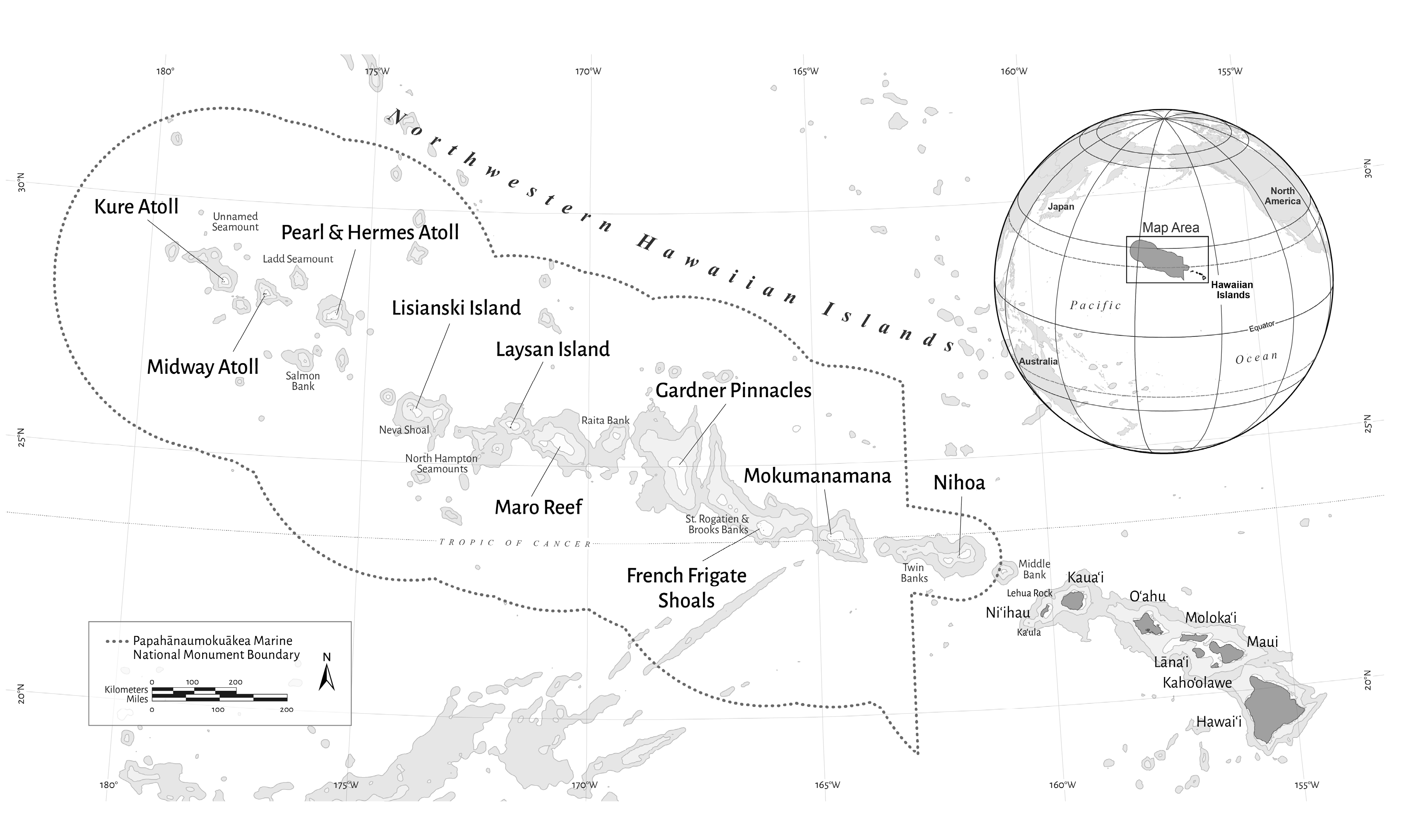
2.1. Fish and Benthic Imagery Surveys
2.2. Generation of 3D Models
2.3. Extraction of Habitat Metrics
2.4. Estimation of Benthic Cover
2.5. Data Analyses
2.5.1. Data Preparation
2.5.2. Univariate Analysis
2.5.3. Multivariate Analysis
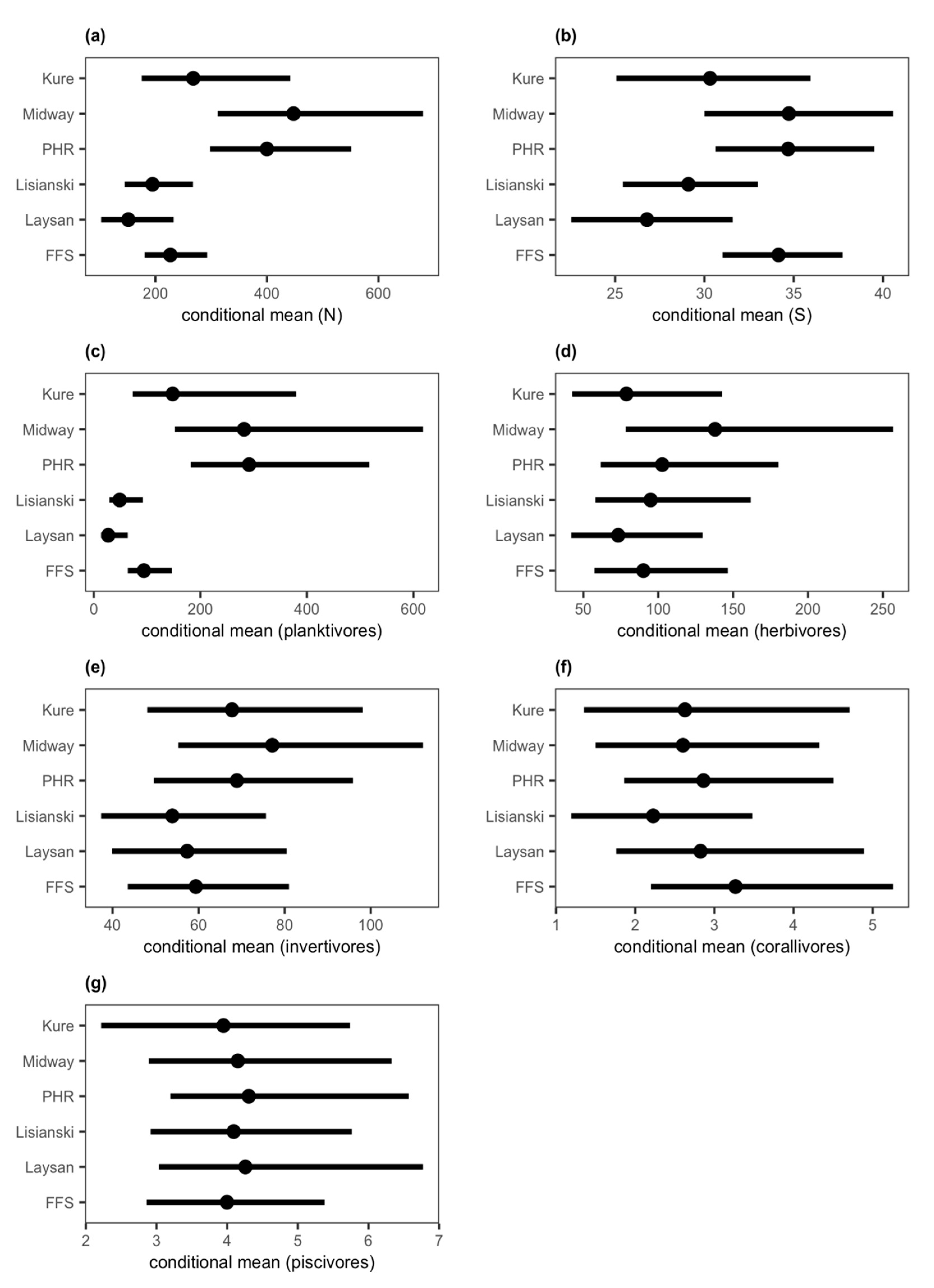
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Depth | VRM 1 cm | VRM Deviation 4 cm | Curvature Range | VRM 1 cm × Curvature Range | Acropora | Pocillopora | sd (loc) | |
|---|---|---|---|---|---|---|---|---|
| total abundance | - | - | - | - | - | - | - | 0.63 (0.25–1.50) |
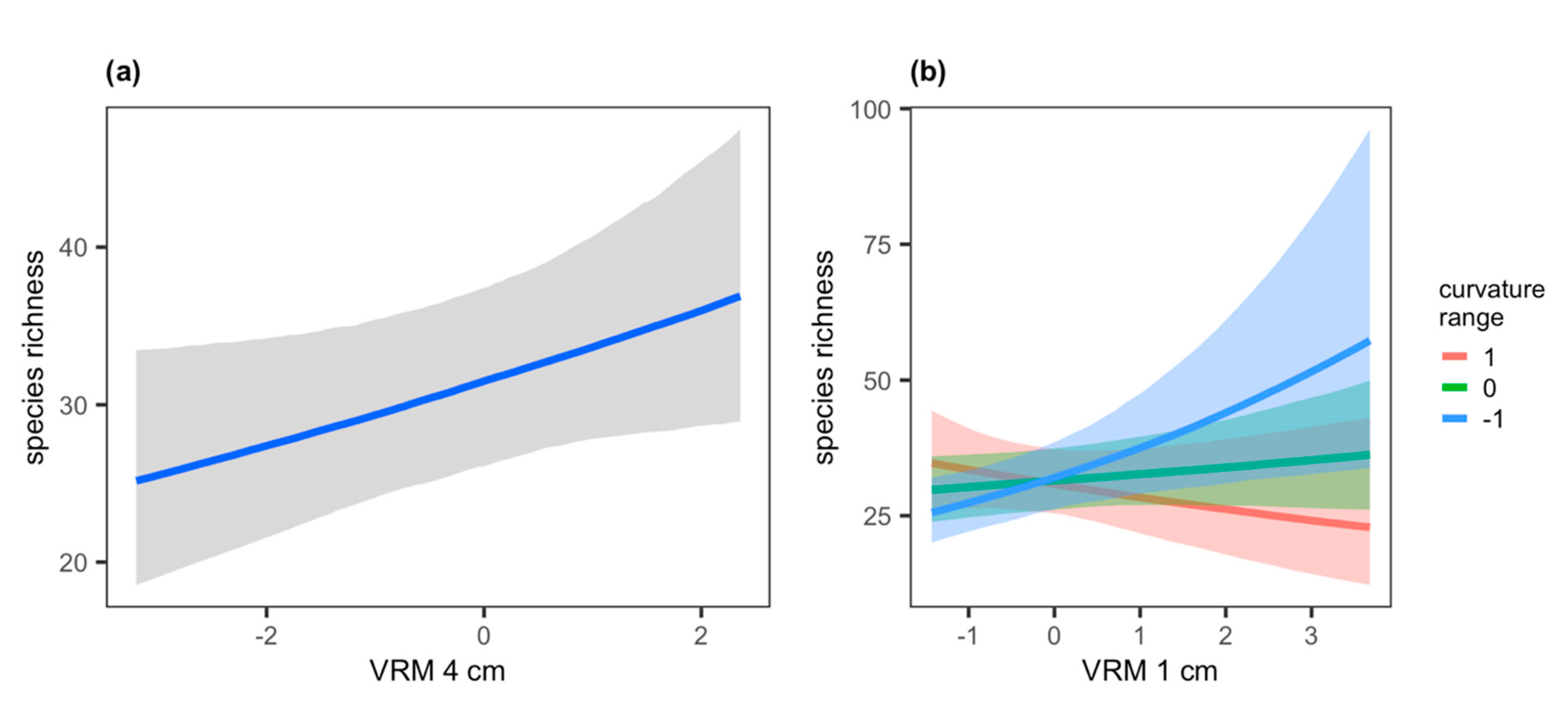
| species richness | - | 0.04 (−0.04–0.11) | 0.07 * (0.00–0.14) | −0.02 (−0.09–0.06) | −0.12 * (−0.24–0.00) | - | - | 0.18 (0.05–0.45) |
| planktivores | - | - | - | - | - | - | - | 1.39 (0.59–3.33) |
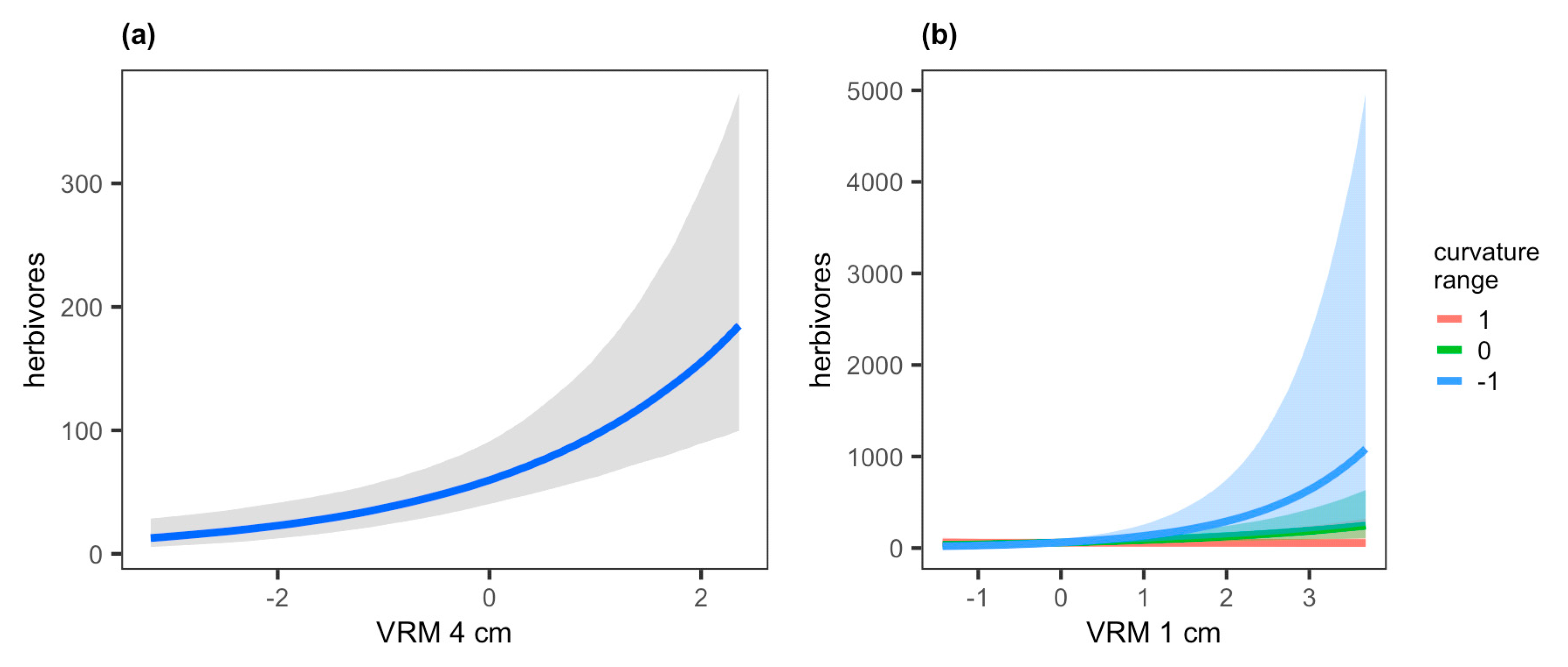
| herbivores | −0.03 * (−0.06–−0.01) | 0.39 * (0.18–0.60) | 0.48 * (0.27–0.71) | −0.03 −0.23–0.18 | −0.39 * (−0.72–−0.06) | - | - | 0.38 (0.04–1.05) |
| invertivores | −0.02 * (−0.04–0.00) | - | - | - | - | - | - | 0.23 (0.03–0.61) |
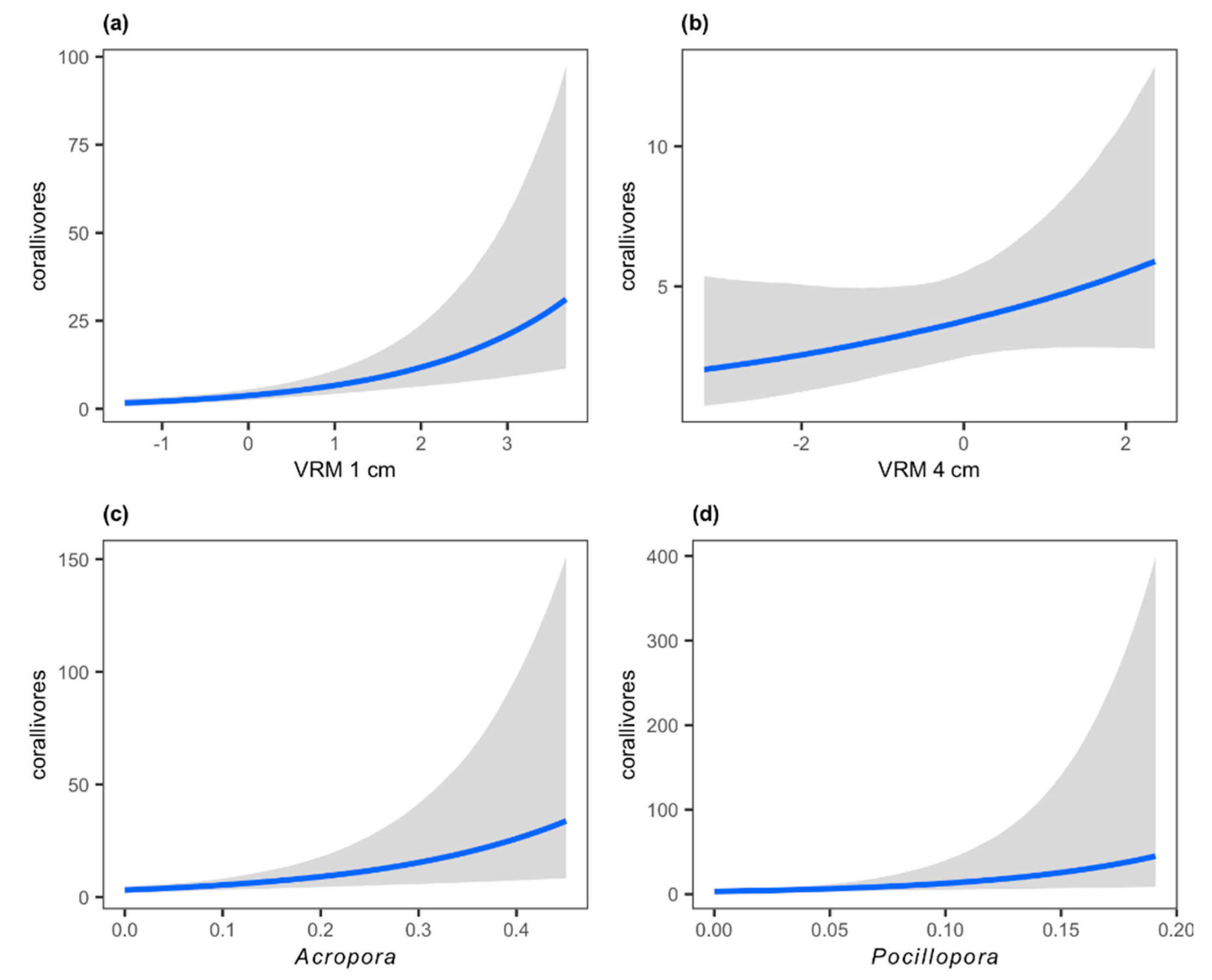
| corallivores | - | 0.58 * (0.31–0.87) | 0.19 (−0.09–0.47) | - | - | 5.33 * (2.17–8.69) | 14.18 * (4.81–25.78) | 0.32 (0.02–0.93) |
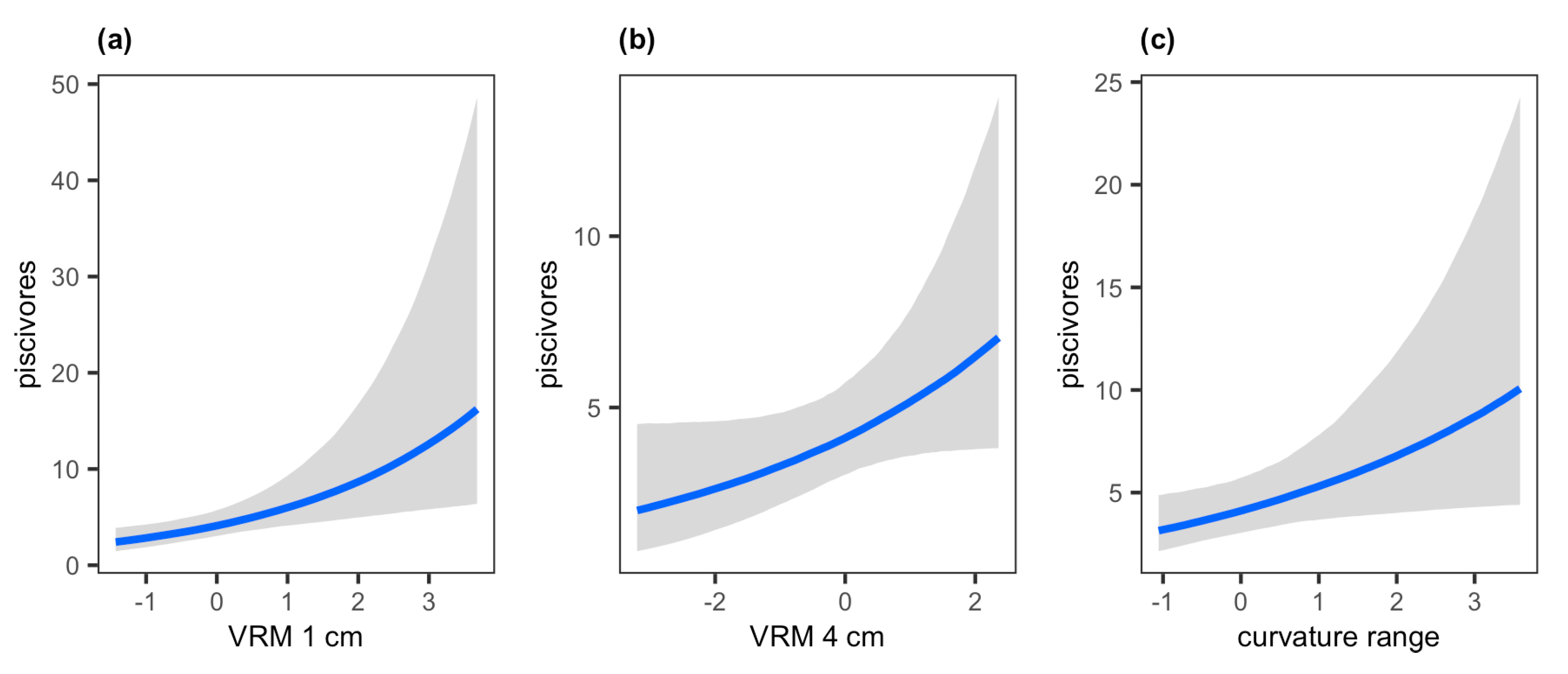
| piscivores | - | 0.38 * (0.13–0.65) | 0.23 (−0.01–0.48) | 0.25 * (0.02–0.48) | - | - | - | 0.19 (0.01–0.67) |
References
- Graham, N.A.J.; Nash, K.L. The importance of structural complexity in coral reef ecosystems. Coral Reefs 2013, 32, 315–326. [Google Scholar] [CrossRef]
- Komyakova, V.; Munday, P.L.; Jones, G.P. Relative importance of coral cover, habitat complexity and diversity in determining the structure of reef fish communities. PLoS ONE 2013, 8, e83178. [Google Scholar] [CrossRef] [PubMed]
