DNA Barcodes Applied to a Rapid Baseline Construction in Biodiversity Monitoring for the Conservation of Aquatic Ecosystems in the Sian Ka’an Reserve (Mexico) and Adjacent Areas
Abstract
1. Introduction
2. Materials and Methods
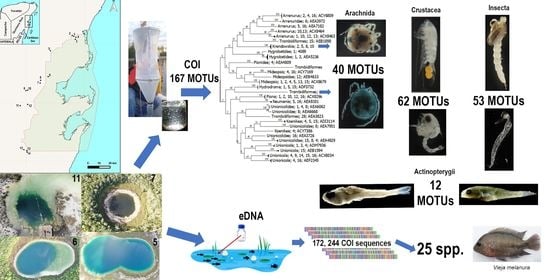
2.1. Sampling
2.2. DNA Extraction and Amplification of Individuals
2.3. Sequencing and Data Analysis
2.4. Metabarcoding and eDNA
2.5. Statistical Analyses
3. Results and Discussion
3.1. DNA Barcoding Baseline
3.2. Metabarcoding and eDNA
3.3. Species Composition Comparison
3.4. General Remarks and Future Biomonitoring
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Olivares, E.A.O.; Torres, S.S.; Jimenez, S.I.B.; Enriquez, J.O.C.; Zignol, F.; Reygadas, Y.; Tiefenbacher, J.P. Climate Change, Land Use/Land Cover Change, and Population Growth as Drivers of Groundwater Depletion in the Central Valleys, Oaxaca, Mexico. Remote Sens. 2019, 11, 1290. [Google Scholar] [CrossRef]
- Adamowicz, S.J.; Purvis, A. How Many Branchiopod Crustacean Species Are There? Quantifying the Components of Underestimation. Glob. Ecol. Biogeogr. 2005, 14, 455–468. [Google Scholar] [CrossRef]
- Elías-Gutiérrez, M.; Martínez-Jerónimo, F.; Ivanova, N.V.; Valdez-Moreno, M. DNA Barcodes for Cladocera and Copepoda from Mexico and Guatemala, Highlights and New Discoveries. Zootaxa 2008, 1849, 1–42. [Google Scholar] [CrossRef]
- Valdez-Moreno, M.; Ivanova, N.V.; Elías-Gutiérrez, M.; Contreras-Balderas, S.; Hebert, P.D.N. Probing Diversity in Freshwater Fishes from Mexico and Guatemala with DNA Barcodes. J. Fish. Biol. 2009, 74, 377–402. [Google Scholar] [CrossRef] [PubMed]
- Ratnasingham, S.; Hebert, P.D.N. Bold: The Barcode of Life Data System (www.Barcodinglife.Org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Ardura, A.; Zaiko, A.; Borrell, Y.J.; Samuiloviene, A.; Garcia-Vazquez, E. Novel Tools for Early Detection of a Global Aquatic Invasive, the Zebra Mussel Dreissena Polymorpha. Aquat. Conserv. Mar. Freshw. Ecosyst. 2017, 27, 165–176. [Google Scholar] [CrossRef]
- Eva, B.; Harmony, P.; Thomas, G.; Francois, G.; Alice, V.; Claude, M.; Tony, D. Trails of River Monsters: Detecting Critically Endangered Mekong Giant Catfish Pangasianodon Gigas Using Environmental DNA. Glob. Ecol. Conserv. 2016, 7, 148–156. [Google Scholar] [CrossRef]
- Valdez-Moreno, M.; Ivanova, N.V.; Elías-Gutiérrez, M.; Pedersen, S.L.; Bessonov, K.; Hebert, P.D.N. Using Edna to Biomonitor the Fish Community in a Tropical Oligotrophic Lake. PLoS ONE 2019, 14, e0215505. [Google Scholar] [CrossRef]
- Moriniere, J.; Balke, M.; Doczkal, D.; Geiger, M.F.; Hardulak, L.A.; Haszprunar, G.; Hausmann, A.; Hendrich, L.; Regalado, L.; Rulik, B.; et al. A DNA Barcode Library for 5200 German Flies and Midges (Insecta: Diptera) and Its Implications for Metabarcoding-Based Biomonitoring. Mol. Ecol. Resour. 2019, 19, 900–928. [Google Scholar] [CrossRef] [PubMed]
- Makino, W.; Maruoka, N.; Nakagawa, M.; Takamura, N. DNA Barcoding of Freshwater Zooplankton in Lake Kasumigaura, Japan. Ecol. Res. 2017, 32, 481–493. [Google Scholar] [CrossRef]
