Abstract
The axial skeleton of all vertebrates is composed of individual units known as vertebrae. Each vertebra has individual anatomical attributes, yet they can be classified in five different groups, namely cervical, thoracic, lumbar, sacral and caudal, according to shared characteristics and their association with specific body areas. Variations in vertebral number, size, morphological features and their distribution amongst the different regions of the vertebral column are a major source of the anatomical diversity observed among vertebrates. In this review I will discuss the impact of those variations on the anatomy of different vertebrate species and provide insights into the genetic origin of some remarkable morphological traits that often serve to classify phylogenetic branches or individual species, like the long trunks of snakes or the long necks of giraffes.
1. Introduction
Despite the remarkable anatomical diversity observed among vertebrate species, they all have an axial skeleton composed of segmental units, the vertebrae. The diversity observed in the number, size and specific attributes of the different vertebrae and in their specific assembly in the vertebral column reflects different adaptation strategies to the wide variety of ecological niches conquered by vertebrates. In this review, I will discuss different aspects of vertebrate body diversity, mostly focusing on the axial skeleton and aiming at providing an account of the possible mechanisms behind the development of some prominent features that often serve as defining hallmarks of a given species.
Somites represent the first sign of segmentation along the main body axis. These are paired epithelial structures at both sides of the developing spinal cord that generate the vertebrae and ribs, the body dermis and the skeletal muscles of both the body and the limbs [1]. Somites are generated sequentially in a rostral to caudal progression through the process of somitogenesis, occurring at the caudal end of the growing body axis. Somitogenesis has been extensively described in many excellent reviews [2,3,4,5] and will not be discussed here.
The total number of somites, and thus vertebrae, produced during embryonic development varies widely among vertebrates but is fairly constant within a given species. This variability represents one of the major sources of morphological diversity across vertebrate phylogeny. It should be noted that in amniotes somitic contribution to vertebral formation starts with somite pair 5, as more anterior somite pairs participate in the development of the occipital bone [6]. In anamniotes only the first three somite pairs contribute to skull elements [7,8]. This is an important consideration as it directly influences evaluation of the impact that differential regional regulation of somite differentiation has on vertebral identity.
Somites are morphologically similar when they are first produced during somitogenesis, but generate vertebrae with distinct individual features that, again, are highly conserved within a given species but vary significantly across vertebrate phylogeny. While in general unique anatomical identities can be attributed to each vertebral unit, subsets of them share features that led to their classification into five different groups, namely cervical, thoracic, lumbar, sacral and caudal, which can be applied to most vertebrate species. The distribution of the vertebrae among these different groups, normally known as the axial formula [9], is another of the major sources of diversity among vertebrate clades.
A large number of expression and genetic studies have shown that both the individual and regional identity of the different vertebrae mostly results from the activity of genes of the Hox family. Given their important role in generating morphological diversity I will provide a brief outline of their main features, but extensive reviews on their function can be found elsewhere [10,11,12].
In vertebrates the different members of the Hox gene family are located in clusters that are thought to have resulted from sequential duplications of the genome during the emergence of vertebrates [13]. The number of clusters vary across vertebrate phylogeny. For instance, mammals, which often serve as the main reference, contain four clusters, named A to D [13], whereas teleost fishes, like the zebrafish (Danio rerio), have seven clusters [14]. The individual Hox genes (39 in mammals) are classified in 13 paralog groups according to sequence homologies and their position within the cluster, distributed from 1 to 13 in a 3’–5’ orientation within the cluster [13]. The genomic arrangement of Hox genes impacts their spatial and temporal expression during embryonic development: lower number paralogs are the first to be activated in anterior embryonic regions, followed almost sequentially by Hox genes of increasing paralog numbers at more posterior embryonic regions as the embryo extends at its caudal end [13]. This type of regulation results in the expression of different combinations of Hox genes at different axial levels, which is thought to play a significant role in the generation of distinct segmental identities along the main body axis [13].
2. The Neck: Different Solutions to Similar Problems
Neck size varies substantially among vertebrate species. The main source of this variation results from the number of vertebral units allocated to this region of the axial skeleton. Amphibians and snakes occupy one end of the spectrum, containing between one and three cervical vertebrae, whereas birds are placed at the other end, containing a variable but always rather large number of cervical vertebrae, championed by swans with their 25 units (Figure 1) [7,15,16,17]. Mammals are located at the middle of the scale, mostly adhering to the seven cervical vertebrae rule with the exception of manatees and sloths [18]. The origin of this variability has been traced to Hox genes. Comparative studies of Hox gene expression in embryos from species with different numbers of neck vertebrae, including snakes (Pantherophis guttatus), lizards (Aspidoscelis uniparens), caecilians (Ichthyophis cf. kohtaoensis), chickens (Gallus gallus), geese (Anser anser), mice (Mus musculus), alligators (Alligator mississippiennsis) and frogs (Xenopus laevis) [15,17,19,20,21], showed a strong correlation between the somite level at which Hox genes of paralog groups 5 and 6 become activated and the anatomical position of the neck to trunk transition in the vertebral column. Experimental support for this hypothesis was provided by genetic experiments in the mouse. In particular, it has been shown that anterior expansion of Hoxb6 expression anticipated the transition into trunk structures with the concomitant reduction of the neck size [22]. Conversely, global inactivation of Hox group 5 or group 6 genes both moved the neck to trunk transition posteriorly by one or two segments [23]. While these null phenotypes look somewhat milder than what might be expected, it is likely that this derives from functional redundancy between genes in these two paralog groups. Indeed, trans-heterozygote animals for the Hoxb6 and Hoxb5 genes indicated their functional cooperation in skeletal patterning processes [24].
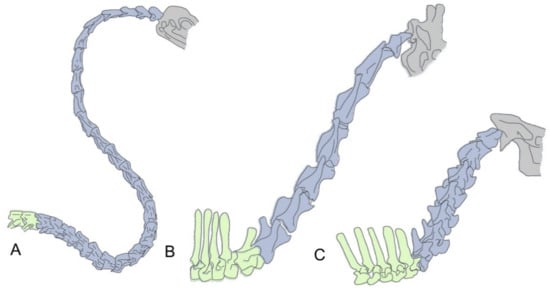
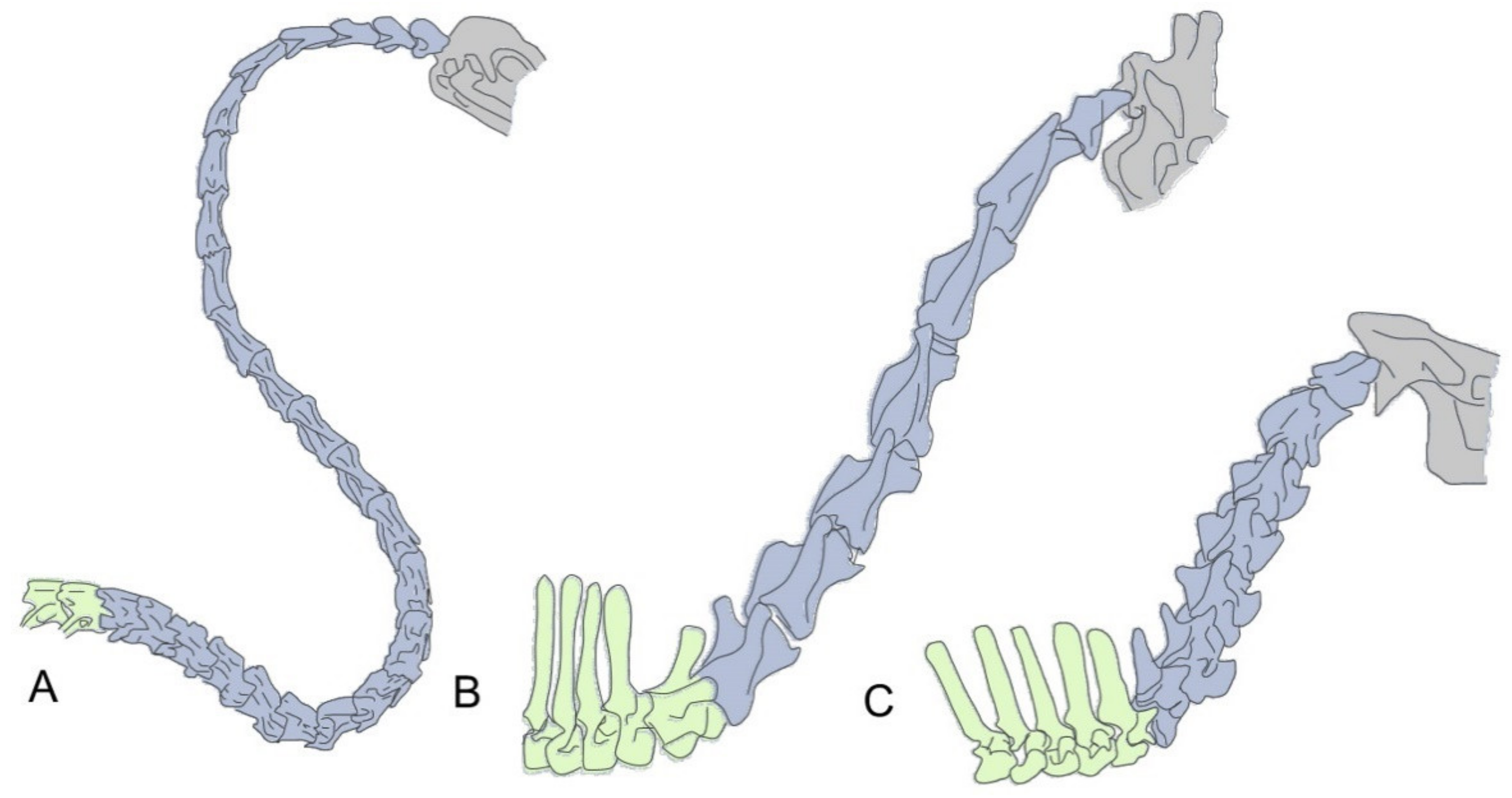
Figure 1.