- Holbrook, S.J.; Brooks, A.J.; Schmitt, R.J. Variation in structural attributes of patch-forming corals and in patterns of abundance of associated fishes. Mar. Freshw. Res. 2002, 53, 1045–1053. [Google Scholar] [CrossRef]
- Floeter, S.R.; Krohling, W.; Gasparini, J.L.; Ferreira, C.E.L.; Zalmon, I.R. Reef fish community structure on coastal islands of the southeastern Brazil: The influence of exposure and benthic cover. Environ. Biol. Fish. 2007, 78, 147–160. [Google Scholar] [CrossRef]
- Hourigan, T.F. Environmental determinants of butterflyfish social systems. Environ. Biol. Fish. 1989, 25, 61–78. [Google Scholar] [CrossRef]
- Burns, J.H.R.; Delparte, D.; Gates, R.D.; Takabayashi, M. Integrating structure-from-motion photogrammetry with geospatial software as a novel technique for quantifying 3D ecological characteristics of coral reefs. PeerJ 2015, 3, e1077. [Google Scholar] [CrossRef]
- Burns, J.H.R.; Delparte, D.; Kapono, L.; Belt, M.; Gates, R.D.; Takabayashi, M. Assessing the impact of acute disturbances on the structure and composition of a coral community using innovative 3D reconstruction techniques. Methods Oceanogr. 2016, 15–16, 49–59. [Google Scholar] [CrossRef]
- Figueira, W.; Ferrari, R.; Weatherby, E.; Porter, A.; Hawes, S.; Byrne, M. Accuracy and precision of habitat structural complexity metrics derived from underwater photogrammetry. Remote Sens. 2015, 7, 16883–16900. [Google Scholar] [CrossRef]
- Bryson, M.; Ferrari, R.; Figueira, W.; Pizarro, O.; Madin, J.; Williams, S.; Byrne, M. Characterization of measurement errors using structure-from-motion and photogrammetry to measure marine habitat structural complexity. Ecol. Evol. 2017, 7, 5669–5681. [Google Scholar] [CrossRef]