- Lagomarsino, L.P.; Frost, L.A. The Central Role of Taxonomy in the Study of Neotropical Biodiversity. Ann. Mo. Bot. Gard. 2020, 105, 405–421. [Google Scholar] [CrossRef]
- Montoliu-Elena, L.; Elias-Gutierrez, M.; Silva-Briano, M. Moina Macrocopa (Straus, 1820): A Species Complex of a Common Cladocera, Highlighted by Morphology and DNA Barcodes. Limnetica 2019, 38, 253–277. [Google Scholar]
- Ratnasingham, S.; Hebert, P.D. A DNA-Based Registry for All Animal Species: The Barcode Index Number (Bin) System. PLoS ONE 2013, 8, e66213. [Google Scholar] [CrossRef]
- Cervantes-Martinez, A.; Elías-Gutiérrez, M.; Suarez-Morales, E. Limnological and Morphometrical Data of Eight Karstic Systems ‘Cenotes’ of the Yucatan Peninsula, Mexico, During the Dry Season (February–May, 2001). Hydrobiologia 2002, 482, 167–177. [Google Scholar] [CrossRef]
- Medina-Moreno, S.; Jimenez-Gonzalez, A.; Gutiérrez-Rojas, M.; Lizardi-Jimenez, M. Hydrocarbon Pollution Studies of Underwater Sinkholes Along Quintana Roo as a Function of Tourism Development in the Mexican Caribbean. Revista Mexicana de Ingenieria Quimica 2014, 13, 509–516. [Google Scholar]
- Elías-Gutiérrez, M.; Valdez-Moreno, M.; Topan, J.; Young, M.R.; Cohuo-Colli, J.A. Improved Protocols to Accelerate the Assembly of DNA Barcode Reference Libraries for Freshwater Zooplankton. Ecol. Evol. 2018, 8, 3002–3018. [Google Scholar] [CrossRef]
- UNESCO. Sian Ka’an. UNESCO Wolrd Heritage Convention. Available online: https://whc.unesco.org/en/list/410/ (accessed on 7 April 2021).
- RAMSAR. Sian Ka’an. Ramsar Sites Information Service. Available online: https://rsis.ramsar.org/ris/1329 (accessed on 27 April 2021).
- Schmitter-Soto, J.J.; Caro, C.I. Distribution of Tilapia, Oreochromis mossambicus (Perciformes: Cichlidae), and Water Body Characteristics in Quintana Roo, Mexico. Rev. Biol. Trop. 1997, 45, 1257–1261. [Google Scholar]
- Mercado-Salas, N.F.; Khodami, S.; Kihara, T.C.; Elías-Gutiérrez, M.; Arbizu, P.M. Genetic Structure and Distributional Patterns of the Genus Mastigodiaptomus (Copepoda) in Mexico, with the Description of a New Species from the Yucatan Peninsula. Arthropod Syst. Phylogeny 2018, 76, 487–507. [Google Scholar]
- Corgosinho, P.H.C.; Mercado-Salas, N.F.; Arbizu, P.M.; Silva, E.N.D.; Kihara, T.C. Revision of the Remaneicaris argentina-Group (Copepoda, Harpacticoida, Parastenocarididae): Supplementary Description of Species, and Description of the First Semi-Terrestrial Remaneicaris from the Tropical Forest of Southeast Mexico. Zootaxa 2017, 4238, 499–530. [Google Scholar] [CrossRef]
- CONABIO, Comisión Nacional Para el Uso y Manejo de la Biodiversidad. 108. Sian Ka’an. CONABIO. Available online: http://www.conabio.gob.mx/conocimiento/regionalizacion/doctos/rhp_108.html (accessed on 27 April 2021).
- CONABIO, Comisión Nacional Para el Conocimiento y Uso de la Biodiversidad. Naturalista. CONABIO. Available online: https://www.naturalista.mx/observations?captive=any&place_id=54190&project_id=1387&taxon_id=47178&verifiable=any&view=species (accessed on 27 April 2021).
- CONANP, Comisión Nacional de Áreas Protegidas. Sian Ka’an. Gobierno de México. Available online: https://simec.conanp.gob.mx/ficha.php?anp=97®=9 (accessed on 27 April 2021).