Two different mechanisms for neck size increase. (A) In birds, neck size is mostly related to the number of cervical vertebrae, illustrated here with the cervical region of a swan (Cygnus olor), which contains up to 25 cervical vertebrae. B,C Most mammals have seven cervical vertebrae and neck size differences depend on the size of their individual cervical vertebrae. Giraffes (B) (e.g., Giraffa camelopardalis) are the paradigmatic example for a long necked mammal. Importantly, comparison with its closely phylogenetically related okapi (Okapia johnstoni) (C) shows that the change in vertebral size is restricted to the cervical region, as their thoracic vertebrae are of equivalent sizes (with the exception of the first thoracic of giraffes). Cervical vertebrae are colored in blue, thoracic vertebrae in green and the head in gray.
The diversity in the number of neck vertebrae might thus reflect species-specific variations in the activation mechanisms of Hox5 and Hox6 paralog genes. So far, a large part of our knowledge about the mechanisms regulating Hox gene expression comes from genetic experiments in mice and embryonic stem cells and, therefore, it is not possible to provide a properly documented account of differential activation mechanisms among species with different neck vertebral counts. However, the identification of regulatory mechanisms involving chromatin structure as a crucial component in the sequential activation of Hox genes [12,25,26,27,28] provides some indications on possible mechanisms for this differential regulation. In particular, it has been shown that the 3’ and 5’ parts of the Hox clusters are embedded within two adjacent topologically associating domains (TADs) [29,30,31], three dimensional chromatin territories marking the genomic space thought to allow interactions between genes and regulatory elements [32,33]. Hox gene activation is controlled by regulatory landscapes within each of the two TADs [12]. Sequential Hox activation thus requires that at a given point transcriptional regulation switches from elements within the 3’ TAD to those in the 5’ TAD [28,30]. This switch might play a relevant role in the Hox5 and Hox6 activation profiles observed in different species. In particular, the border between the two Hox-associated TADs is located at the level of the Hox5 and Hox6 paralogs, and it has been shown that the integrity of this border is important to guarantee proper Hox gene regulation [29,34]. Detailed experiments in mice also showed that activation of Hoxa5 and Hoxa6 is associated with a change in Hox regulation from WNT- to Cdx-dependent mechanisms [35]. It is thus possible that the difference in the number of cervical vertebrae results from variations in the mechanisms eliciting the switch between TAD regulatory landscapes, which could result from distinct intrinsic properties of Cdx or WNT-related factors, or of their downstream effectors. Understanding whether these or other mechanisms are involved in this process will require direct evaluation in different species.
Not all neck size variabilities are associated with a different number of vertebrae allocated to this segment of the axial skeleton. Mammals, for instance, contain a fixed number of cervical vertebrae, but their relative neck size varies extensively among clades. A study including 352 species revealed that with few exceptions, the relative size of the mammalian cervical column correlates inversely with the animal’s body mass [36]. Giraffes represent the best known exception to this inverse allometry and provide a paradigmatic example to illustrate the generation of long necks through a substantial size increase of their cervical vertebral units [37]. Importantly, the cervical vertebrae are the only elements of the giraffe axial skeleton whose size deviates significantly from that observed in the homologous bones from related species with shorter necks [37] (Figure 1), indicating a very precise regional restriction of the mechanisms responsible for the increased vertebral size. Despite the fascination that the long necks of giraffes have generated over the years, we are still far from providing a convincing account of the mechanisms that generate this remarkable anatomical feature. Some possible explanations have been suggested from the genomic sequence analysis of two giraffe species (Giraffa camelopardalis and Giraffa tippelskirchi) [38,39]. One such hypotheses stemmed from the significant giraffe-specific nucleotide changes observed in some members of the NOTCH, WNT and FGF pathways. Given the substantial involvement of these pathways in the temporal dynamics of somitogenesis and, eventually, somite size [4], it was suggested that the regulatory balance between these pathways during giraffe somitogenesis would produce larger somites. Specific attention was given to FGF signaling because experimental alterations in its activity during chicken somitogenesis were paralleled by significant changes in somite size [40]. This suggested the possibility that functional modifications in Fgfrl1, one of the most divergent genes in giraffe [38], could have readjusted the dynamics of somitogenesis in a way that favors production of larger somites. Recent genetic data, however, linked the giraffe-specific Fgfrl1 features not to the size of cervical vertebrae but to their remarkable resistance to high blood pressure [39]. An alternative mechanism for the production of the large vertebrae involves differential growth of the skeletal elements derived from otherwise similarly sized neck somites.
Irrespective from the specific mechanism, however, full understanding of the origin of giraffe’s large cervical vertebrae must also account for its precise regional specificity. Hox genes, and most particularly those of paralog groups 3 and 4, are prime candidates to fulfill this role, as they have been shown to shape the anatomy and size of the cervical vertebrae in the mouse [41,42,43]. Significant sequence changes in giraffe Hox genes when compared to okapi (Okapia johnstoni) were only found in Hoxb3 [38], thus making it unlikely that intrinsic properties of these genes justify the production of the large cervical vertebrae. Hox genes might still control selective growth of cervical vertebrae through significant functional changes in their downstream effectors. For instance, considering that the NOTCH, WNT and FGF signaling pathways are also involved in skeletal growth and remodeling processes [44,45,46], it would be possible that the giraffe specific changes in these pathways enhance their capacity to promote skeletal growth. If any of these factors are part of specific regulatory networks downstream of Hox3 or Hox4 proteins, the activity of these Hox genes in the giraffe embryo could then promote a regional specific increase of the cervical vertebrae. This or other possible hypotheses for the origin of the skeletal neck pattern of giraffes will require direct experimental validation.
Neck vertebral development in cetaceans took a route opposite to that of giraffes: they also have seven vertebral units, but are very small and fused [18], a structural characteristic thought to facilitate swimming by providing stability at the expense of head mobility. The origin of this cervical pattern is essentially unknown but could be related to variations in similar processes as those producing long cervical elements in giraffes but with an opposite value. Independent of the mechanisms, the presence of the same number of cervical vertebrae in giraffes and cetaceans, regardless of their totally different relative size in the adult animal, highlights the inability of mammals to adapt to environmental pressures requiring changes in neck size by modifying the number of vertebral elements, relying instead on alternative mechanisms. Indeed, the seven cervical rule seems to be broken only by manatees and sloths [18,47], and even for sloths, recent data suggest that although they have 8–10 vertebrae before the first segment associated with a sternal rib, ossification criteria indicate that only the first seven vertebrae fit the cervical parameters [48], which would place sloths back into adhering to the seven cervical rule.
There are two main lines of thought to explain this remarkable developmental constrain. One of them links it to the presence of a muscular diaphragm, a mammalian-specific feature playing a fundamental role in their respiratory mechanics and thought to have allowed the development of distinctive mammalian physiological traits, including elevated body temperature and high resting metabolic rates [47,49]. The developmental module that generates and guides migratory pathways of the muscle precursors for the diaphragm are tightly coordinated with those guiding forelimb muscle development, involving adjacent somites of the prospective cervical region [50]. It has been suggested that the tight functional links between these two processes reduces the capacity of the system to allow significant changes without fatally interfering with fundamental features of mammalian physiology [49]. The observation that the diaphragm of manatees differs in its position from those in other mammals, likely associated with the unique position of their lungs along the animals back [51], seems to provide some support to the diaphragm-based hypothesis. The other line of thought stems from the observation of increased incidence of malformations potentially linked to altered Hox gene expression, like ectopic cervical ribs, in children suffering from congenital cancers [52]. It was then proposed that in mammals changes in Hox gene expression required to modify the number of cervical vertebrae would be associated with an increased production of malignances that would interfere with fixing the new developmental traits, an effect less likely to happen in birds or reptiles, given their apparently increased resistance to cancer compared to mammals [52].
It should be noted that, while neck length variation in birds derives to a large extent from the number of cervical vertebrae, which vary from 10 to 25, it has also been shown that the size of the individual elements also contributes to the neck length diversity observed across avian phylogeny, which, contrary to what is observed in mammals, shows general positive allometry with body size [53].
3. Trunk Size and Shape: Extreme Differences
In general terms, it can be considered that the trunk is the body region holding most of the organs of the cardiovascular, digestive, excretory and reproductive systems, roughly corresponding to the thoracolumbar and sacral regions of the axial skeleton. Trunk length varies widely among vertebrates, from spanning the length of just a few vertebrae in frogs to more than 300 in some snakes.