- Raoult, V.; Reid-Anderson, S.; Ferri, A.; Williamson, J.E. How reliable is structure from motion (SfM) over time and between observers? A case study using coral reef bommies. Remote Sens. 2017, 9, 740. [Google Scholar]
- Fukunaga, A.; Burns, J.H.R.; Craig, B.K.; Kosaki, R.K. Integrating three-dimensional benthic habitat characterization techniques into ecological monitoring of coral reefs. J. Mar. Sci. Eng. 2019, 7, 27. [Google Scholar] [CrossRef]
- Risk, M.J. Fish diversity on a coral reef in the Virgin Islands. Atoll Res. Bull. 1972, 153, 1–4. [Google Scholar] [CrossRef]
- Walbridge, S.; Slocum, N.; Pobuda, M.; Wright, D.J. Unified geomorphological analysis workflows with Benthic Terrain Modeler. Geosciences 2018, 8, 94. [Google Scholar] [CrossRef]
- Young, G.C.; Dey, S.; Rogers, A.D.; Exton, D. Cost and time-effective method for multi-scale measures of rugosity, fractal dimension, and vector dispersion from coral reef 3D models. PLoS ONE 2017, 12, e0175341. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.H.R.; Fukunaga, A.; Pascoe, K.H.; Runyan, A.; Craig, B.K.; Talbot, J.; Pugh, A.; Kosaki, R.K. 3D habitat complexity of coral reefs in the Northwestern Hawaiian Islands is driven by coral assemblage structure. In Underwater 3D Recording & Modelling: A Tool for Modern Applications and CH Recording; The International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences: Limassol, Cyprus, 2019; Volume XLII-2/W10, pp. 61–67. [Google Scholar]
- Fukunaga, A.; Burns, J.H.R.; Pascoe, K.H.; Kosaki, R.K. Associations between benthic cover and habitat complexity metrics obtained from 3D reconstruction of coral reefs at different resolutions. Remote Sens. 2020, 12, 1011. [Google Scholar] [CrossRef]