- Valdez-Moreno, M.; Elías-Gutiérrez, M. Programa De Detección Temprana Piloto De Especies Acuáticas Invasoras a Través De Los Métodos De Código De Barras De La Vida Y Análisis De ADN Ambiental En La Reserva De La Biosfera Sian Ka´An. Proyecto 00089333 Aumentar Las Capacidades De México Para Manejar Especies Exóticas Invasoras a Través De La Implementación De La Estrategia Nacional De Especies Invasoras; PNUD México (Programa de Naciones Unidas para el Desarrollo), Ed.; El Colegio de la Frontera Sur: Chetumal, Mexico, 2019; p. 65. [Google Scholar]
- Montes-Ortiz, L.; Elías-Gutiérrez, M. Faunistic Survey of the Zooplankton Community in an Oligotrophic Sinkhole, Cenote Azul (Quintana Roo, Mexico), Using Different Sampling Methods, and Documented with DNA Barcodes. J. Limnol. 2018, 77, 428–440. [Google Scholar] [CrossRef]
- Porco, D.; Rougerie, R.; Deharveng, L.; Hebert, P. Coupling Non-Destructive DNA Extraction and Voucher Retrieval for Small Soft-Bodied Arthropods in a High-Throughput Context: The Example of Collembola. Mol. Ecol. Resour. 2010, 10, 942–945. [Google Scholar] [CrossRef] [PubMed]
- Elías-Gutiérrez, M.; Suárez-Morales, E.; Gutiérrez-Aguirre, M.; Silva-Briano, M.; Granados-Ramirez, J.G.; Garfias-Espejo, T. Guía Ilustrada De Los Microcrustáceos (Cladocera Y Copepoda) De Las Aguas Continentales De México; Universidad Nacional Autónoma de México: Mexico City, Mexico, 2008. [Google Scholar]
- Ivanova, N.V.; DeWaard, J.R.; Hebert, P.D.N. An Inexpensive, Automation-Friendly Protocol for Recovering High-Quality DNA. Mol. Ecol. Notes 2006, 6, 998–1002. [Google Scholar] [CrossRef]
- Hajibabaei, M.; DeWaard, J.R.; Ivanova, N.V.; Ratnasingham, S.; Dooh, R.; Kirk, S.L.; Mackie, P.M.; Hebert, P.D.N. Critical Factors for Assembling a High Volume of DNA Barcodes. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2005, 360, 1959–1967. [Google Scholar] [CrossRef]
- Kimura, M. A Simple Method of Estimating Evolutionary Rate of Base Substitutions through Comparative Studies. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Mutanen, M.; Kivela, S.M.; Vos, R.A.; Doorenweerd, C.; Ratnasingham, S.; Hausmann, A.; Huemer, P.; Dinca, V.; van Nieukerken, E.J.; Lopez-Vaamonde, C.; et al. Species-Level Para- and Polyphyly in DNA Barcode Gene Trees: Strong Operational Bias in European Lepidoptera. Syst. Biol. 2016, 65, 1024–1040. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, N.V.; Fazekas, A.J.; Hebert, P.D.N. Semi-Automated, Membrane-Based Protocol for DNA Isolation from Plants. Plant. Mol. Biol. Report. 2008, 26, 186–198. [Google Scholar] [CrossRef]
- Ivanova, N.V.; Clare, E.L.; Borisenko, A.V. DNA Barcoding in Mammals. Methods Mol. Biol. 2012, 858, 153–182. [Google Scholar]
- Whittaker, R.H. Vegetation of the Siskiyou Mountains, Oregon and California. Ecol. Monogr. 1960, 30, 279–338. [Google Scholar] [CrossRef]
- Jost, L. Independence of Alpha and Beta Diversities. Ecology 2010, 91, 1969–1974. [Google Scholar] [CrossRef]
- Schroeder, P.J.; Jenkins, D.G. How Robust Are Popular Beta Diversity Indices to Sampling Error? Ecosphere 2018, 9, e02100. [Google Scholar] [CrossRef]