In general, the neck to trunk transition in the axial skeleton is marked by the position of the first vertebra containing a moveable rib attached to the sternum. Therefore, the position of the first trunk vertebra and neck length are two sides of the same process. The new question is then what determines where the trunk ends. As discussed for the neck size, the correlation between activation of specific Hox paralog groups and anatomical transitions in the axial skeleton, together with the role of these genes in specifying segmental identities, suggested their possible involvement as key determinants of trunk size variability among vertebrates. These correlations also brought these genes to center stage in the quest for the origin of the snake body plan, which has been a major driver of research on the mechanisms behind anatomical diversity among vertebrates. Indeed, the finding that activation of Hox paralog groups 10–13 was strongly delayed in snakes compared to other vertebrates, correlating with the end of trunk structures [17,20], was consistent with this hypothesis. Similarly, a more posterior activation of the same Hox paralogs in mutant mice with extended trunks was also indicative of a connection between Hox genes and trunk size [54,55,56]. However, extensive genetic evidence in mice seems to argue against a causal role of Hox genes in this process, as major changes in Hox gene expression, either through their single or combined inactivation or by gain of function approaches, failed to produce significant changes in trunk size, as estimated by the distance between the fore and hind limb buds, despite generating in some cases major vertebral identity changes along the anterior posterior axis [10].
Contrary to what was observed for Hox genes, genetic inactivation of various factors involved in the TGFβ signaling pathway resulted in significant expansions of the trunk region, often at the expense of the development of more caudal areas of the body (Figure 2). Collective analyses of those experiments identified Gdf11, acting through the Tgfbr1 and Acvr2b receptors, as the key activator of the trunk to tail transition and, thus, a major player in the establishment of trunk length [54,55,57,58,59,60,61,62]. While most genetic experiments were performed in mice, recent data indicate that the role of Gdf11 to control the end of trunk development is likely a general feature across vertebrate phylogeny. In particular, it has been shown that its expression closely correlates with the position of the transition to tail development in different vertebrate species [56,63]. In addition, experimental modulation of Gdf11 activity was able to change the position of the trunk to tail transition in animals other than mice [63,64].
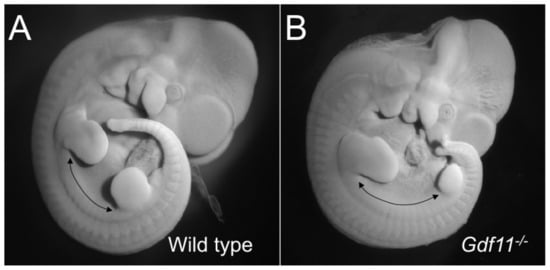
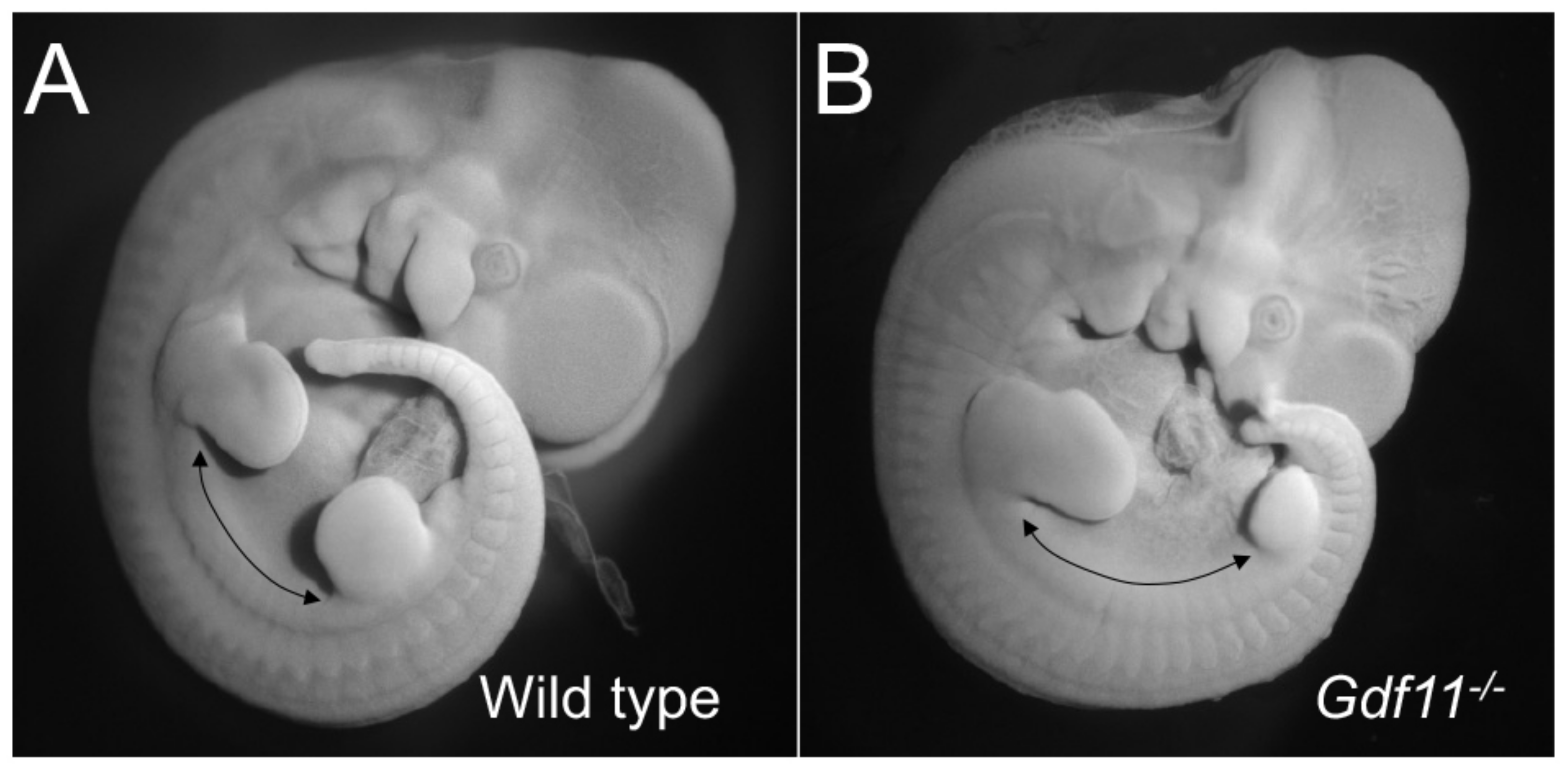
Figure 2.
Trunk elongation in the absence of Gdf11 signaling. The figure shows a mouse wild type (A) and a Gdf11-/- embryo (B) at embryonic stage 11.5, to illustrate the increase in size of the interlimb region (as estimated by the number of somites), which represents the trunk region, in Gdf11 mutant embryos.
An extension of the trunk region has also been reported for mice lacking all three miR196 genes [65]. Interestingly, experimental miR196 up- or downregulation during zebrafish embryonic development produced small trunk contractions or expansions, respectively, further linking miR196s to trunk size regulation [66]. miR196 genes are located within the Hox clusters and have been reported to be able to control expression of some Hox genes [67]. Hox genes also showed abnormal patterns in miR196 global mutants that correlate with the changes in the axial skeleton [65]. However, it is not clear whether these changes in Hox expression patterns derive from direct regulation by miR196 and if changes in Hox gene expression are the cause or the consequence of the extended trunk region in the miR196 mutants. In this regard, there are some indications suggesting that miR196s could control the Gdf11 pathway, as the global miR196 mutant phenotype resembles features observed in some Gdf11 heterozygous mice and Gdf11 expression was significantly reduced in miR196 mutant embryos [65]. Direct experimental analyses will be required to determine whether or not miR196 and Gdf11 are indeed functionally connected and the nature of such an eventual connection.
While interference with Gdf11 signaling or inactivation of the miR196 family changed trunk size, they had no significant effect on overall extension of the body axis [54,65]. These observations indicate that changes in these activities are not sufficient to generate the typical snake body plan. Indeed, a combination of experimental and modeling approaches led to the identification of two additional processes that might play significant roles in the production of the elongated trunks of snakes. One of these processes consist of an extended period of Oct4 (Pou5f1) activity in the epiblast of snake embryos, most likely resulting from major changes in the regulation of its expression [56], which might be the key to guarantee the continuous production of tissue required to extend the body axis through the trunk area. The possibility of direct experimental intervention in snake embryos is very reduced, thus limiting the extent of direct validation that can be done with these embryos, but transgenic experiments in mice provided some support for this hypothesis [56]. A second process that might have contributed to the large vertebral count characteristic of snakes is the accelerated somitogenesis that results in the generation of a larger number of segments forming trunk vertebrae [16]. These somites are smaller than those of embryos in other vertebrate species but generate vertebrae with dimensions equivalent to those of other vertebrates of similar size, and therefore, the expansion of each individual segment during differentiation results in a significant global elongation of their main body axis.
The basic body layout generated by Oct4 and Gdf11 signaling (the involvement of additional factors cannot be ruled out) leads to the production of body structures through the coordinated action of downstream factors that guarantee the production of the tissues appropriate to the different body areas. This is the stage where the contribution of Hox genes is essential by regulating the layout of vertebral patterns fitting the specific requirements of the different body regions along the main body axis [68]. The specific attributes of the trunk-associated skeleton are not uniform, varying significantly across vertebrate phylogeny and often adjusted to facilitate particular characteristics of the animal physiology or behavior. Mammals provide a good example for this. Their trunk skeleton is the most clearly regionalized among vertebrates, typically composed of a closed rib cage (thoracic region) consisting of ribs connecting the vertebra with the sternum in the ventral midline, often followed by a ribless lumbar region before the sacrum that marks the end of trunk structures. The number of thoracolumbar vertebrae in mammals is not as strict as that in their neck but still stays within a relatively short range, from 19 in even-toed ungulates to 24 in African elephants [69]. Interestingly, it has been argued that the specific anatomy and evolutionary conservation of the vertebral structure of this area results from adaptation to the type of locomotion of the species. It is thought, for instance, that the presence of a ribless and flexible lumbar region is important to allow fast and agile movements, features that are not so important in slower and sturdier animals [69]. Indeed, elephants lack the ribless lumbar domain characteristic of most other mammals.