- Williams, I.D.; Couch, C.S.; Beijbom, O.; Oliver, T.A.; Vargas-Angel, B.; Schumacher, B.D.; Brainard, R.E. Leveraging automated image analysis tools to transform our capacity to assess status and trends of coral reefs. Front. Mar. Sci. 2019, 6, 222. [Google Scholar] [CrossRef]
- González-Rivero, M.; Beijbom, O.; Rodriguez-Ramirez, A.; Bryant, D.E.P.; Ganase, A.; Gonzalez-Marrero, Y.; Herrera-Reveles, A.; Kennedy, E.V.; Kim, C.J.S.; Lopez-Marcano, S.; et al. Monitoring of coral reefs using artificial intelligence: A feasible and cost-effective approach. Remote Sens. 2020, 12, 489. [Google Scholar] [CrossRef]
- Hopkinson, B.M.; King, A.C.; Owen, D.P.; Johnson-Roberson, M.; Long, M.H.; Bhandarkar, S.M. Automated classification of three-dimensional reconstructions of coral reefs using convolutional neural networks. PLoS ONE 2020, 15, e0230671. [Google Scholar] [CrossRef]
- Williams, I.D.; Baum, J.K.; Heenan, A.; Hanson, K.M.; Nadon, M.O.; Brainard, R.E. Human, oceanographic and habitat drivers of central and western Pacific coral reef fish assemblages. PLoS ONE 2015, 10, e0120516. [Google Scholar] [CrossRef]
- Bohnsack, J.A.; Bannerot, S.P. A Stationary Visual Census Technique for Quantitatively Assessing Community Structure of Coral Reef Fishes; NOAA Technical Report NMFS 41; U.S. Department of Commerce, National Oceanic and Atmospheric Administration, National Marine Fisheries Service: Silver Spring, MD, USA, 1986; p. 15. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing, 3.6.2; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Beijbom, O.; Edmunds, P.J.; Roelfsema, C.; Smith, J.; Kline, D.I.; Neal, B.P.; Dunlap, M.J.; Moriarty, V.; Fan, T.-Y.; Tan, C.-J.; et al. Towards automated annotation of benthic survey images: Variability of human experts and operational modes of automation. PLoS ONE 2015, 10, e0130312. [Google Scholar] [CrossRef]