- Baselga, A.; Orme, C.D.L. Betapart: An R Package for the Study of Beta Diversity. Methods Ecol. Evol. 2012, 3, 808–812. [Google Scholar] [CrossRef]
- Hamerlik, L.; Wojewodka, M.; Zawisza, E.; Duran, S.C.; Macario-Gonzalez, L.; Perez, L.; Szeroczynska, K. Subfossil Chironomidae (Diptera) in Surface Sediments of the Sinkholes (Cenotes) of the Yucatan Peninsula: Diversity and Distribution. J. Limnol. 2018, 77, 213–219. [Google Scholar] [CrossRef]
- Smirnov, N.N.; Elías-Gutiérrez, M. Biocenotic Characteristics of Some Yucatan Lentic Water Bodies Based on Invertebrate Remains in Sediments. Inland Water Biol. 2011, 4, 211–217. [Google Scholar] [CrossRef]
- Prosser, S.; Martínez-Arce, A.; Elías-Gutiérrez, M. A New Set of Primers for Coi Amplification from Freshwater Microcrustaceans. Mol. Ecol. Resour. 2013, 13, 1151–1155. [Google Scholar] [CrossRef]
- Montes-Ortiz, L.; Elías-Gutiérrez, M. Water Mite Diversity (Acariformes: Prostigmata: Parasitengonina: Hydrachnidiae) from Karst Ecosystems in Southern of Mexico: A Barcoding Approach. Diversity 2020, 12, 329. [Google Scholar] [CrossRef]
- Goldschmidt, T.; Helson, J.E.; Williams, D.D. Ecology of Water Mite Assemblages in Panama—First Data on Water Mites (Acari, Hydrachnidia) as Bioindicators in the Assessment of Biological Integrity of Neotropical Streams. Limnologica 2016, 59, 63–77. [Google Scholar] [CrossRef]
- Wojewodka, M.; Sinev, A.Y.; Zawisza, E. A Guide to the Identification of Subfossil Non-Chydorid Cladocera (Crustacea: Branchiopoda) from Lake Sediments of Central America and the Yucatan Peninsula, Mexico: Part I. J. Paleolimnol. 2020, 63, 269–282. [Google Scholar] [CrossRef]
- Wojewodka, M.; Sinev, A.Y.; Zawisza, E.; Stanczak, J. A Guide to the Identification of Subfossil Chydorid Cladocera (Crustacea: Branchiopoda) from Lake Sediments of Central America and the Yucatan Peninsula, Mexico: Part II. J. Paleolimnol. 2020, 63, 37–64. [Google Scholar] [CrossRef]
- Elías-Gutiérrez, M.; Montes-Ortiz, L. Present Day Kwnoledge on Diversity of Freshwater Zooplancton (Invertebrates) of the Yucatan Peninsula, Using Integrated Taxonomy. Teoria Y Praxis 2018, 14, 31–48. [Google Scholar]
- Yoo, H.; Cohuo, S.; Macario-Gonzalez, L.; Karanovic, I. A New Freshwater Ostracod Genus from the Northern Neotropical Region and Its Phylogenetic Position in the Family Cyprididae (Podocopida). Zool. Anz. 2017, 266, 196–215. [Google Scholar] [CrossRef][Green Version]
- Macario-Gonzalez, L.; Cohuo, S.; Elías-Gutiérrez, M.; Vences, M.; Perez, L.; Schwalb, A. Integrative Taxonomy of Freshwater Ostracodes (Crustacea: Ostracoda) of the Yucatan Peninsula, Implications for Paleoenvironmental Reconstructions in the Northern Neotropical Region. Zool. Anz. 2018, 275, 20–36. [Google Scholar] [CrossRef]
- Cohuo-Durán, S.; Elías-Gutiérrez, M.; Karanovic, I. On Three New Species of Cypretta Vávra, 1895 (Crustacea: Ostracoda) from the Yucatan Peninsula, Mexico. Zootaxa 2013, 3636, 501–524. [Google Scholar] [CrossRef]
- Karanovic, T.; Krajicek, M. When Anthropogenic Translocation Meets Cryptic Speciation Globalized Bouillon Originates; Molecular Variability of the Cosmopolitan Freshwater Cyclopoid Macrocyclops albidus (Crustacea: Copepoda). Ann. Limnol. Int. J. Limnol. 2012, 48, 63–80. [Google Scholar] [CrossRef][Green Version]