Birds also have a unique configuration of their trunk axial skeleton, adapted to provide their wings and legs the strong support required to facilitate their flying and bipedal walking mechanics. It is composed of a compact rib cage with a reduced number of ribs followed by the synsacrum, a structure formed by the fusion of their lumbar, sacral and the last thoracic units [70], which also fuse with the pelvic girdle to provide the strong attachment for the hindlimbs required for a bipedal walk with a gravity center rostral to the leg position.
Snakes show a totally different type of trunk axial skeleton, consisting of a large number of rib-containing vertebrae extending all the way to the anal region. The number of trunk vertebrae varies both among snake species and often within a given species, showing sexual dimorphism, with females normally containing a larger number of trunk vertebrae that produce longer trunks, a trait thought to favor fecundity [71]. Importantly, snakes do not have a sternum. Therefore, contrary to mammals, birds or limbed lizards, snakes lack a closed rib cage and their ribs end freely within the soft tissues at the ventral part of the body, a feature that might be essential to allow the extreme body expansion required for their feeding strategies.
Subregionalization of the trunk axial skeleton depends, to a large extent, on differential Hox gene expression and most specifically from genes of paralogs 7–11, as revealed by genetic experiments [10,11]. A paradigmatic example is provided by Hox10 genes that are essential for generating the ribless lumbar domain of mammals [72,73]. Matching expression data with skeletal patterns in non-mammalian species suggests a conservation of Hox gene involvement in subregionalization of the trunk axial skeleton [17,20] beyond mammals. An unpredicted illustration of this idea was provided by experiments in snakes, where precise morphometric analyses of individual vertebrae revealed the existence of subtle subregionalization along the anterior posterior axis, roughly corresponding to specific domains of Hox gene expression, thus challenging the classical conception of absent regionalization in the snake axial skeleton [74]. However, snake experiments also revealed that Hox patterning activity of skeletal elements cannot be just assumed from their expression and that additional hidden layers might shape their function, which can be rather divergent among species. In particular, the snake Hoxa10 and Hoxc10 genes were found to be expressed well within the somitic area generating rib-containing vertebrae of the trunk [17,20], an observation at odds with the known role of Hox10 genes in the genesis of ribless lumbar vertebrae revealed by genetic experiments in mice [72,73]. This apparent discrepancy was shown to result from a single nucleotide polymorphism in an essential Hox-responsive enhancer that made it insensitive to the rib-blocking activity of Hox10 proteins [75], thus allowing the vertebral bodies of the caudal trunk area to acquire lumbar-type features, while still being attached to full grown ribs. Interestingly, the same polymorphism in the homologous enhancer of elephants seems to be in the origin of the presence of ribs in the vertebral region corresponding to the lumbar area in other mammals [75].
While it is assumed that Hox genes can also play an important role in the production of the bird-specific trunk axial skeleton [15], its clear deviation from mouse patterns makes it difficult to understand what this role would be. Genetic experiments in mice showed the essential role for Hox paralog group 11 to generate the characteristic fusion between sacral vertebrae in mammals [72]. This would suggest the involvement of these genes in the generation of the synsacrum. While expression data in chicken embryos with well-developed hindlimbs placed Hox11 gene activation posterior to the somite levels generating the lumbosacral region [15], at earlier stages their expression overlaps with the region that will give rise to the vertebrae contributing to the synsacrum [76]. In the mouse, it has been shown that patterning activity of some Hox genes is provided in the presomitic mesoderm when somites are being formed rather than in the somites themselves [73]. Therefore, if the same principle applies to Hox11 genes in chicken embryos, these genes could also play a significant role in the formation of the synsacrum.
Turtles deserve special mention in the context of the trunk axial skeleton, as their carapace represents an extreme modification of their ribs, their vertebral bodies and their associated muscle and dermal derivatives of the somites [77]. In particular, during early stages of turtle somite differentiation, their rib primordia grow laterally, likely driven by signals from a specialized structure in the lateral wall of the embryo, eventually covering the developing limb buds dorsally [77,78]. Development of the muscle precursors is also affected, probably because of a turtle specific modification in the Myf5 gene [79], and the intercostal muscles between the ribs are replaced by a calcified dermal structure that completes the carapace structure [80].
4. The Tail Is Not Just a Tail
The tail represents the body region extending from the main body axis posterior to the anus. When considering the axial skeleton, the basic components of the tail are the caudal vertebrae. In amphibians, mammals and reptiles tail size is directly related to their caudal vertebral count, which can vary from just a few fused elements, like the coccyx of humans, to more than one hundred in some lizard species, like the long tailed grass lizard (Takydromus sexlineatus). However, differently to the neck or trunk, tail size is not always a direct read out of the extension of their vertebral column but depends on other features. In birds, for instance, caudal feathers are the main determinant of tail size and shape diversity, often including sexual dimorphism, adapted to facilitate the flying mechanics of the different species, and being often involved in mating strategies [81]. This diversity is not a consequence of variations in the size of their tail axial skeleton, which is rather similar among species and composed of a few small caudal vertebrae followed by the pygostyle, a skeletal element made of 4–7 fused vertebral elements [70] that provides the attachment point for the caudal feathers.
Another source of tail size and shape variation is provided by the caudal fin of fishes, an appendage extending the body axis beyond the position of the last caudal vertebra representing itself a major source of anatomical diversity among fish clades [82]. In general, caudal fins contain a skeleton composed of several fin rays, bony or cartilaginous elements attached in different ways to the caudal vertebrae. Fin rays vary widely among clades in number, size, shape, stiffness and their connection to the axial skeleton [82,83]. There are three major types of caudal fins on the basis of their general structure [84]. Sharks have a heterocercal type of caudal fin, i.e., its dorsal lobe is bigger than the ventral lobe. In this configuration the vertebral column is bent dorsally entering the dorsal lobe [82]. Teleost, like the zebrafish, have a homocercal type of caudal fin, meaning that they have dorsal and ventral fin lobes of a similar size. While the homocercal fin is externally symmetric, its vertebral skeleton is still dorsally bent at the caudal end, becoming associated with a series of new skeletal elements mostly of ventral origin, which also vary among species, that serve as one of the main supporting structures for the fin rays [82,85,86]. Finally, lungfish have a diphycercal type of fin, in which lobes are truly symmetric, as they arise from dorsal and ventral sides of the axis [84]. Even within these three main types of caudal fins the diversity among species is very high in both the internal structure and mobility, adapted to facilitate the distinct swimming profiles of different species [82,83].
Aquatic mammals, including sirenians and cetaceans, also have a fan-shaped structure at the posterior end of the tail, the fluke, which, similar to the caudal fin of fishes, facilitates the animal’s propulsion in an aquatic medium. Despite their shared function, flukes and caudal fins are not homologous structures. They are oriented differently relative to the body: the caudal fin extends vertically along the dorsoventral body axis, whereas the flukes are lateral (horizontal) extensions of the tail. Structurally, the fluke is composed of dense fibrous connective tissue [87], lacking the intrinsic skeletal or muscle elements that can be observed in the different types of caudal fins [88,89]. These differences might actually result from their developmental identity. The developmental origin and the growth and patterning processes involved in caudal fin formation are similar to those of the other fish medial and lateral fins, making them bona fide non-paired appendages of fishes [89]. The control of the fluke’s formation is essentially unknown. They are not modified hindlimbs. Indeed, while adult sirenians and cetaceans do not have hindlimbs, a limb bud is actually induced in the embryo at a more anterior position relative to the fluke, but have their development halted, leaving in place just a small hip bone within the muscular tissue at the end of the abdominal cavity [90,91]. So far, the few descriptions of fluke’s development do not show any of the elements known to regulate limb development that would classify them together with vertebrate paired appendages [92], thus fitting with the absence of any skeletal or muscle tissue typical of those appendages. Therefore, caudal fins and flukes cannot be considered homologous structures, and considering the phylogenetic distances between sirenians and cetaceans, it is possible that their flukes also resulted from two independent events. Caudal fins and flukes thus represent a prime example of convergent evolution to adapt the animal’s mobility to an aquatic medium.
During embryonic development, axial extension through the trunk and tail regions differs in several ways. A major difference between the two stages of axial extension is the association of neck and trunk development with the process of gastrulation and tail elongation with the activity of the tail bud [93,94,95]. In fish, tail development is also associated with caudal fin morphogenesis [82,85], starting with the formation of a fin bud in the ventral part of the developing tail, which repositions to the posterior end of the tail as it grows and differentiates [96]. The caudal fin primordium contains signaling centers homologous to those guiding fin and limb development [89,96] that provide dorsoventral and lateral polarity to the fin. On the basis of the localization of such centers, it has been suggested that the dorsal margin of the caudal fin primordium is equivalent to the anterior margin of paired fins or of the limb bud, generating a dorsoventral polarity, whose implementation could be a key determinant of caudal fin morphological diversity [89,96].