- Pebesma, E.J.; Bivand, R.S. Classes and methods for spatial data in R. R News 2005, 5, 9–13. [Google Scholar]
- Bivand, R.S.; Pebesma, E.; Gómez-Rubio, V. Applied Spatial Data Analysis with R, 2nd ed.; Springer: New York, NY, USA, 2013. [Google Scholar]
- Hijmans, R.J. Raster: Geographic Data Analysis and Modeling. R Package Version 3.0-7. 2019. Available online: https://CRAN.R-project.org/package=raster (accessed on 12 May 2020).
- Bivand, R.; Rundel, C. Rgeos: Interface to Geometry Engine—Open Source (GEOS). R Package Version 0.5-2. 2019. Available online: https://CRAN.R-project.org/package=rgeos (accessed on 12 May 2020).
- Fukunaga, A.; Burns, J.H.R. Metrics of coral reef structural complexity extracted from 3D mesh models and Digital Elevation Models. Remote Sens. 2020, 12, 2676. [Google Scholar] [CrossRef]
- Sappington, J.M.; Longshore, K.M.; Thompson, D.B. Quantifying landscape ruggedness for animal habitat analysis: A case study using bighorn sheep in the Mojave Desert. J. Wildl. Manage. 2007, 71, 1419–1426. [Google Scholar] [CrossRef]
- Zevenbergen, L.W.; Thorne, C.R. Quantitative analysis of land surface topography. Earth Surf. Process. Landf. 1987, 12, 47–56. [Google Scholar] [CrossRef]
- Randall, J.E. Reef and Shore Fishes of the Hawaiian Islands; Sea Grant College Program, University of Hawaii: Honolulu, HI, USA, 2007; p. 546. [Google Scholar]
- Fukunaga, A.; Kosaki, R.K. Use of multivariate control charts to assess the status of reef fish assemblages in the Northwestern Hawaiian Islands. PeerJ 2017, 5, e3651. [Google Scholar] [CrossRef] [PubMed]
- Bürkner, P.-C. brms: An R package for Bayesian multilevel models using Stan. J. Stat. Softw. 2017, 80, 1–28. [Google Scholar] [CrossRef]
- Bürkner, P.-C. Advanced Bayesian multilevel modeling with the R package brms. R J. 2018, 10, 395–411. [Google Scholar] [CrossRef]