- Fiers, F.; Ghenne, V.; Suárez-Morales, E. New Species of Continental Cyclopoid Copepods (Crustacea, Cyclopoida) from the Yucatan Peninsula, Mexico. Stud. Neotrop. Fauna Environ. 2000, 35, 209–251. [Google Scholar] [CrossRef]
- Barba Macias, E. Faunistic Analysis of the Caridean Shrimps Inhabiting Seagrasses Along the Nw Coast of the Gulf of Mexico and Caribbean Sea. Rev. Biol. Trop. 2012, 60, 1161–1175. [Google Scholar] [CrossRef]
- Moriniere, J.; Hendrich, L.; Balke, M.; Beermann, A.J.; Konig, T.; Hess, M.; Koch, S.; Muller, R.; Leese, F.; Hebert, P.D.N.; et al. A DNA Barcode Library for Germanys Mayflies, Stoneflies and Caddisflies (Ephemeroptera, Plecoptera and Trichoptera). Mol. Ecol. Resour. 2017, 17, 1293–1307. [Google Scholar] [CrossRef]
- Vinogradova, E.M. Six New Species of Polypedilum Kieffer, 1912, from the Yucatan Peninsula (Insecta, Diptera, Chironomidae). Spixiana 2008, 31, 277–288. [Google Scholar]
- Vinogradova, E.M.; Riss, W. Chironomids of the Yucatán Peninsula. Chironomus J. Chironomidae Res. 2007, 20, 32–35. [Google Scholar] [CrossRef]
- Contreras-Ramos, A.; Andersen, T. A Survey of the Chironomidae (Diptera) of Calakmul Biosphere Reserve. Mexico. Chironomus 1999, 12, 3–5. [Google Scholar]
- Messing, J. New M13 Vectors for Cloning. Methods Enzymol. 1983, 101, 20–78. [Google Scholar]
- Schmitter-Soto, J.J. Catálogo De Los Peces Continentales De Quintana Roo; El Colegio de la Frontera Sur: San Cistóbal de las Casas, Chiapas, Mexico, 1998. [Google Scholar]
- Cervantes-Martínez, A.; Elías-Gutiérrez, M.; Gutiérrez-Aguirre, M.A.; Kotov, A.A. Ecological Remarks on Mastigodiaptomus nesus Bowman, 1986 (Copepoda: Calanoida) in a Mexican Karstic Sinkhole. Hydrobiologia 2005, 542, 95–102. [Google Scholar] [CrossRef]
- Cervantes-Martinez, A.; Gutiérrez-Aguirre, M.A. Physicochemistry and Zooplankton of Two Karstic Sinkholes in the Yucatan Peninsula, Mexico. J. Limnol. 2015, 74, 382–393. [Google Scholar] [CrossRef]
- Gutiérrez-Aguirre, M.A.; Cervantes-Martinez, A.; Elías-Gutiérrez, M.; Lugo-Vazquez, A. Remarks on Mastigodiaptomus (Calanoida: Diaptomidae) from Mexico Using Integrative Taxonomy, with a Key of Identification and Three New Species. PeerJ 2020, 8, e8416. [Google Scholar] [CrossRef]
- Bauer-Gottwein, P.; Gondwe, B.R.N.; Charvet, G.; Marín, L.E.; Rebolledo-Vieyra, M.; Merediz-Alonso, G. Review: The Yucatán Peninsula Karst Aquifer, Mexico. Hydrogeol. J. 2011, 19, 507–524. [Google Scholar] [CrossRef]
- Gondwe, B.R.N.; Hong, S.H.; Wdowinski, S.; Bauer-Gottwein, P. Hydrologic Dynamics of the Ground-Water-Dependent Sian Ka’an Wetlands, Mexico, Derived from Insar and Sar Data. Wetlands 2010, 30, 1–13. [Google Scholar] [CrossRef]
- CONABIO. Capital Natural De México, Vol I: Conocimiento Actual De La Biodiversidad; Comisión Nacional para el Conocimiento y Uso de la Biodiversidad, México: Mexico City, Mexico, 2008; Volume I. [Google Scholar]
