Characterization of a Pseudokeronopsis Strain (Ciliophora, Urostylida) and Its Bacterial Endosymbiont “Candidatus Trichorickettsia” (Alphaproteobacteria, Rickettsiales)
Abstract
1. Introduction
2. Materials and Methods
3. Results
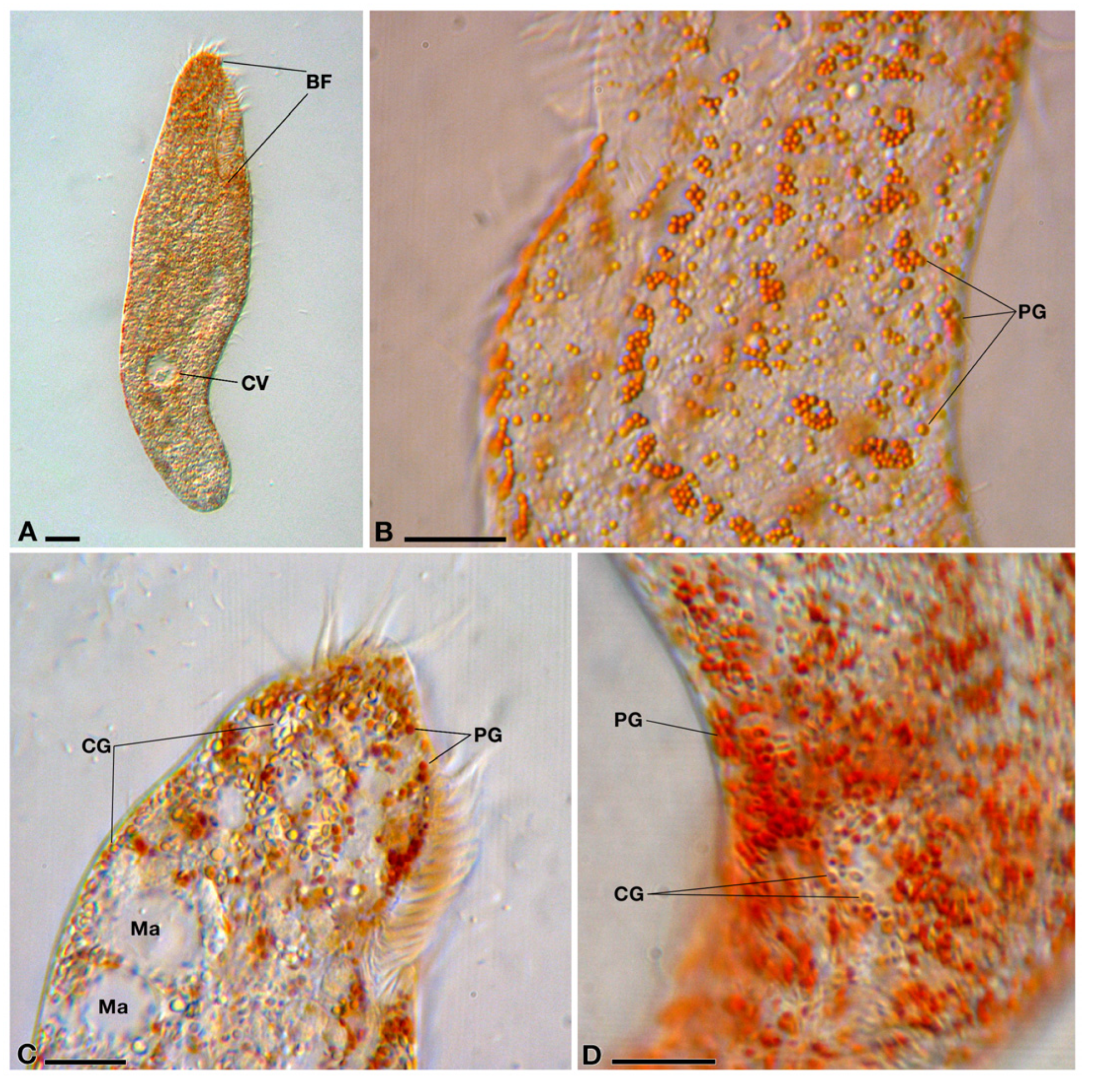
3.1. Characterization and Identification of the Host
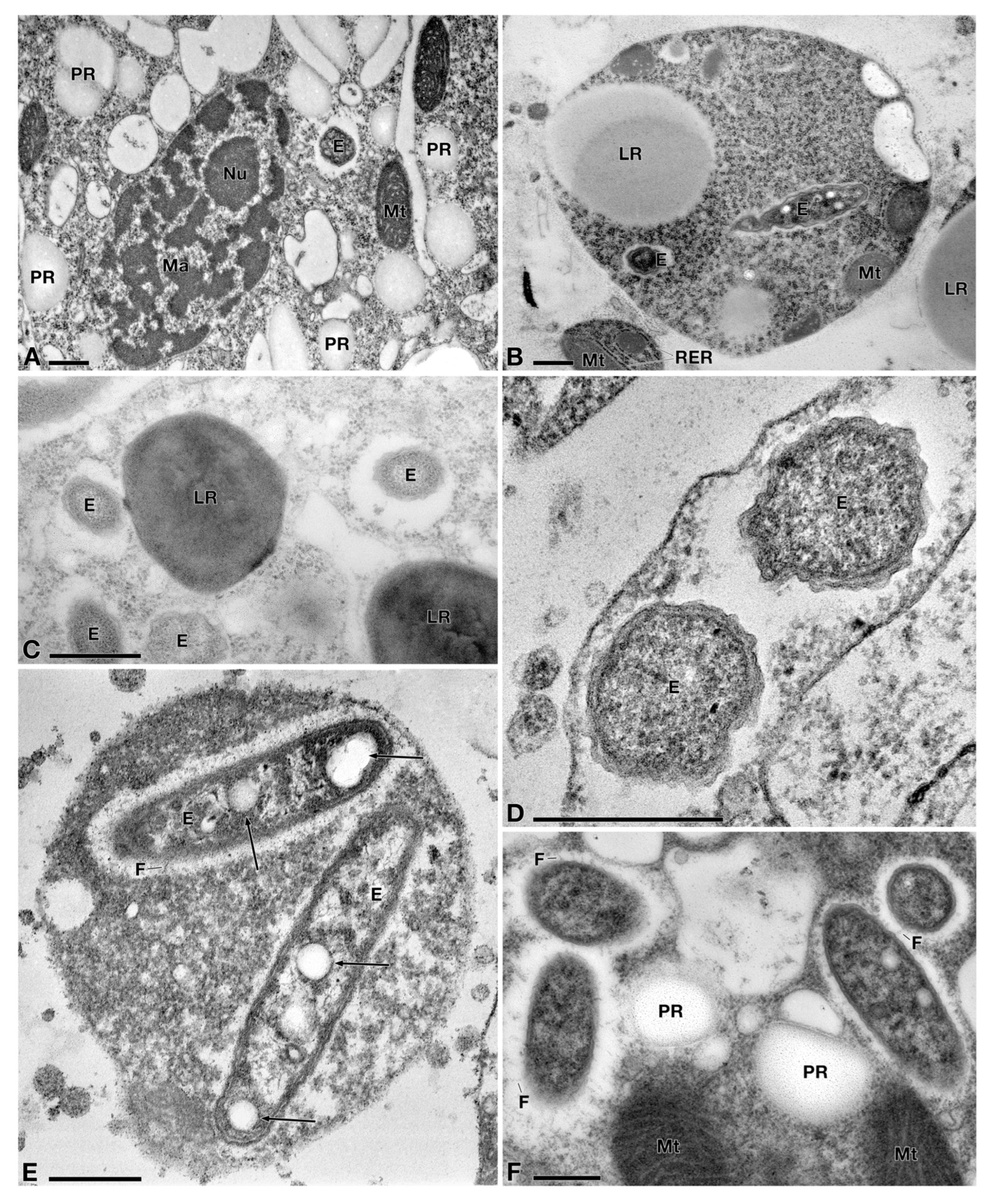
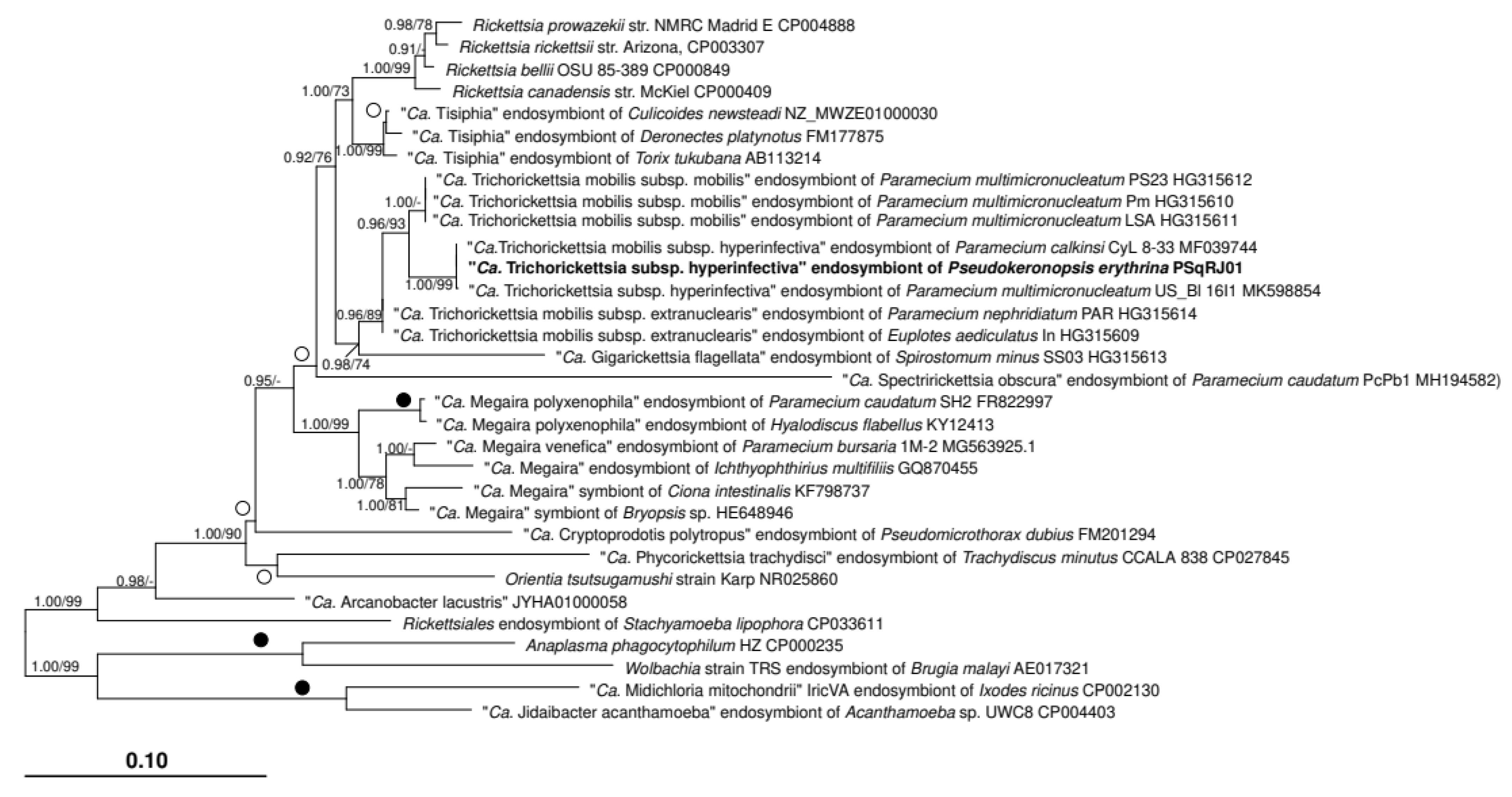
3.2. Characterization of the Bacterial Symbiont
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Husnik, F.; Tashyreva, D.; Boscaro, V.; George, E.E.; Lukeš, J.; Keeling, P.J. Bacterial and Archaeal Symbioses with Protists. Curr. Biol. 2021, 31, R862–R877. [Google Scholar] [CrossRef] [PubMed]
- Nowack, E.C.M.; Melkonian, M. Endosymbiotic Associations within Protists. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Dziallas, C.; Allgaier, M.; Monaghan, M.T.; Grossart, H.-P. Act Together—Implications of Symbioses in Aquatic Ciliates. Front. Microbiol. 2012, 3, 288. [Google Scholar] [CrossRef]
- Serra, V.; Gammuto, L.; Nitla, V.; Castelli, M.; Lanzoni, O.; Sassera, D.; Bandi, C.; Sandeep, B.V.; Verni, F.; Modeo, L.; et al. Morphology, Ultrastructure, Genomics, and Phylogeny of Euplotes Vanleeuwenhoeki Sp. Nov. and Its Ultra-Reduced Endosymbiont “Candidatus Pinguicoccus Supinus” Sp. Nov. Sci. Rep. 2020, 10, 20311. [Google Scholar] [CrossRef] [PubMed]
- Castelli, M.; Sassera, D.; Petroni, G. Biodiversity of “Non-Model” Rickettsiales and Their Association with Aquatic Organisms. In Rickettsiales; Thomas, S., Ed.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 59–91. [Google Scholar]
- Collingro, A.; Köstlbacher, S.; Mussmann, M.; Stepanauskas, R.; Hallam, S.J.; Horn, M. Unexpected Genomic Features in Widespread Intracellular Bacteria: Evidence for Motility of Marine Chlamydiae. ISME J. 2017, 11, 2334–2344. [Google Scholar] [CrossRef]
- Hugoson, E.; Guliaev, A.; Ammunét, T.; Guy, L. Host Adaptation in Legionellales Is 1.9 Ga, Coincident with Eukaryogenesis. Mol. Biol. Evol. 2022, 39, msac037. [Google Scholar] [CrossRef]
- Watanabe, K.; Nakao, R.; Fujishima, M.; Tachibana, M.; Shimizu, T.; Watarai, M. Ciliate Paramecium Is a Natural Reservoir of Legionella pneumophila. Sci. Rep. 2016, 6, 24322. [Google Scholar] [CrossRef]
- Richards, A.M.; Von Dwingelo, J.E.; Price, C.T.; Abu Kwaik, Y. Cellular Microbiology and Molecular Ecology of Legionella-Amoeba Interaction. Virulence 2013, 4, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Mungroo, M.R.; Siddiqui, R.; Khan, N.A. War of the Microbial World: Acanthamoeba Spp. Interactions with Microorganisms. Folia Microbiol. 2021, 66, 689–699. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, M. Comparative Genomic Analysis of Acanthamoeba Endosymbionts Highlights the Role of Amoebae as a “Melting Pot” Shaping the Rickettsiales Evolution. Genome Biol. Evol. 2017, 9, 3214–3224. [Google Scholar] [CrossRef]