- Eddelbuettel, D.; François, R. Rcpp: Seamless R and C++ integration. J. Stat. Softw. 2011, 40, 1–18. [Google Scholar] [CrossRef]
- Eddelbuettel, D. Seamless R and C++ Integration with Rcpp; Springer: New York, NY, USA, 2013; p. 220. [Google Scholar]
- Eddelbuettel, D.; Balamuta, J.J. Extending R with C++: A brief introduction to Rcpp. PeerJ Preprints 2017, 5, e3188v3181. [Google Scholar] [CrossRef]
- Hiatt, R.W.; Strasburg, D.W. Ecological relationships of the fish fauna on coral reefs of the Marshall Islands. Ecol. Monogr. 1960, 30, 65–127. [Google Scholar] [CrossRef]
- Hobson, E.S. Feeding relationships of teleostean fishes on coral reefs in Kona, Hawaii. Fish. Bull. 1974, 72, 915–1031. [Google Scholar]
- Hoover, J.P. Hawaii’s Fish; Mutual Publishing: Honolulu, HI, USA, 1993; p. 183. [Google Scholar]
- Legendre, P.; Anderson, M.J. Distance-based redundancy analysis: Testing multispecies responses in multifactorial ecological experiments. Ecol. Monogr. 1999, 69, 1–24. [Google Scholar] [CrossRef]
- McArdle, B.H.; Anderson, M.J. Fitting multivariate models to community data: A comment on distance-based redundancy analysis. Ecology 2001, 82, 290–297. [Google Scholar] [CrossRef]
- Anderson, M.J.; Robinson, J. Generalized discriminant analysis based on distances. Aust. N. Z. J. Stat. 2003, 45, 3301–3318. [Google Scholar] [CrossRef]
- Anderson, M.J.; Willis, T.J. Canonical analysis of principal coordinates: A useful method of constrained ordination for ecology. Ecology 2003, 84, 511–525. [Google Scholar] [CrossRef]
- Pascoe, K.H.; Fukunaga, A.; Kosaki, R.K.; Burns, J.H.R. 3D assessment of a coral reef in the Northwestern Hawaiian Islands reveals varying responses of habitat metrics following a catastrophic hurricane. (manuscript in preparation).