- Gutiérrez, M.F.; Molina, F.R.; Frau, D.; Mayora, G.; Battauz, Y. Interactive Effects of Fish Predation and Sublethal Insecticide Concentrations on Freshwater Zooplankton Communities. Ecotoxicol. Environ. Saf. 2020, 196, 110497. [Google Scholar] [CrossRef]
- Almeida, R.; Formigo, N.E.; Sousa-Pinto, I.; Antunes, S.C. Contribution of Zooplankton as a Biological Element in the Assessment of Reservoir Water Quality. Limnetica 2020, 39, 245–261. [Google Scholar]
- Dixon, J.; Hamilton, K.; Pagiola, S.; Segnestam, L. Tourism and the Environment in the Caribbean: An Economic Framework. In Environmental Economic Series; World Bank: Washington, DC, USA, 2001; p. 68. [Google Scholar]
- Yanez-Montalvo, A.; Gomez-Acata, S.; Aguila, B.; Hernandez-Arana, H.; Falcon, L.I. The Microbiome of Modern Microbialites in Bacalar Lagoon, Mexico. PLoS ONE 2020, 15, e0230071. [Google Scholar] [CrossRef]








| Number | Name | Coordinates | Zone | Location in Sian Ka’an | Municipality |
|---|---|---|---|---|---|
| Latitude N | Longitude W | ||||
| 1 | Laguna Muyil 1 | 20.0686 | 87.5944 | Buffer zone | Felipe Carrillo Puerto |
| 2 | Laguna Muyil 2 | 20.0753 | 87.6073 | ||
| 3 | Chunyaxché 1 | 20.0422 | 87.5807 | Buffer zone | Felipe Carrillo Puerto |
| 4 | Chunyaxché 2 | 20.0601 | 87.5757 | Buffer zone | |
| 5 | Km 48 | 19.9431 | 97.7938 | Influence area | Felipe Carrillo Puerto |
| 6 | Del Padre sinkhole | 19.6038 | 88.0028 | Influence area | Felipe Carrillo Puerto |
| 7 | Tres Reyes 1 sinkhole | 19.6682 | 87.8812 | Influence area | Felipe Carrillo Puerto |
| 8 | Tres Reyes sinkhole 2 | 19.6916 | 87.8774 | Influence area | Felipe Carrillo Puerto |
| 9 | Santa Teresa sinkhole | 19.7240 | 87.8130 | Buffer zone | Felipe Carrillo Puerto |
| 10 | Minicenote sinkhole | 19.6070 | 87.9887 | Influence area | Felipe Carrillo Puerto |
| 11 | Sijil Noh Há sinkhole | 19.4746 | 88.0516 | Influence area | Felipe Carrillo Puerto |
| 12 | Chancah Veracruz sinkhole | 19.4855 | 87.9879 | Influence area | Felipe Carrillo Puerto |
| 13 | El Toro sinkhole | 19.0981 | 88.0206 | Influence area | Bacalar |
| 14 | Pucté Cafetal sinkhole | 19.0788 | 87.9943 | Influence area | Bacalar |
| 15 | Pucté 2 sinkhole | 19.0915 | 87.9942 | Influence area | Bacalar |
| Primer Name | Direction | Primer Sequence | Reference |
|---|---|---|---|
| AquaF2_t1 | F | [M13F]ATCACRACCATCATYAAYATRAARCC | [34] |
| AquaF3_t1 | F | [M13F]CCAGCCATTTCNCARTACCARACRCC | [20] |
| C_FishR1 cocktail: | Cocktail primers (FR1d: FishR2; 1:1) | [35] | |
| FR1d_t1 | R | [M13R]ACCTCAGGGTGTCCGAARAAYCARAA | |
| FishR2_t1 | R | [M13R]ACTTCAGGGTGACCGAAGAATCAGAA | |
| M13-tails | |||
| M13F | F | TGTAAAACGACGGCCAGT | [58] |
| M13R | R | CAGGAAACAGCTATGAC | [58] |
| NGS-fusion | |||
| IonA-M13F-ion1-96 | F | CCATCTCATCCCTGCGTGTCTCC[GACT][IonExpress-MID][M13F] | Ion Torrent, Thermo Fisher Scientific |
| trP1-M13R | R | CCTCTCTATGGGCAGTCGGTGAT [M13R] | Ion Torrent, Thermo Fisher Scientific |
| Identification | Chunyaxché 2 | Muyil 1 | Km 48 | Santa Teresa | Tres Reyes 2 | Tres Reyes 1 | Minicenote | Del Padre | El Toro | Sijil Noh Há | Pucté 2 | Chacah Veracruz | Pucté Cafetal 1 |
| Aramides cajaneus | 13 | ||||||||||||
| Megaceryle torquata | 1887 | ||||||||||||
| Meleagris gallopavo | 10 | ||||||||||||