- Barker, J.; Brown, M.R. Trojan Horses of the Microbial World: Protozoa and the Survival of Bacterial Pathogens in the Environment. Microbiology 1994, 140 Pt 6, 1253–1259. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Higuchi, Y.; Shimmura, M.; Tachibana, M.; Fujishima, M.; Shimizu, T.; Watarai, M. Peculiar Paramecium Peculiar Hosts Fail to Establish a Stable Intracellular Relationship with Legionella pneumophila. Front. Microbiol. 2020, 11, 596731. [Google Scholar] [CrossRef] [PubMed]
- Fujishima, M.; Kodama, Y. Mechanisms for Establishing Primary and Secondary Endosymbiosis in Paramecium. J. Eukaryot. Microbiol. 2022, 69, e12901. [Google Scholar] [CrossRef] [PubMed]
- Siegmund, L.; Schweikert, M.; Fischer, M.S.; Wöstemeyer, J. Bacterial Surface Traits Influence Digestion by Tetrahymena Pyriformis and Alter Opportunity to Escape from Food Vacuoles. J. Eukaryot. Microbiol. 2018, 65, 600–611. [Google Scholar] [CrossRef] [PubMed]
- Siegmund, L.; Burmester, A.; Fischer, M.S.; Wöstemeyer, J. A Model for Endosymbiosis: Interaction between Tetrahymena Pyriformis and Escherichia Coli. Eur. J. Protistol. 2013, 49, 552–563. [Google Scholar] [CrossRef] [PubMed]
- Vannini, C.; Boscaro, V.; Ferrantini, F.; Benken, K.A.; Mironov, T.I.; Schweikert, M.; Görtz, H.-D.; Fokin, S.I.; Sabaneyeva, E.V.; Petroni, G. Flagellar Movement in Two Bacteria of the Family Rickettsiaceae: A Re-Evaluation of Motility in an Evolutionary Perspective. PLoS ONE 2014, 9, e87718. [Google Scholar] [CrossRef]
- Mironov, T.; Sabaneyeva, E. A Robust Symbiotic Relationship Between the Ciliate Paramecium multimicronucleatum and the Bacterium Ca. Trichorickettsia Mobilis. Front. Microbiol. 2020, 11, 603335. [Google Scholar] [CrossRef]
- Sabaneyeva, E.; Castelli, M.; Szokoli, F.; Benken, K.; Lebedeva, N.; Salvetti, A.; Schweikert, M.; Fokin, S.; Petroni, G. Host and Symbiont Intraspecific Variability: The Case of Paramecium calkinsi and “Candidatus Trichorickettsia Mobilis”. Eur. J. Protistol. 2018, 62, 79–94. [Google Scholar] [CrossRef]
- Davison, H.R.; Pilgrim, J.; Wybouw, N.; Parker, J.; Pirro, S.; Hunter-Barnett, S.; Campbell, P.M.; Blow, F.; Darby, A.C.; Hurst, G.D.D.; et al. Genomic Diversity across the Rickettsia and “Candidatus Megaira” Genera and Proposal of Genus Status for the Torix Group. Nat. Commun. 2022, 13, 2630. [Google Scholar] [CrossRef]
- Blanton, L.S. The Rickettsioses: A Practical Update. Infect. Dis. Clin. N. Am. 2019, 33, 213–229. [Google Scholar] [CrossRef]
- Dumler, J.S.; Walker, D.H. Rickettsiales. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; p. 1. [Google Scholar]