- Hixon, M.A.; Beets, J.P. Predation, prey refuges, and the structure of coral-reef fish assemblages. Ecol. Monogr. 1993, 63, 77–101. [Google Scholar] [CrossRef]
- Alvarez-Filip, L.; Gill, J.A.; Dulvy, N.K. Complex reef architecture supports more small-bodied fishes and longer food chains on Caribbean reefs. Ecosphere 2011, 2, 1–17. [Google Scholar] [CrossRef]
- Gochfeld, D.J. Energetics of a predator-prey interaction: Corals and coral-feeding fishes. Pac. Sci. 1991, 45, 246–256. [Google Scholar]
- Tricas, T.C. Prey selection by coral-feeding butterflyfishes: Strategies to maximize the profit. Environ. Biol. Fish. 1989, 25, 171–185. [Google Scholar] [CrossRef]
- Hourigan, T.F.; Tricas, T.C.; Reese, E.S. Coral reef fishes as indicators of environmental stress in coral reefs. In Marine Organisms as Indicators; Soule, D.F., Kleppel, G.S., Eds.; Springer: New York, NY, USA, 1988; pp. 107–135. [Google Scholar]
- Reese, E.S. Predation on corals by fishes of the family Chaetodontidae: Implications for conservation and management of coral reef ecosystems. Bull. Mar. Sci. 1981, 31, 594–604. [Google Scholar]
- Irons, D.K. Temporal and areal feeding behavior of the butterflyfish, Chaetodon trifascialis, at Johnston Atoll. Environ. Biol. Fish. 1989, 25, 187–193. [Google Scholar] [CrossRef]
- Franklin, E.C. Vagrancy in paradise: Documentation of the chevron butterflyfish Chaetodon trifascialis in Kaneohe Bay, Oahu, Hawaiian islands. Mar. Biodivers. Rec. 2017, 10, 22. [Google Scholar] [CrossRef]
- Jones, R.S. Ecological relationships in Hawaiian and Johnston Island Acanthuridae (Surgeonfishes). Micronesica 1968, 4, 309–361. [Google Scholar]
- Howard, K.G.; Schumacher, B.D.; Parrish, J.D. Community structure and habitat associations of parrotfishes on Oahu, Hawaii. Environ. Biol. Fish. 2009, 85, 175–186. [Google Scholar] [CrossRef]
- DeMartini, E.; Jokiel, P.; Beets, J.; Stender, Y.; Storlazzi, C.; Minton, D.; Conklin, E. Terrigenous sediment impact on coral recruitment and growth affects the use of coral habitat by recruit parrotfishes (F. Scaridae). J. Coast. Conserv. 2013, 17, 417–429. [Google Scholar] [CrossRef]
- Ortiz, D.M.; Tissot, B.N. Evaluating ontogenetic patterns of habitat use by reef fish in relation to the effectiveness of marine protected areas in West Hawaii. J. Exp. Mar. Biol. Ecol. 2012, 432–433, 83–93. [Google Scholar] [CrossRef]
- Randall, J.E. A contribution to the biology of the convict surgeonfish of the Hawaiian Islands, Acanthurus triostegus sandoicensis. Pac. Sci. 1961, 15, 215–272. [Google Scholar]
- Sale, P.F. Pertinent stimuli for habitat selection by the juvenile manini, Acanthurus triostegus sandvicensis. Ecology 1969, 50, 616–623. [Google Scholar] [CrossRef]