In most vertebrate species tail size in the adult animal is directly associated with the embryo’s capacity to keep extending the body axis and generating somites after finishing the production of trunk structures. Frogs provide a notable exception to this general rule: during the tadpole stage it contains a long and fully developed tail that shares many structural characteristics with the tail of other tetrapods but adapted to facilitate swimming. In the transition to the adult body pattern during metamorphosis, the tail is completely lost, generating a tail-less adult frog [7].
In mammals, the switch from the primitive streak to tail bud-dependent development is triggered by Gdf11 acting on the Tgfbr1 receptor [57,97]. While trunk and tail development share several of the growth control mechanisms, including those involving T (Brachyury), the Cdx genes or FGF and WNT signaling [98,99], the switch from the primitive streak to tail bud development is associated with a change in the factors at the top of the regulatory hierarchy, eventually impacting the final tail size. Contrary to trunk development, tail bud growth is Oct4-independent, relying instead on Lin28/let7 driven mechanisms [100,101,102]. Indeed, persistent Lin28 expression in the tail bud produced significant extension of the tail, whereas its inactivation resulted in tail shortening. The switch from trunk to tail development in zebrafish, and possibly also in other anamniotes, seems to depend on a different set of control factors, including a shift to a BMP-dependent network [103,104].
Genetic experiments in mice indicate that Hox genes of paralog group 13 might be a central component of the network bringing tail growth to an end, at least in part by down-regulating Lin28 activity in tail bud progenitors [101,105,106]. Importantly, both Lin28/let7 and Hox13 activities seem to affect extension through the tail bud but to have little effect on trunk extension [100,101], indicating different intrinsic characteristics of the trunk and tail progenitors. Whether the mechanisms triggering the end of tail extension identified in mice also operate in other vertebrate species remains to be determined. Studies in chicken embryos showed that expression of Hox13 genes follows patterns resembling those observed in mice and that these genes are able to slow down axial extension when prematurely activated in the progenitor zone of chicken embryos [76]. It is thus possible that the mechanisms observed in mice are also operative at least in chickens. However, recent work showed a positive involvement of Hox13 genes in the axial elongation of zebrafish embryos [107], which seems to be in contradiction with the data from the mouse and chicken. The origin of this discrepancy is not totally clear, but it possibly reflects differences among species. In particular, while in zebrafish, axial truncation was observed upon combined downregulation of Hoxd13a and Hoxa13b [107], in the mouse, double inactivation of Hoxa13 and Hoxd13 genes truncated the limb autopod, but no alterations in axial development were reported [108,109]. Conversely, Hoxb13 inactivation in the mouse resulted in tail extension [106], indicating its involvement in stopping rather than promoting axial elongation, also fitting with the tail truncation observed upon premature activation Hoxb13 in the mouse progenitor zone [101]. It is therefore possible that mouse and zebrafish embryos use different regulatory networks to extend their tail bud, which could reflect the reported differences in axial extension mechanisms between mice and zebrafish [104]. Indeed, while tail extension in mouse and zebrafish embryos share some regulatory features, like the requirement of Brachyury or the Cdx genes [110,111,112], genetic data suggest that in what concerns Hox13 genes zebrafish tail development is more akin to distal limb formation than to axial extension in mammals. Interestingly, it has been described that Hoxb13 becomes activated during the initial stages of axolotl (Ambystoma mexicanum) tail regeneration [113], compatible with a role in promoting tail extension rather than with being a stop signal. It is therefore possible that Hox13 genes influence axial extension differently in different vertebrate clades.
5. Concluding Remarks
The diversity of morphological patterns in the axial skeleton observed across vertebrate phylogeny essentially derives from two main factors: a differential period of active axial elongation and specificity in the mechanisms regulating somite differentiation. I should point out that somite differentiation routes are also in the origin of sources of diversity in the axial skeleton that had not been included in this review, like anatomical features typical of fish vertebrae that are not present in tetrapods [114]. As discussed in this review, some of the key regulators of axial length and morphological diversity of the vertebral elements have been identified. However, we still know very little about how these factors operate to produce highly reproducible and distinct species-specific morphological patterns involving both shape and size. Hox genes are a good example for this: we know that they play central roles in the control of segmental identity but, after decades of work, how they actually regulate those processes is still largely unknown. In addition, direct analysis of the emergence of specific traits is often hampered by the total or partial inability to access the relevant tissues or by experimental limitations. The neck vertebrae of giraffes are a good example of such constraints. The standard method to bypass these limitations include testing relevant hypothesis using other vertebrate species accessible to complex genetic or grafting techniques. The limitations of such approaches are obvious and became further highlighted by recent reports showing the essential contribution of the global cellular environment to the activity of key regulators of developmental processes [115,116]. For instance, using in vitro systems, it has been shown that conserved processes like the segmentation clock or motor neuron induction run at different paces in mouse or human tissues. Swapping the key regulator of the process under evaluation between mice and humans did not affect the global behavior of the cells. This indicates that intrinsic properties of key control genes might play a limited role in the regulation of species-specific features, showing instead that they can adapt to a heterologous cell environment and acquire functional characteristics resembling those of the host species. If this is a general biological principle, it might indicate that, for instance, a potential regulator of the size of the giraffe’s cervical vertebrae might not reproduce the phenotype when introduced into mouse embryos. It is therefore likely that understanding the origin of morphological diversity among species will require fundamental changes in experimental paradigms allowing direct experimental evaluation in the cellular context of the species under analysis. It is possible that the incorporation of emerging alternative experimental approaches, like in vitro surrogate systems that recapitulate complex embryological processes, often to a remarkable level of detail [117,118,119], might open opportunities to explore developmental processes in species currently not susceptible of direct experimental analyses.
Funding
Research in the author’s laboratory was funded by FUNDAÇÃO PARA A CIÊNCIA E A TECNOLOGIA (Portugal), grant number PTDC/BIA-BID/30254/2017.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
I would like to thank the members of my group for their comments on the manuscript.
Conflicts of Interest
The author declares no conflict of interest. The funder had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish this manuscript.
References
- Mallo, M. The Axial Musculoskeletal System. In Kaufman’s Atlas of Mouse Development Supplement; Baldock, R., Bard, J., Davidson, D.R., Morriss-Kay, G., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 165–175. ISBN 9780128000434. [Google Scholar]
- Bénazéraf, B.; Pourquié, O. Formation and Segmentation of the Vertebrate Body Axis. Annu. Rev. Cell Dev. Biol. 2013, 29, 1–26. [Google Scholar] [CrossRef]