- Boscaro, V.; Schrallhammer, M.; Benken, K.A.; Krenek, S.; Szokoli, F.; Berendonk, T.U.; Schweikert, M.; Verni, F.; Sabaneyeva, E.V.; Petroni, G. Rediscovering the Genus Lyticum, Multiflagellated Symbionts of the Order Rickettsiales. Sci. Rep. 2013, 3, 3305. [Google Scholar] [CrossRef] [PubMed]
- Sassera, D.; Lo, N.; Epis, S.; D’Auria, G.; Montagna, M.; Comandatore, F.; Horner, D.; Peretó, J.; Luciano, A.M.; Franciosi, F.; et al. Phylogenomic Evidence for the Presence of a Flagellum and cbb(3) Oxidase in the Free-Living Mitochondrial Ancestor. Mol. Biol. Evol. 2011, 28, 3285–3296. [Google Scholar] [CrossRef] [PubMed]
- Schön, M.E.; Martijn, J.; Vosseberg, J.; Köstlbacher, S.; Ettema, T.J.G. The Evolutionary Origin of Host Association in the Rickettsiales. Nat. Microbiol. 2022, 7, 1189–1199. [Google Scholar] [CrossRef] [PubMed]
- Castelli, M.; Sabaneyeva, E.; Lanzoni, O.; Lebedeva, N.; Floriano, A.M.; Gaiarsa, S.; Benken, K.; Modeo, L.; Bandi, C.; Potekhin, A.; et al. Deianiraea, an Extracellular Bacterium Associated with the Ciliate Paramecium, Suggests an Alternative Scenario for the Evolution of Rickettsiales. ISME J. 2019, 13, 2280–2294. [Google Scholar] [CrossRef] [PubMed]
- Schulz, F.; Martijn, J.; Wascher, F.; Lagkouvardos, I.; Kostanjšek, R.; Ettema, T.J.G.; Horn, M. A Rickettsiales Symbiont of Amoebae with Ancient Features. Environ. Microbiol. 2016, 18, 2326–2342. [Google Scholar] [CrossRef] [PubMed]
- Floriano, A.M.; Biffignandi, G.B.; Castelli, M.; Olivieri, E.; Clementi, E.; Comandatore, F.; Rinaldi, L.; Opara, M.; Plantard, O.; Palomar, A.M.; et al. The Origin and Evolution of Mitochondrial Tropism in Midichloria Bacteria. bioRxiv 2022. [Google Scholar] [CrossRef]
- Anesi, A.; Buonanno, F.; di Giuseppe, G.; Ortenzi, C.; Guella, G. Metabolites from the Euryhaline CiliatePseudokeronopsis Erythrina. Eur. J. Org. Chem. 2016, 2016, 1330–1336. [Google Scholar] [CrossRef]
- Guella, G.; Frassanito, R.; Mancini, I.; Sandron, T.; Modeo, L.; Verni, F.; Dini, F.; Petroni, G. Keronopsamides, a New Class of Pigments from Marine Ciliates. Eur. J. Org. Chem. 2010, 2010, 427–434. [Google Scholar] [CrossRef]
- Medlin, L.; Elwood, H.J.; Stickel, S.; Sogin, M.L. The Characterization of Enzymatically Amplified Eukaryotic 16S-like rRNA-Coding Regions. Gene 1988, 71, 491–499. [Google Scholar] [CrossRef]
- Petroni, G.; Dini, F.; Verni, F.; Rosati, G. A Molecular Approach to the Tangled Intrageneric Relationships Underlying Phylogeny in Euplotes (Ciliophora, Spirotrichea). Mol. Phylogenet. Evol. 2002, 22, 118–130. [Google Scholar] [CrossRef]