- Barlow, G.W. Extraspecific imposition of social grouping among surgeonfishes (Pisces: Acanthuridae). J. Zool. 1974, 174, 333–340. [Google Scholar] [CrossRef]
- Thresher, R.E. Environmental correlates of the distribution of planktivorous fishes in the One Tree reef lagoon. Mar. Ecol. Prog. Ser. 1983, 10, 137–145. [Google Scholar] [CrossRef]
- Gajdzik, L.; Parmentier, E.; Sturaro, N.; Frédérich, B. Trophic specializations of damselfishes are tightly associated with reef habitats and social behaviours. Mar. Biol. 2016, 163, 249. [Google Scholar] [CrossRef]
- Agudo-Adriani, E.A.; Cappelletto, J.; Cavada-Blanco, F.; Cróquer, A. Structural complexity and benthic cover explain reef-scale variability of fish assemblages in Los Roques National Park, Venezuela. Front. Mar. Sci. 2019, 6, 690. [Google Scholar] [CrossRef]
- Richardson, L.E.; Graham, N.A.J.; Pratchett, M.S.; Eurich, J.G.; Hoey, A.S. Mass coral bleaching causes biotic homogenization of reef fish assemblages. Glob. Change Biol. 2018, 24, 3117–3129. [Google Scholar] [CrossRef] [PubMed]
- Hughes, T.P.; Kerry, J.T.; Baird, A.H.; Connolly, S.R.; Dietzel, A.; Eakin, C.M.; Heron, S.F.; Hoey, A.S.; Hoogenboom, M.O.; Liu, G.; et al. Global warming transforms coral reef assemblages. Nature 2018, 556, 492–496. [Google Scholar] [CrossRef]
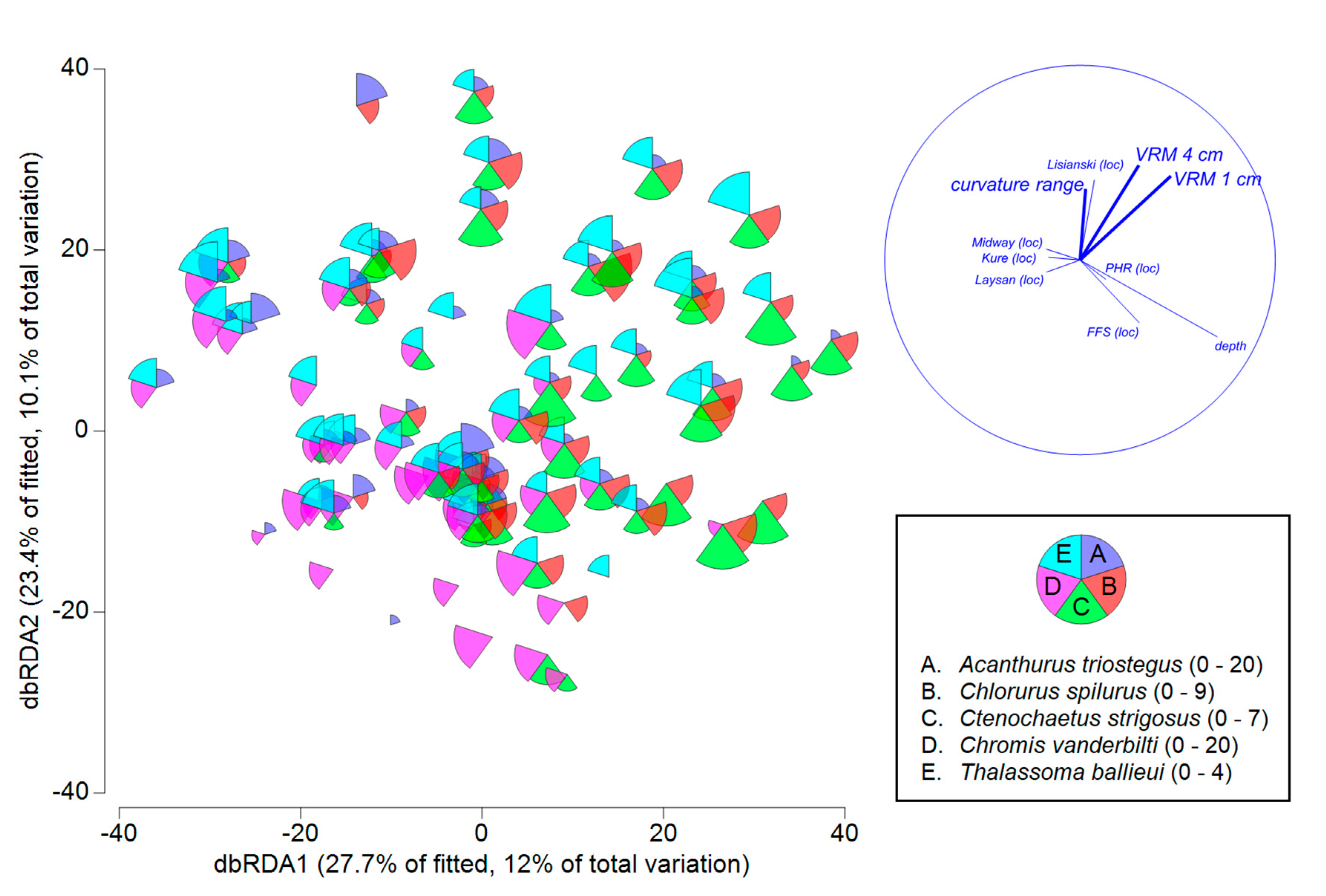
| Variables | Explained SS | Pseudo-F | P | R2 |
|---|---|---|---|---|
| location | 42,474 | 4.650 | 0.0002 | 0.239 |
| +depth | 16,720 | 10.302 | 0.0002 | 0.333 |
| +standardized VRM 1 cm | 3680 | 2.308 | 0.0060 | 0.354 |
| +standardized VRM deviation 4 cm | 10,597 | 7.221 | 0.0002 | 0.414 |
| +standardized curvature range | 3202 | 2.220 | 0.0026 | 0.432 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fukunaga, A.; Kosaki, R.K.; Pascoe, K.H.; Burns, J.H.R. Fish Assemblage Structure in the Northwestern Hawaiian Islands Is Associated with the Architectural Complexity of Coral-Reef Habitats. Diversity 2020, 12, 430. https://doi.org/10.3390/d12110430
Fukunaga A, Kosaki RK, Pascoe KH, Burns JHR. Fish Assemblage Structure in the Northwestern Hawaiian Islands Is Associated with the Architectural Complexity of Coral-Reef Habitats. Diversity. 2020; 12(11):430. https://doi.org/10.3390/d12110430
Chicago/Turabian StyleFukunaga, Atsuko, Randall K. Kosaki, Kailey H. Pascoe, and John H. R. Burns. 2020. "Fish Assemblage Structure in the Northwestern Hawaiian Islands Is Associated with the Architectural Complexity of Coral-Reef Habitats" Diversity 12, no. 11: 430. https://doi.org/10.3390/d12110430
APA StyleFukunaga, A., Kosaki, R. K., Pascoe, K. H., & Burns, J. H. R. (2020). Fish Assemblage Structure in the Northwestern Hawaiian Islands Is Associated with the Architectural Complexity of Coral-Reef Habitats. Diversity, 12(11), 430. https://doi.org/10.3390/d12110430