| Artibeus lituratus | 5 | ||||||||||||
| Lampronycteris brachyotis | 47 | ||||||||||||
| Lonchorhina aurita | 95 | ||||||||||||
| Oryzomys couesi | 5063 | ||||||||||||
| Pteronotus parnellii | 3 | ||||||||||||
| Kinosternon acutum | 3 | ||||||||||||
| Trachemys sp. | 610 | ||||||||||||
| Atherinella sp.+ | 290 | 77 | |||||||||||
| Belonesox belizanus | 287 | 3 | |||||||||||
| Bramocharax-Astyanax *+ | 321 | 2 | 2 | 2 | 3 | 4678 | |||||||
| Cribroheros robertsoni | 23 | 130 | 83 | ||||||||||
| Cryptoheros chetumalensis | 2 | 3033 | 13265 | ||||||||||
| Cyprinodon beltrani-simus *+ | 969 | ||||||||||||
| Dormitator maculatus+ | 66 | ||||||||||||
| Gambusia sexradiata | 4 | 2840 | 1563 | 816 | |||||||||
| Gambusia yucatana + | 2 | 31 | 4 | 9 | 2 | ||||||||
| Gerres cinereus | 71 | 381 | |||||||||||
| Gobiomorus dormitor | 9 | ||||||||||||
| Hyphessobrycon compressus | 1332 | ||||||||||||
| Garmanella pulchra+ | 411 | ||||||||||||
| Lutjanus griseus | 108 | ||||||||||||
| Mayaheros urophthalmus+ | 69 | 10 | 16 | 18 | 29 | 1723 | |||||||
| Ophisternon | 233 | ||||||||||||
| Parachromis friedrichsthalii | 25 | ||||||||||||
| Petenia splendida | 919 | 3927 | |||||||||||
| Poecilia mexicana | 9 | 2 | 4 | 28 | 6 | 2 | 528 | 2 | |||||
| Rhamdia quelen | 2 | 8 | 2 | 15 | 4 | 4 | 156 | 6 | |||||
| Rocio octofasciata | 42 | ||||||||||||
| Thorichthys helleri | 15 | ||||||||||||
| Thorichthys meeki+ | 4 | 7 | |||||||||||
| Trichromis salvini+ | 6 | 26 | 2 | 3 | 39 | 6 | 20 | 2780 | |||||
| Vieja melanura+ | 2 | 4 | 2196 | ||||||||||
| Aves | |||||||||||||
| Mammalia | |||||||||||||
| Reptilia | |||||||||||||
| Actinopterygii |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valdez-Moreno, M.; Mendoza-Carranza, M.; Rendón-Hernández, E.; Alarcón-Chavira, E.; Elías-Gutiérrez, M. DNA Barcodes Applied to a Rapid Baseline Construction in Biodiversity Monitoring for the Conservation of Aquatic Ecosystems in the Sian Ka’an Reserve (Mexico) and Adjacent Areas. Diversity 2021, 13, 292. https://doi.org/10.3390/d13070292
Valdez-Moreno M, Mendoza-Carranza M, Rendón-Hernández E, Alarcón-Chavira E, Elías-Gutiérrez M. DNA Barcodes Applied to a Rapid Baseline Construction in Biodiversity Monitoring for the Conservation of Aquatic Ecosystems in the Sian Ka’an Reserve (Mexico) and Adjacent Areas. Diversity. 2021; 13(7):292. https://doi.org/10.3390/d13070292
Chicago/Turabian StyleValdez-Moreno, Martha, Manuel Mendoza-Carranza, Eduardo Rendón-Hernández, Erika Alarcón-Chavira, and Manuel Elías-Gutiérrez. 2021. "DNA Barcodes Applied to a Rapid Baseline Construction in Biodiversity Monitoring for the Conservation of Aquatic Ecosystems in the Sian Ka’an Reserve (Mexico) and Adjacent Areas" Diversity 13, no. 7: 292. https://doi.org/10.3390/d13070292
APA StyleValdez-Moreno, M., Mendoza-Carranza, M., Rendón-Hernández, E., Alarcón-Chavira, E., & Elías-Gutiérrez, M. (2021). DNA Barcodes Applied to a Rapid Baseline Construction in Biodiversity Monitoring for the Conservation of Aquatic Ecosystems in the Sian Ka’an Reserve (Mexico) and Adjacent Areas. Diversity, 13(7), 292. https://doi.org/10.3390/d13070292