- Rosati, G.; Modeo, L.; Melai, M.; Petroni, G.; Verni, F. A Multidisciplinary Approach to Describe Protists: A Morphological, Ultrastructural, and Molecular Study on Peritromus kahli Villeneuve-Brachon, 1940 (Ciliophora, Heterotrichea). J. Eukaryot. Microbiol. 2004, 51, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Boscaro, V.; Fokin, S.I.; Verni, F.; Petroni, G. Survey of Paramecium duboscqui Using Three Markers and Assessment of the Molecular Variability in the Genus Paramecium. Mol. Phylogenet. Evol. 2012, 65, 1004–1013. [Google Scholar] [CrossRef] [PubMed]
- Vannini, C.; Rosati, G.; Verni, F.; Petroni, G. Identification of the Bacterial Endosymbionts of the Marine Ciliate Euplotes magnicirratus (Ciliophora, Hypotrichia) and Proposal of “Candidatus Devosia Euplotis”. Int. J. Syst. Evol. Microbiol. 2004, 54, 1151–1156. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Manz, W.; Amann, R.; Ludwig, W.; Wagner, M.; Schleifer, K.-H. Phylogenetic Oligodeoxynucleotide Probes for the Major Subclasses of Proteobacteria: Problems and Solutions. Syst. Appl. Microbiol. 1992, 15, 593–600. [Google Scholar] [CrossRef]
- Amann, R.I.; Binder, B.J.; Olson, R.J.; Chisholm, S.W.; Devereux, R.; Stahl, D.A. Combination of 16S rRNA-Targeted Oligonucleotide Probes with Flow Cytometry for Analyzing Mixed Microbial Populations. Appl. Environ. Microbiol. 1990, 56, 1919–1925. [Google Scholar] [CrossRef] [PubMed]
- Modeo, L.; Salvetti, A.; Rossi, L.; Castelli, M.; Szokoli, F.; Krenek, S.; Serra, V.; Sabaneyeva, E.; Di Giuseppe, G.; Fokin, S.I.; et al. Trichorickettsia Mobilis", a Bacterium, Can Be Transiently Transferred from the Unicellular Eukaryote to the Planarian. PeerJ 2020, 8, e8977. [Google Scholar] [CrossRef] [PubMed]
- Westram, R.; Bader, K.; Prüsse, E.; Kumar, Y.; Meier, H.; Glöckner, F.O.; Ludwig, W. ARB: A Software Environment for Sequence Data. In Handbook of Molecular Microbial Ecology I: Metagenomics and Complementary Approaches; Wiley-Blackwell: Hoboken, NJ, USA, 2011; pp. 399–406. [Google Scholar]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More Models, New Heuristics and Parallel Computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Gascuel, O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Clamp, J.C.; Song, W. Phylogeny and Systematic Revision of the Family Pseudokeronopsidae (Protista, Ciliophora, Hypotricha), with Description of a New Estuarine Species of Pseudokeronopsis. Zool. Scr. 2011, 40, 659–671. [Google Scholar] [CrossRef]
- Yurchenko, T.; Ševčíková, T.; Přibyl, P.; El Karkouri, K.; Klimeš, V.; Amaral, R.; Zbránková, V.; Kim, E.; Raoult, D.; Santos, L.M.A.; et al. A Gene Transfer Event Suggests a Long-Term Partnership between Eustigmatophyte Algae and a Novel Lineage of Endosymbiotic Bacteria. ISME J. 2018, 12, 2163–2175. [Google Scholar] [CrossRef] [PubMed]