- Gridley, T. The long and short of it: Somite formation in mice. Dev. Dyn. 2006, 235, 2330–2336. [Google Scholar] [CrossRef] [PubMed]
- Hubaud, A.; Pourquie, O. Signalling dynamics in vertebrate segmentation. Nat. Rev. Mol. Cell Biol. 2014, 15, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Mallo, M. Revisiting the involvement of signaling gradients in somitogenesis. FEBS J. 2016, 283, 1430–1437. [Google Scholar] [CrossRef] [PubMed]
- Couly, G.F.; Coltey, P.M.; Le Douarin, N.M. The triple origin of skull in higher vertebrates: A study in quail-chick chimeras. Development 1993, 117, 409–429. [Google Scholar] [CrossRef]
- Handrigan, G.R.; Wassersug, R.J. The anuran Bauplan: A review of the adaptive, developmental, and genetic underpinnings of frog and tadpole morphology. Biol. Rev. 2007, 82, 1–25. [Google Scholar] [CrossRef]
- Morin-Kensicki, E.M.; Melancon, E.; Eisen, J.S. Segmental relationship between somites and vertebral column in zebrafish. Development 2002, 129, 3851–3860. [Google Scholar] [CrossRef]
- Mallo, M.; Vinagre, T.; Carapuço, M. The road to the vertebral formula. Int. J. Dev. Biol. 2009, 53, 1469–1481. [Google Scholar] [CrossRef]
- Mallo, M.; Wellik, D.M.; Deschamps, J. Hox genes and regional patterning of the vertebrate body plan. Dev. Biol. 2010, 344, 7–15. [Google Scholar] [CrossRef]
- Wellik, D.M. Hox patterning of the vertebrate axial skeleton. Dev. Dyn. 2007, 236, 2454–2463. [Google Scholar] [CrossRef]
- Deschamps, J.; Duboule, D. Embryonic timing, axial stem cells, chromatin dynamics, and the Hox clock. Genes Dev. 2017, 31, 1406–1416. [Google Scholar] [CrossRef]
- Krumlauf, R. Hox genes in vertebrate development. Cell 1994, 78, 191–201. [Google Scholar] [CrossRef]
- Amores, A.; Force, A.; Yan, Y.-L.; Joly, L.; Amemiya, C.; Fritz, A.; Ho, R.K.; Langeland, J.; Prince, V.; Wang, Y.-L.; et al. Zebrafish hox Clusters and Vertebrate Genome Evolution. Science 1998, 282, 1711–1714. [Google Scholar] [CrossRef]
- Burke, A.C.; Nelson, C.E.; Morgan, B.A.; Tabin, C. Hox genes and the evolution of vertebrate axial morphology. Development 1995, 121, 333–346. [Google Scholar] [CrossRef]
- Gomez, C.; Özbudak, E.M.; Wunderlich, J.; Baumann, D.; Lewis, J.; Pourquie, O. Control of segment number in vertebrate embryos. Nature 2008, 454, 335–339. [Google Scholar] [CrossRef]
- Woltering, J.M.; Vonk, F.J.; Müller, H.; Bardine, N.; Tuduce, I.L.; de Bakker, M.A.G.; Knöchel, W.; Sirbu, I.O.; Durston, A.J.; Richardson, M.K. Axial patterning in snakes and caecilians: Evidence for an alternative interpretation of the Hox code. Dev. Biol. 2009, 332, 82–89. [Google Scholar] [CrossRef]
- Narita, Y.; Kuratani, S. Evolution of the vertebral formulae in mammals: A perspective on developmental constraints. J. Exp. Zool. Part B Mol. Dev. Evol. 2005, 304, 91–106. [Google Scholar] [CrossRef]
- Gaunt, S.J. Conservation in the Hox code during morphological evolution. Int. J. Dev. Biol. 1994, 38, 549–552. [Google Scholar]
- Di-Poï, N.; Montoya-Burgos, J.I.; Miller, H.; Pourquie, O.; Milinkovitch, M.C.; Duboule, D. Changes in Hox genes’ structure and function during the evolution of the squamate body plan. Nature 2010, 464, 99–103. [Google Scholar] [CrossRef]
- Mansfield, J.H.; Abzhanov, A. Hox expression in the American alligator and evolution of archosaurian axial patterning. J. Exp. Zool. Part B Mol. Dev. Evol. 2010, 314B, 629–644. [Google Scholar] [CrossRef]
- Vinagre, T.; Moncaut, N.; Carapuço, M.; Nóvoa, A.; Bom, J.; Mallo, M. Evidence for a Myotomal Hox/Myf Cascade Governing Nonautonomous Control of Rib Specification within Global Vertebral Domains. Dev. Cell 2010, 18, 655–661. [Google Scholar] [CrossRef]
- McIntyre, D.C.; Rakshit, S.; Yallowitz, A.R.; Loken, L.; Jeannotte, L.; Capecchi, M.R.; Wellik, D.M. Hox patterning of the vertebrate rib cage. Development 2007, 134, 2981–2989. [Google Scholar] [CrossRef]
- Rancourt, D.E.; Tsuzuki, T.; Capecchi, M.R. Genetic interaction between hoxb-5 and hoxb-6 is revealed by nonallelic noncomplementation. Genes Dev. 1995, 9, 108–122. [Google Scholar] [CrossRef]
- Noordermeer, D.; Leleu, M.; Schorderet, P.; Joye, E.; Chabaud, F.; Duboule, D. Temporal dynamics and developmental memory of 3D chromatin architecture at Hox gene loci. eLife 2014, 2014, e02557. [Google Scholar] [CrossRef]
- Mallo, M.; Alonso, C.R. The regulation of Hox gene expression during animal development. Development 2013, 140, 3951–3963. [Google Scholar] [CrossRef]
- Soshnikova, N.; Duboule, D. Epigenetic Temporal Control of Mouse Hox Genes in Vivo. Science 2009, 324, 1320–1323. [Google Scholar] [CrossRef]
- Berlivet, S.; Paquette, D.; Du Mouchel, A.; Langlais, D.; Dostie, J.; Kmita, M. Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of HoxA Genes in Developing Limbs. PLoS Genet. 2013, 9, e1004018. [Google Scholar] [CrossRef]
- Narendra, V.; Rocha, P.P.; An, D.; Raviram, R.; Skok, J.A.; Mazzoni, E.O.; Reinberg, D. CTCF establishes discrete functional chromatin domains at theHoxclusters during differentiation. Science 2015, 347, 1017–1021. [Google Scholar] [CrossRef]
- Andrey, G.; Montavon, T.; Mascrez, B.; Gonzalez, F.; Noordermeer, D.; Leleu, M.; Trono, D.; Spitz, F.; Duboule, D. A Switch Between Topological Domains Underlies HoxD Genes Collinearity in Mouse Limbs. Science 2013, 340, 1234167. [Google Scholar] [CrossRef]
- Woltering, J.M.; Noordermeer, D.; Leleu, M.; Duboule, D. Conservation and Divergence of Regulatory Strategies at Hox Loci and the Origin of Tetrapod Digits. PLoS Biol. 2014, 12, e1001773. [Google Scholar] [CrossRef]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Nora, E.P.; Lajoie, B.R.; Schulz, E.G.; Giorgetti, L.; Okamoto, I.; Servant, N.; Piolot, T.; van Berkum, N.L.; Meisig, J.; Sedat, J.; et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 2012, 485, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Narendra, V.; Bulajic, M.; Dekker, J.; Mazzoni, E.O.; Reinberg, D. CTCF-mediated topological boundaries during development foster appropriate gene regulation. Genes Dev. 2016, 30, 2657–2662. [Google Scholar] [CrossRef] [PubMed]
- Neijts, R.; Amin, S.; van Rooijen, C.; Deschamps, J. Cdx is crucial for the timing mechanism driving colinear Hox activation and defines a trunk segment in the Hox cluster topology. Dev. Biol. 2017, 422, 146–154. [Google Scholar] [CrossRef]
- Arnold, P.; Amson, E.; Fischer, M.S. Differential scaling patterns of vertebrae and the evolution of neck length in mammals. Evolution 2017, 71, 1587–1599. [Google Scholar] [CrossRef]
- Badlangana, N.L.; Adams, J.W.; Manger, P.R. The giraffe (Giraffa camelopardalis) cervical vertebral column: A heuristic example in understanding evolutionary processes? Zool. J. Linn. Soc. 2009, 155, 736–757. [Google Scholar] [CrossRef]
- Agaba, M.; Ishengoma, E.; Miller, W.C.; McGrath, B.C.; Hudson, C.N.; Reina, O.C.B.; Ratan, A.; Burhans, R.; Chikhi, R.; Medvedev, R.C.P.; et al. Giraffe genome sequence reveals clues to its unique morphology and physiology. Nat. Commun. 2016, 7, 11519. [Google Scholar] [CrossRef]
- Liu, C.; Gao, J.; Cui, X.; Li, Z.; Chen, L.; Yuan, Y.; Zhang, Y.; Mei, L.; Zhao, L.; Cai, D.; et al. A towering genome: Experimentally validated adaptations to high blood pressure and extreme stature in the giraffe. Sci. Adv. 2021, 7, eabe9459. [Google Scholar] [CrossRef]
- Dubrulle, J.; McGrew, M.J.; Pourquie, O. FGF Signaling Controls Somite Boundary Position and Regulates Segmentation Clock Control of Spatiotemporal Hox Gene Activation. Cell 2001, 106, 219–232. [Google Scholar] [CrossRef]
- Condie, B.G.; Capecchi, M.R. Mice with targeted disruptions in the paralogous genes hoxa-3 and hoxd-3 reveal synergistic interactions. Nature 1994, 370, 304–307. [Google Scholar] [CrossRef]
- Condie, B.G.; Capecchi, M.R. Mice homozygous for a targeted disruption of Hoxd-3 (Hox-4.1) exhibit anterior transformations of the first and second cervical vertebrae, the atlas and the axis. Development 1993, 119, 579–595. [Google Scholar] [CrossRef]
- Horan, G.S.B.; Ramírez-Solis, R.; Featherstone, M.S.; Wolgemuth, D.J.; Bradley, A.; Behringer, R.R. Compound mutants for the paralogous hoxa-4, hoxb-4, and hoxd-4 genes show more complete homeotic transformations and a dose-dependent increase in the number of vertebrae transformed. Genes Dev. 1995, 9, 1667–1677. [Google Scholar] [CrossRef]