- Martijn, J.; Schulz, F.; Zaremba-Niedzwiedzka, K.; Viklund, J.; Stepanauskas, R.; Andersson, S.G.E.; Horn, M.; Guy, L.; Ettema, T.J.G. Single-Cell Genomics of a Rare Environmental Alphaproteobacterium Provides Unique Insights into Rickettsiaceae Evolution. ISME J. 2015, 9, 2373–2385. [Google Scholar] [CrossRef] [PubMed]
- Lanzoni, O.; Sabaneyeva, E.V.; Modeo, L.; Castelli, M.; Lebedeva, N.; Verni, F.; Schrallhammer, M.; Potekhin, A.; Petroni, G. Diversity and Environmental Distribution of the Cosmopolitan Endosymbiont “Candidatus Megaira”. Sci. Rep. 2019, 9, 1179. [Google Scholar] [CrossRef] [PubMed]
- Castelli, M.; Serra, V.; Senra, M.V.X.; Basuri, C.K.; Soares, C.A.G.; Fokin, S.I.; Modeo, L.; Petroni, G. The Hidden World of Rickettsiales Symbionts: “Candidatus Spectririckettsia Obscura,” a Novel Bacterium Found in Brazilian and Indian Paramecium caudatum. Microb. Ecol. 2019, 77, 748–758. [Google Scholar] [CrossRef]
- Muñoz-Gómez, S.A.; Hess, S.; Burger, G.; Lang, B.F.; Susko, E.; Slamovits, C.H.; Roger, A.J. An Updated Phylogeny of the Reveals That the Parasitic and Have Independent Origins. Elife 2019, 8, e42535. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.-S.; Park, S.-C.; Chun, J. Towards a Taxonomic Coherence between Average Nucleotide Identity and 16S rRNA Gene Sequence Similarity for Species Demarcation of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Mironov, T.; Yakovlev, A.; Sabaneyeva, E. Together Forever: Inseparable Partners of the Symbiotic System Paramecium multimicronucleatum/Ca. Trichorickettsia Mobilis. Symbiosis 2022, 87, 19–30. [Google Scholar] [CrossRef]
- Hess, S. Description of Hyalodiscus flabellus Sp. Nov. (Vampyrellida, Rhizaria) and Identification of Its Bacterial Endosymbiont, “Candidatus Megaira Polyxenophila” (Rickettsiales, Alphaproteobacteria). Protist 2017, 168, 109–133. [Google Scholar] [CrossRef]
- George, E.E.; Husnik, F.; Tashyreva, D.; Prokopchuk, G.; Horák, A.; Kwong, W.K.; Lukeš, J.; Keeling, P.J. Highly Reduced Genomes of Protist Endosymbionts Show Evolutionary Convergence. Curr. Biol. 2020, 30, 925–933.e3. [Google Scholar] [CrossRef]
- Boscaro, V.; Manassero, V.; Keeling, P.J.; Vannini, C. Single-Cell Microbiomics Unveils Distribution and Patterns of Microbial Symbioses in the Natural Environment. Microb. Ecol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Boscaro, V.; Husnik, F.; Vannini, C.; Keeling, P.J. Symbionts of the Ciliate Euplotes: Diversity, Patterns and Potential as Models for Bacteria-Eukaryote Endosymbioses. Proc. Biol. Sci. 2019, 286, 20190693. [Google Scholar] [CrossRef]
- Berger, H. Monograph of the Urostyloidea (Ciliophora, Hypotricha); Springer Science & Business Media: Berlin/Heidelberg, Germany, 2007; ISBN 9781402052736. [Google Scholar]
- Li, J.; Xu, K. Morphology and Taxonomy of Pseudokeronopsis rubra (Ehrenberg, 1836) and Pseudokeronopsis parasongi Sp. Nov. (Ciliophora, Hypotrichia, Urostylida) from the Yellow Sea. Eur. J. Protistol. 2020, 76, 125737. [Google Scholar] [CrossRef]
- Li, J.; Zhan, Z.; Xu, K. Systematics and Molecular Phylogeny of the Ciliate Genus Pseudokeronopsis (Ciliophora, Hypotrichia). J. Eukaryot. Microbiol. 2017, 64, 850–872. [Google Scholar] [CrossRef]
- Chen, X.; Li, J.; Xu, K. Insights into the Phylogeny of Three Systematically Controversial Subfamilies of Urostylid Ciliates Based on rDNA. Zool. Scr. 2021, 50, 383–395. [Google Scholar] [CrossRef]
- Xu, Y.; Shen, Z.; Gentekaki, E.; Xu, J.; Yi, Z. Comparative Transcriptome Analyses during the Vegetative Cell Cycle in the Mono-Cellular Organism Pseudokeronopsis erythrina (Alveolata, Ciliophora). Microorganisms 2020, 8, 108. [Google Scholar] [CrossRef] [PubMed]
- Boscaro, V.; Carducci, D.; Barbieri, G.; Senra, M.V.X.; Andreoli, I.; Erra, F.; Petroni, G.; Verni, F.; Fokin, S.I. Focusing on Genera to Improve Species Identification: Revised Systematics of the Ciliate Spirostomum. Protist 2014, 165, 527–541. [Google Scholar] [CrossRef] [PubMed]
- Melekhin, M.; Yakovleva, Y.; Lebedeva, N.; Nekrasova, I.; Nikitashina, L.; Castelli, M.; Mayén-Estrada, R.; Romanovich, A.E.; Petroni, G.; Potekhin, A. Cryptic Diversity in Revealed with a Polyphasic Approach. Microorganisms 2022, 10, 974. [Google Scholar] [CrossRef]
- Obert, T.; Vďačný, P. Delimitation of Five Astome Ciliate Species Isolated from the Digestive Tube of Three Ecologically Different Groups of Lumbricid Earthworms, Using the Internal Transcribed Spacer Region and the Hypervariable D1/D2 Region of the 28S rRNA Gene. BMC Evol. Biol. 2020, 20, 37. [Google Scholar] [CrossRef]
- Zhang, T.; Shao, C.; Zhang, T.; Song, W.; Vd’ačný, P.; Al-Farraj, S.A.; Wang, Y. Multi-Gene Phylogeny of the Ciliate Genus Trachelostyla (Ciliophora, Hypotrichia), With Integrative Description of Two Species, Trachelostyla multinucleata Spec. Nov. and T. Pediculiformis (Cohn, 1866). Front. Microbiol. 2022, 12, 775570. [Google Scholar] [CrossRef]
- Shazib, S.U.A.; Vďačný, P.; Kim, J.H.; Jang, S.W.; Shin, M.K. Molecular Phylogeny and Species Delimitation within the Ciliate Genus Spirostomum (Ciliophora, Postciliodesmatophora, Heterotrichea), Using the Internal Transcribed Spacer Region. Mol. Phylogenet. Evol. 2016, 102, 128–144. [Google Scholar] [CrossRef] [PubMed]
- Belart, P.; Habib, R.; Raposo, D.; Clemente, I.; Alves Martins, M.V.; Frontalini, F.; Figueiredo, M.S.L.; Lorini, M.L.; Laut, L. Seasonal Dynamics of Benthic Foraminiferal Biocoenosis in the Tropical Saquarema Lagoonal System (Brazil). Estuaries Coast. 2019, 42, 822–841. [Google Scholar] [CrossRef]