- Su, N.; Jin, M.; Chen, L. Role of FGF/FGFR signaling in skeletal development and homeostasis: Learning from mouse models. Bone Res. 2014, 2, 14003. [Google Scholar] [CrossRef]
- Regard, J.B.; Zhong, Z.; Williams, B.O.; Yang, Y. Wnt Signaling in Bone Development and Disease: Making Stronger Bone with Wnts. Cold Spring Harb. Perspect. Biol. 2012, 4, a007997. [Google Scholar] [CrossRef]
- Zieba, J.T.; Chen, Y.-T.; Lee, B.H.; Bae, Y. Notch Signaling in Skeletal Development, Homeostasis and Pathogenesis. Biomolecules 2020, 10, 332. [Google Scholar] [CrossRef]
- Buchholtz, E.A.; Bailin, H.G.; Laves, S.A.; Yang, J.T.; Drozd, L.E.; Chan, M.-Y. Fixed cervical count and the origin of the mammalian diaphragm. Evol. Dev. 2012, 14, 399–411. [Google Scholar] [CrossRef]
- Hautier, L.; Weisbecker, V.; Sanchez-Villagra, M.R.; Goswami, A.; Asher, R.J. Skeletal development in sloths and the evolution of mammalian vertebral patterning. Proc. Natl. Acad. Sci. USA 2010, 107, 18903–18908. [Google Scholar] [CrossRef]
- Buchholtz, E.A. Crossing the frontier: A hypothesis for the origins of meristic constraint in mammalian axial patterning. Zoology 2014, 117, 64–69. [Google Scholar] [CrossRef][Green Version]
- Vasyutina, E.; Birchmeier, C. The development of migrating muscle precursor cells. Anat. Embryol. 2006, 211, 37–41. [Google Scholar] [CrossRef]
- Rommel, S.; Reynolds, J.E. Diaphragm structure and function in the Florida manatee (Trichechus manatus latirostris). Anat. Rec. Adv. Integr. Anat. Evol. Biol. 2000, 259, 41–51. [Google Scholar] [CrossRef]
- Galis, F. Why do almost all mammals have seven cervical vertebrae? Developmental constraints, Hox genes, and cancer. J. Exp. Zool. 1999, 285, 19–26. [Google Scholar] [CrossRef]
- Marek, R.D.; Falkingham, P.L.; Benson, R.B.J.; Gardiner, J.D.; Maddox, T.W.; Bates, K.T. Evolutionary versatility of the avian neck. Proc. R. Soc. B Boil. Sci. 2021, 288, 20203150. [Google Scholar] [CrossRef]
- McPherron, A.C.; Lawler, A.M.; Lee, S.-J. Regulation of anterior/posterior patterning of the axial skeleton by growth/differentiation factor 11. Nat. Genet. 1999, 22, 260–264. [Google Scholar] [CrossRef]
- Szumska, D.; Pieles, G.; Essalmani, R.; Bilski, M.; Mesnard, D.; Kaur, K.; Franklyn, A.; El Omari, K.; Jefferis, J.; Bentham, J.; et al. VACTERL/caudal regression/Currarino syndrome-like malformations in mice with mutation in the proprotein convertase Pcsk5. Genes Dev. 2008, 22, 1465–1477. [Google Scholar] [CrossRef]
- Aires, R.; Jurberg, A.D.; Leal, F.; Nóvoa, A.; Cohn, M.J.; Mallo, M. Oct4 Is a Key Regulator of Vertebrate Trunk Length Diversity. Dev. Cell 2016, 38, 262–274. [Google Scholar] [CrossRef]
- Jurberg, A.D.; Aires, R.; Varela-Lasheras, I.; Nóvoa, A.; Mallo, M. Switching Axial Progenitors from Producing Trunk to Tail Tissues in Vertebrate Embryos. Dev. Cell 2013, 25, 451–462. [Google Scholar] [CrossRef]
- McPherron, A.C.; Huynh, T.V.; Lee, S.-J. Redundancy of myostatin and growth/differentiation factor 11 function. BMC Dev. Biol. 2009, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.P.; Li, E. The signaling pathway mediated by the type IIB activin receptor controls axial patterning and lateral asymmetry in the mouse. Genes Dev. 1997, 11, 1812–1826. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.P.; Yeo, C.-Y.; Lee, Y.; Schrewe, H.; Whitman, M.; Li, E. Activin type IIA and IIB receptors mediate Gdf11 signaling in axial vertebral patterning. Genes Dev. 2002, 16, 2749–2754. [Google Scholar] [CrossRef] [PubMed]
- Essalmani, R.; Zaid, A.; Marcinkiewicz, J.; Chamberland, A.; Pasquato, A.; Seidah, N.G.; Prat, A. In vivo functions of the proprotein convertase PC5/6 during mouse development: Gdf11 is a likely substrate. Proc. Natl. Acad. Sci. USA 2008, 105, 5750–5755. [Google Scholar] [CrossRef] [PubMed]
- Andersson, O.; Reissmann, E.; Ibáñez, C.F. Growth differentiation factor 11 signals through the transforming growth factor-β receptor ALK5 to regionalize the anterior–posterior axis. EMBO Rep. 2006, 7, 831–837. [Google Scholar] [CrossRef]
- Matsubara, Y.; Hirasawa, T.; Egawa, S.; Hattori, A.; Suganuma, T.; Kohara, Y.; Nagai, T.; Tamura, K.; Kuratani, S.; Kuroiwa, A.; et al. Anatomical integration of the sacral–hindlimb unit coordinated by GDF11 underlies variation in hindlimb positioning in tetrapods. Nat. Ecol. Evol. 2017, 1, 1392–1399. [Google Scholar] [CrossRef]
- Ho, D.M.; Yeo, C.-Y.; Whitman, M. The role and regulation of GDF11 in Smad2 activation during tailbud formation in the Xenopus embryo. Mech. Dev. 2010, 127, 485–495. [Google Scholar] [CrossRef]
- Wong, S.F.L.; Agarwal, V.; Mansfield, J.H.; Denans, N.; Schwartz, M.G.; Prosser, H.M.; Pourquie, O.; Bartel, D.P.; Tabin, C.J.; McGlinn, E. Independent regulation of vertebral number and vertebral identity by microRNA-196 paralogs. Proc. Natl. Acad. Sci. USA 2015, 112, E4884–E4893. [Google Scholar] [CrossRef]
- He, X.; Yan, Y.-L.; Eberhart, J.K.; Herpin, A.; Wagner, T.U.; Schartl, M.; Postlethwait, J.H. miR-196 regulates axial patterning and pectoral appendage initiation. Dev. Biol. 2011, 357, 463–477. [Google Scholar] [CrossRef]
- Asli, N.S.; Kessel, M. Spatiotemporally restricted regulation of generic motor neuron programs by miR-196-mediated repression of Hoxb8. Dev. Biol. 2010, 344, 857–868. [Google Scholar] [CrossRef]
- Mallo, M. Reassessing the Role of Hox Genes during Vertebrate Development and Evolution. Trends Genet. 2018, 34, 209–217. [Google Scholar] [CrossRef]
- Galis, F.; Carrier, D.R.; van Alphen, J.; van der Mije, S.D.; van Dooren, T.J.M.; Metz, J.A.J.; Broek, C.M.A.T. Fast running restricts evolutionary change of the vertebral column in mammals. Proc. Natl. Acad. Sci. USA 2014, 111, 11401–11406. [Google Scholar] [CrossRef]
- Nickel, R.; Schummer, A.; Seiferle, E.; Siller, W.G.; Wight, P.A.L. Anatomy of the Domestic Birds; Springer: Heidelberg/Berlin, Germany, 1977. [Google Scholar]
- Shine, R. Vertebral numbers in male and female snakes: The roles of natural, sexual and fecundity selection. J. Evol. Biol. 2000, 13, 455–465. [Google Scholar] [CrossRef]
- Wellik, D.M.; Capecchi, M.R. Hox10 and Hox11 Genes Are Required to Globally Pattern the Mammalian Skeleton. Science 2003, 301, 363–367. [Google Scholar] [CrossRef]
- Carapuço, M.; Nóvoa, A.; Bobola, N.; Mallo, M. Hox genes specify vertebral types in the presomitic mesoderm. Genes Dev. 2005, 19, 2116–2121. [Google Scholar] [CrossRef]
- Head, J.J.; Polly, P.D. Evolution of the snake body form reveals homoplasy in amniote Hox gene function. Nature 2015, 520, 86–89. [Google Scholar] [CrossRef]
- Guerreiro, I.; Nunes, A.; Woltering, J.M.; Casaca, A.; Nóvoa, A.; Vinagre, T.; Hunter, M.E.; Duboule, D.; Mallo, M. Role of a polymorphism in a Hox/Pax-responsive enhancer in the evolution of the vertebrate spine. Proc. Natl. Acad. Sci. USA 2013, 110, 10682–10686. [Google Scholar] [CrossRef]
- Denans, N.; Iimura, T.; Pourquié, O. Hox genes control vertebrate body elongation by collinear Wnt repression. eLife 2015, 4, 4379. [Google Scholar] [CrossRef]
- Hirasawa, T.; Pascual-Anaya, J.; Kamezaki, N.; Taniguchi, M.; Mine, K.; Kuratani, S. The evolutionary origin of the turtle shell and its dependence on the axial arrest of the embryonic rib cage. J. Exp. Zool. Part B Mol. Dev. Evol. 2015, 324, 194–207. [Google Scholar] [CrossRef]
- Kawashima-Ohya, Y.; Narita, Y.; Nagashima, H.; Usuda, R.; Kuratani, S. Hepatocyte growth factor is crucial for development of the carapace in turtles. Evol. Dev. 2011, 13, 260–268. [Google Scholar] [CrossRef]
- Ohya, Y.K.; Usuda, R.; Kuraku, S.; Nagashima, H.; Kuratani, S. Unique features of Myf-5 in turtles: Nucleotide deletion, alternative splicing, and unusual expression pattern. Evol. Dev. 2006, 8, 415–423. [Google Scholar] [CrossRef]
- Hirasawa, T.; Nagashima, H.; Kuratani, S. The endoskeletal origin of the turtle carapace. Nat. Commun. 2013, 4, 2107. [Google Scholar] [CrossRef]
- Thomas, A.L.R. On the Tails of Birds. Bioscience 1997, 47, 215–225. [Google Scholar] [CrossRef]
- Desvignes, T.; Carey, A.; Postlethwait, J.H. Evolution of caudal fin ray development and caudal fin hypural diastema complex in spotted gar, teleosts, and other neopterygian fishes. Dev. Dyn. 2018, 247, 832–853. [Google Scholar] [CrossRef]