- Carrier, T.J.; Leigh, B.A.; Deaker, D.J.; Devens, H.R.; Wray, G.A.; Bordenstein, S.R.; Byrne, M.; Reitzel, A.M. Microbiome Reduction and Endosymbiont Gain from a Switch in Sea Urchin Life History. Proc. Natl. Acad. Sci. USA 2021, 118, e2022023118. [Google Scholar] [CrossRef]
- Kwan, J.C.; Schmidt, E.W. Bacterial Endosymbiosis in a Chordate Host: Long-Term Co-Evolution and Conservation of Secondary Metabolism. PLoS ONE 2013, 8, e80822. [Google Scholar] [CrossRef] [PubMed]
- Castelli, M.; Nardi, T.; Gammuto, L.; Bellinzona, G.; Sabaneyeva, E.; Potekhin, A.; Serra, V.; Petroni, G.; Sassera, D. Host Association and Intracellularity Evolved Multiple Times Independently in the Rickettsiales. bioRxiv 2022. [Google Scholar] [CrossRef]
- Diepold, A.; Armitage, J.P. Type III Secretion Systems: The Bacterial Flagellum and the Injectisome. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20150020. [Google Scholar] [CrossRef]
- Gillespie, J.J.; Kaur, S.J.; Rahman, M.S.; Rennoll-Bankert, K.; Sears, K.T.; Beier-Sexton, M.; Azad, A.F. Secretome of Obligate Intracellular Rickettsia. FEMS Microbiol. Rev. 2015, 39, 47–80. [Google Scholar]
- Eicher, S.C.; Dehio, C. Bartonella Entry Mechanisms into Mammalian Host Cells. Cell. Microbiol. 2012, 14, 1166–1173. [Google Scholar] [CrossRef]
- Perlman, S.J.; Hunter, M.S.; Zchori-Fein, E. The Emerging Diversity of Rickettsia. Proc. Biol. Sci. 2006, 273, 2097–2106. [Google Scholar] [CrossRef]
- Parola, P.; Paddock, C.D.; Socolovschi, C.; Labruna, M.B.; Mediannikov, O.; Kernif, T.; Abdad, M.Y.; Stenos, J.; Bitam, I.; Fournier, P.-E.; et al. Update on Tick-Borne Rickettsioses around the World: A Geographic Approach. Clin. Microbiol. Rev. 2013, 26, 657–702. [Google Scholar] [CrossRef]

| Organism | 1. | 2. | 3. | 4. | 5. | 6. | 7. | 8. |
|---|---|---|---|---|---|---|---|---|
| 1. “Ca. Trichorickettsia mobilis subsp. mobilis”—Paramecium multimicronucleatum PS23 HG315612 | 100 | 100 | 100 | 97.7 | 97.7 | 97.6 | 98.4 | 98.4 |
| 2. “Ca. Trichorickettsia mobilis subsp. mobilis”—Paramecium multimicronucleatum Pm HG315610 | 100 | 100 | 97.7 | 97.7 | 97.6 | 98.4 | 98.4 | |
| 3. “Ca. Trichorickettsia mobilis subsp. mobilis”—Paramecium multimicronucleatum LSA HG315611 | 100 | 97.7 | 97.7 | 97.6 | 98.4 | 98.4 | ||
| 4. “Ca. Trichorickettsia mobilis subsp. hyperinfectiva”—Paramecium calkinsi CyL 8-33 MF039744 | 100 | 100 | 100 | 97.3 | 97.3 | |||
| 5. “Ca. Trichorickettsia mobilis subsp. hyperinfectiva”—Pseudokeronopsis erythrina PSqRJ01 | 100 | 100 | 97.3 | 97.3 | ||||
| 6. “Ca. Trichorickettsia mobilis subsp. hyperinfectiva”—Paramecium multimicronucleatum US_Bl 16I1 MK598854 | 100 | 97.2 | 97.2 | |||||
| 7. “Ca. Trichorickettsia mobilis subsp. extranuclearis”—Paramecium nephridiatum PAR13 HG315614 | 100 | 100 | ||||||
| 8. “Ca. Trichorickettsia mobilis subsp. extranuclearis”—Euplotes aediculatus In HG315609 | 100 |
| Organism | 1. | 2. | 3. | 4. | 5. | 6. | 7. | 8. |
|---|---|---|---|---|---|---|---|---|
| 1. “Ca. Trichorickettsia mobilis subsp. mobilis”—Paramecium multimicronucleatum PS23 HG315612 | 100 | 100 | 100 | 99.5 | 99.5 | 99.5 | 99.7 | 99.7 |
| 2. “Ca. Trichorickettsia mobilis subsp. mobilis”—Paramecium multimicronucleatum Pm HG315610 | 100 | 100 | 99.5 | 99.5 | 99.5 | 99.7 | 99.7 | |
| 3. “Ca. Trichorickettsia mobilis subsp. mobilis”—Paramecium multimicronucleatum LSA HG315611 | 100 | 99.5 | 99.5 | 99.5 | 99.7 | 99.7 | ||
| 4. “Ca. Trichorickettsia mobilis subsp. hyperinfectiva”—Paramecium calkinsi CyL 8-33 MF039744 | 100 | 100 | 100 | 99.5 | 99.5 | |||
| 5. “Ca. Trichorickettsia mobilis subsp. hyperinfectiva”—Pseudokeronopsis erythrina PSqRJ01 | 100 | 100 | 99.5 | 99.5 | ||||
| 6. “Ca. Trichorickettsia mobilis subsp. hyperinfectiva”—Paramecium multimicronucleatum US_Bl 16I1 MK598854 | 100 | 99.5 | 99.5 | |||||
| 7. “Ca. Trichorickettsia mobilis subsp. extranuclearis”—Paramecium nephridiatum PAR13 HG315614 | 100 | 100 | ||||||
| 8. “Ca. Trichorickettsia mobilis subsp. extranuclearis”—Euplotes aediculatus In HG315609 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castelli, M.; Serra, V.; Gammuto, L.; Senra, M.V.X.; Modeo, L.; Petroni, G. Characterization of a Pseudokeronopsis Strain (Ciliophora, Urostylida) and Its Bacterial Endosymbiont “Candidatus Trichorickettsia” (Alphaproteobacteria, Rickettsiales). Diversity 2022, 14, 1032. https://doi.org/10.3390/d14121032
Castelli M, Serra V, Gammuto L, Senra MVX, Modeo L, Petroni G. Characterization of a Pseudokeronopsis Strain (Ciliophora, Urostylida) and Its Bacterial Endosymbiont “Candidatus Trichorickettsia” (Alphaproteobacteria, Rickettsiales). Diversity. 2022; 14(12):1032. https://doi.org/10.3390/d14121032
Chicago/Turabian StyleCastelli, Michele, Valentina Serra, Leandro Gammuto, Marcus V. X. Senra, Letizia Modeo, and Giulio Petroni. 2022. "Characterization of a Pseudokeronopsis Strain (Ciliophora, Urostylida) and Its Bacterial Endosymbiont “Candidatus Trichorickettsia” (Alphaproteobacteria, Rickettsiales)" Diversity 14, no. 12: 1032. https://doi.org/10.3390/d14121032
APA StyleCastelli, M., Serra, V., Gammuto, L., Senra, M. V. X., Modeo, L., & Petroni, G. (2022). Characterization of a Pseudokeronopsis Strain (Ciliophora, Urostylida) and Its Bacterial Endosymbiont “Candidatus Trichorickettsia” (Alphaproteobacteria, Rickettsiales). Diversity, 14(12), 1032. https://doi.org/10.3390/d14121032