- Lauder, G.V. Caudal Fin Locomotion in Ray-finned Fishes: Historical and Functional Analyses. Am. Zool. 1989, 29, 85–102. [Google Scholar] [CrossRef]
- Metscher, B.D.; Ahlberg, P.E. Origin of the teleost tail: Phylogenetic frameworks for developmental studies. In Major Events in Early Vertebrate Evolution: Palaeontology, Phylogeny, Genetics and Development; Ahlberg, P.E., Ed.; Taylor & Francis: London, UK; New York, NY, USA, 2001; Volume 61, pp. 333–349. [Google Scholar]
- Bird, N.C.; Mabee, P.M. Developmental morphology of the axial skeleton of the zebrafish, Danio rerio (Ostariophysi: Cyprinidae). Dev. Dyn. 2003, 228, 337–357. [Google Scholar] [CrossRef] [PubMed]
- Woltering, J.M.; Holzem, M.; Schneider, R.F.; Nanos, V.; Meyer, A. The skeletal ontogeny of Astatotilapia burtoni—A direct-developing model system for the evolution and development of the teleost body plan. BMC Dev. Biol. 2018, 18, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Felts, W.J.L. Some functional and structural characteristics of cetacean flippers and flukes. In Whales, Dolphins, and Porpoises; Norris, K.S., Ed.; University of California Press: Berkeley, CA, USA, 1966; pp. 255–276. [Google Scholar]
- Flammang, B.E. The fish tail as a derivation from axial musculoskeletal anatomy: An integrative analysis of functional morphology. Zoology 2014, 117, 86–92. [Google Scholar] [CrossRef]
- Marí-Beffa, M.; Murciano, C. Dermoskeleton morphogenesis in zebrafish fins. Dev. Dyn. 2010, 239, 2779–2794. [Google Scholar] [CrossRef]
- Thewissen, J.G.M.; Cohn, M.J.; Stevens, L.S.; Bajpai, S.; Heyning, J.; Horton, W.E.; Thewissen, J.G.M.; Cohn, M.J.; Stevens, L.S.; Bajpai, S.; et al. Developmental basis for hind-limb loss in dolphins and origin of the cetacean bodyplan. Proc. Natl. Acad. Sci. USA 2006, 103, 8414–8418. [Google Scholar] [CrossRef]
- Thewissen, J.G.M. Highlights of Cetacean Embryology. Aquat. Mamm. 2018, 44, 591–602. [Google Scholar] [CrossRef]
- Ogawa, T. On the presence and disappearance of the hind limb in the cetacean embryos. Sci. Rep. Whales Res. Inst. 1953, 8, 127–132. [Google Scholar]
- Wilson, V.; Olivera-Martinez, I.; Storey, K.G. Stem cells, signals and vertebrate body axis extension. Development 2009, 136, 1591–1604. [Google Scholar] [CrossRef]
- Mallo, M. The vertebrate tail: A gene playground for evolution. Cell. Mol. Life Sci. 2020, 77, 1021–1030. [Google Scholar] [CrossRef]
- Holmdahl, D.E. Experimentelle Untersuchungen uber die Lage der Grenze primarer und sekundarer Korperentwicklung beim Huhn. Anat. Anz. 1925, 59, 393–396. [Google Scholar]
- Hadzhiev, Y.; Lele, Z.; Schindler, S.; Wilson, S.W.; Ahlberg, P.; Strähle, U.; Müller, F. Hedgehog signaling patterns the outgrowth of unpaired skeletal appendages in zebrafish. BMC Dev. Biol. 2007, 7, 75. [Google Scholar] [CrossRef]
- Dias, A.; Lozovska, A.; Wymeersch, F.J.; Nóvoa, A.; Binagui-Casas, A.; Sobral, D.; Martins, G.G.; Wilson, V.; Mallo, M. A Tgfbr1/Snai1-dependent developmental module at the core of vertebrate axial elongation. eLife 2020, 9, e56615. [Google Scholar] [CrossRef]
- Henrique, D.; Abranches, E.; Verrier, L.; Storey, K.G. Neuromesodermal progenitors and the making of the spinal cord. Development 2015, 142, 2864–2875. [Google Scholar] [CrossRef]
- Steventon, B.; Arias, A.M. Evo-engineering and the cellular and molecular origins of the vertebrate spinal cord. Dev. Biol. 2017, 432, 3–13. [Google Scholar] [CrossRef]
- Robinton, D.A.; Chal, J.; da Rocha, E.L.; Han, A.; Yermalovich, A.V.; Oginuma, M.; Schlaeger, T.M.; Sousa, P.; Rodriguez, A.; Urbach, A.; et al. The Lin28/let-7 Pathway Regulates the Mammalian Caudal Body Axis Elongation Program. Dev. Cell 2019, 48, 396–405. [Google Scholar] [CrossRef]
- Aires, R.; de Lemos, L.; Nóvoa, A.; Jurberg, A.D.; Mascrez, B.; Duboule, D.; Mallo, M. Tail Bud Progenitor Activity Relies on a Network Comprising Gdf11, Lin28, and Hox13 Genes. Dev. Cell 2019, 48, 383–395.e8. [Google Scholar] [CrossRef]
- DeVeale, B.; Brokhman, I.; Mohseni, P.; Babak, T.; Yoon, C.; Lin, A.; Onishi, K.; Tomilin, A.; Pevny, L.; Zandstra, P.W.; et al. Oct4 Is Required∼E7.5 for Proliferation in the Primitive Streak. PLoS Genet. 2013, 9, e1003957. [Google Scholar] [CrossRef]
- Agathon, A.; Thisse, C.; Thisse, B. The molecular nature of the zebrafish tail organizer. Nature 2003, 424, 448–452. [Google Scholar] [CrossRef]
- Holley, S.A. Anterior-posterior differences in vertebrate segments: Specification of trunk and tail somites in the zebrafish blastula. Genes Dev. 2006, 20, 1831–1837. [Google Scholar] [CrossRef]
- Young, T.; Rowland, J.E.; van de Ven, C.; Bialecka, M.; Novoa, A.; Carapuco, M.; van Nes, J.; de Graaff, W.; Duluc, I.; Freund, J.-N.; et al. Cdx and Hox Genes Differentially Regulate Posterior Axial Growth in Mammalian Embryos. Dev. Cell 2009, 17, 516–526. [Google Scholar] [CrossRef]
- Economides, K.D.; Capecchi, M.R. Hoxb13 is required for normal differentiation and secretory function of the ventral prostate. Development 2003, 130, 2061–2069. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Kimelman, D. hox13 genes are required for mesoderm formation and axis elongation during early zebrafish development. Development 2020, 147, 185298. [Google Scholar] [CrossRef]
- Fromental-Ramain, C.; Warot, X.; Lakkaraju, S.; Favier, B.; Haack, H.; Birling, C.; Dierich, A.; Dolle, P.; Chambon, P. Specific and redundant functions of the paralogous Hoxa-9 and Hoxd-9 genes in forelimb and axial skeleton patterning. Development 1996, 122, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Warot, X.; Fromental-Ramain, C.; Fraulob, V.; Chambon, P.; Dolle, P. Gene dosage-dependent effects of the Hoxa-13 and Hoxd-13 mutations on morphogenesis of the terminal parts of the digestive and urogenital tracts. Development 1997, 124, 4781–4791. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Bae, Y.-K.; Muraoka, O.; Hibi, M. Interaction of Wnt and caudal-related genes in zebrafish posterior body formation. Dev. Biol. 2005, 279, 125–141. [Google Scholar] [CrossRef] [PubMed]
- Hammerschmidt, M.; Pelegri, F.; Mullins, M.C.; Kane, D.A.; Brand, M.; van Eeden, F.J.M.; Furutani-Seiki, M.; Granato, M.; Haffter, P.; Heisenberg, C.P.; et al. Mutations affecting morphogenesis during gastrulation and tail formation in the zebrafish, Danio rerio. Development 1996, 123, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Griffin, K.J.; Kimelman, D. Interplay between FGF, one-eyed pinhead, and T-box transcription factors during zebrafish posterior development. Dev. Biol. 2003, 264, 456–466. [Google Scholar] [CrossRef]
- Carlson, M.R.J.; Komine, Y.; Bryant, S.V.; Gardiner, D.M. Expression of Hoxb13 and Hoxc10 in Developing and Regenerating Axolotl Limbs and Tails. Dev. Biol. 2001, 229, 396–406. [Google Scholar] [CrossRef]
- Galbusera, F.; Bassani, T. The Spine: A Strong, Stable, and Flexible Structure with Biomimetics Potential. Biomimetics 2019, 4, 60. [Google Scholar] [CrossRef]
- Matsuda, M.; Hayashi, H.; Garcia-Ojalvo, J.; Yoshioka-Kobayashi, K.; Kageyama, R.; Yamanaka, Y.; Ikeya, M.; Toguchida, J.; Alev, C.; Ebisuya, M. Species-specific segmentation clock periods are due to differential biochemical reaction speeds. Science 2020, 369, 1450–1455. [Google Scholar] [CrossRef]
- Rayon, T.; Stamataki, D.; Perez-Carrasco, R.; Garcia-Perez, L.; Barrington, C.; Melchionda, M.; Exelby, K.; Lazaro, J.; Tybulewicz, V.L.J.; Fisher, E.M.C.; et al. Species-specific pace of development is associated with differences in protein stability. Science 2020, 369, eaba7667. [Google Scholar] [CrossRef]
- Beccari, L.; Moris, N.; Girgin, M.; Turner, D.A.; Baillie-Johnson, P.; Cossy, A.-C.; Lutolf, M.P.; Duboule, D.; Arias, A.M. Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids. Nature 2018, 562, 272–276. [Google Scholar] [CrossRef]
- Veenvliet, J.V.; Bolondi, A.; Kretzmer, H.; Haut, L.; Scholze-Wittler, M.; Schifferl, D.; Koch, F.; Guignard, L.; Kumar, A.S.; Pustet, M.; et al. Mouse embryonic stem cells self-organize into trunk-like structures with neural tube and somites. Science 2020, 370, eaba4937. [Google Scholar] [CrossRef]
- O’Hara-Wright, M.; Gonzalez-Cordero, A. Retinal organoids: A window into human retinal development. Development 2020, 147, 189746. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).