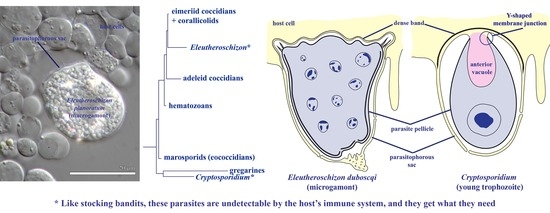
Morphological and Phylogenetic Study of Protococcidians Sheds Light on the Evolution of Epicellular Parasitism in Sporozoa (Apicomplexa), with the Description of Eleutheroschizon planoratum sp. nov
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Polychaete Hosts and Isolation of Parasites
2.2. Light Microscopy
2.3. Electron Microscopy
2.4. Molecular Phylogenetic Analysis
3. Results
3.1. E. duboscqi Brasil, 1906, Emend
3.1.1. Occurrence
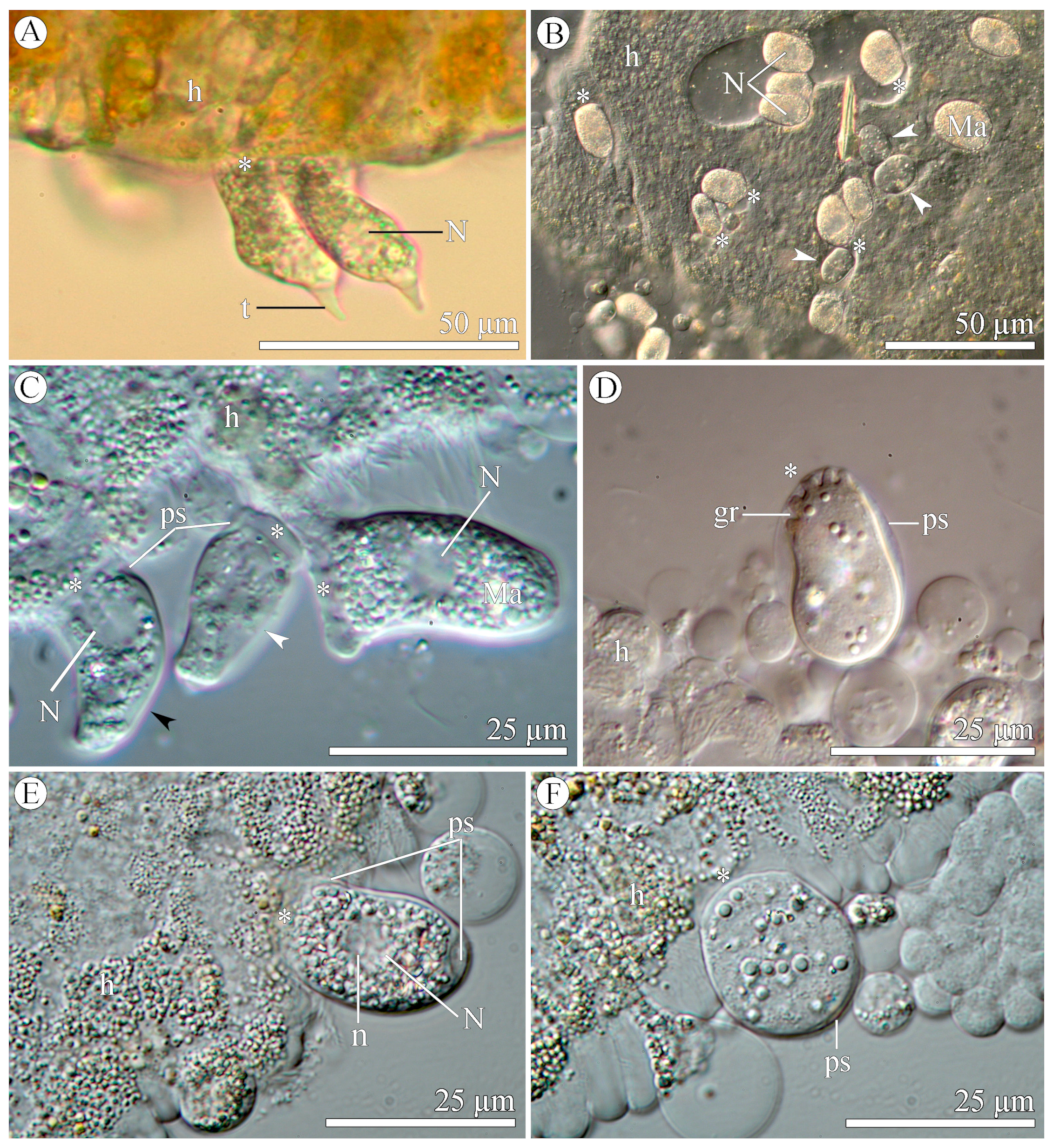
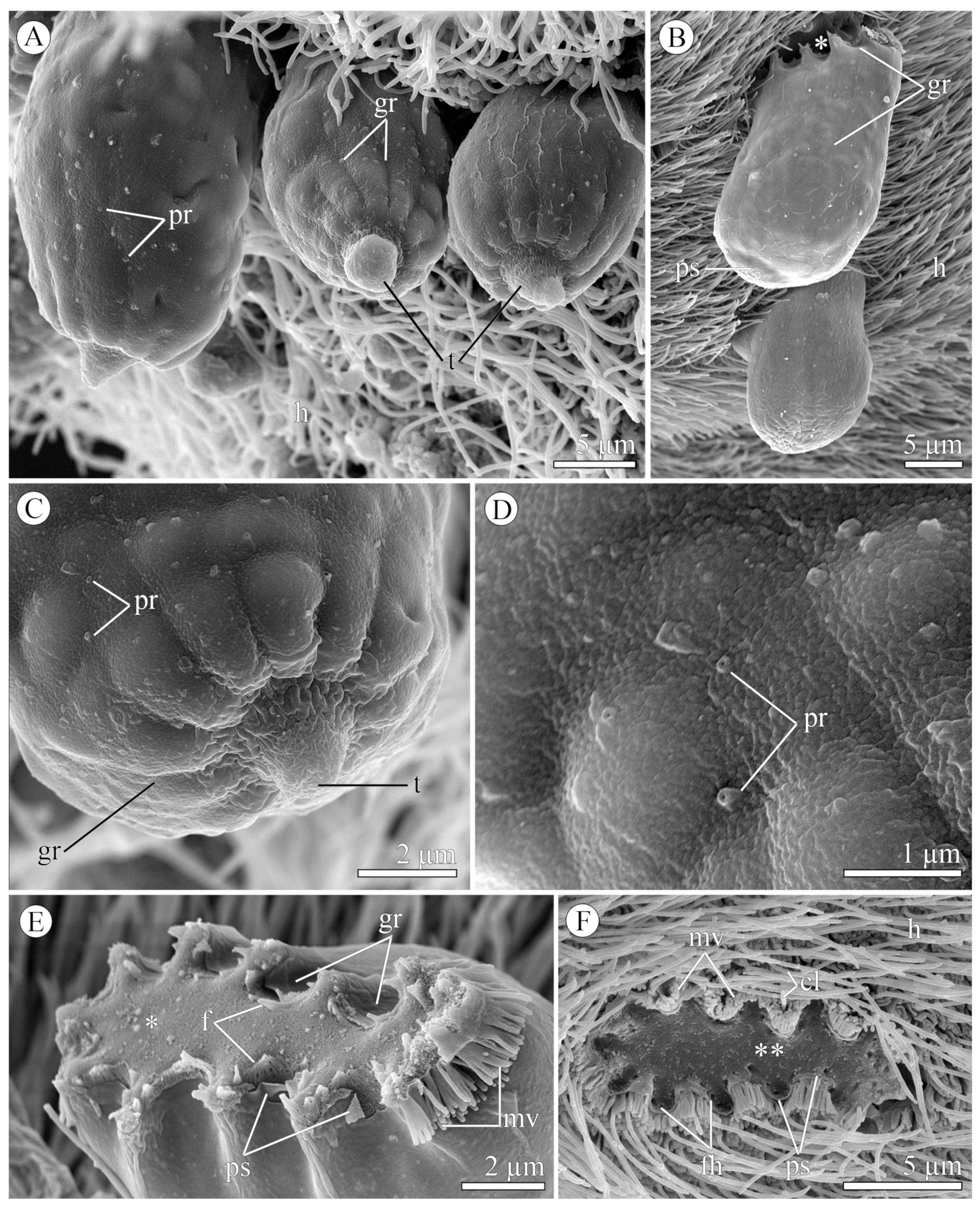
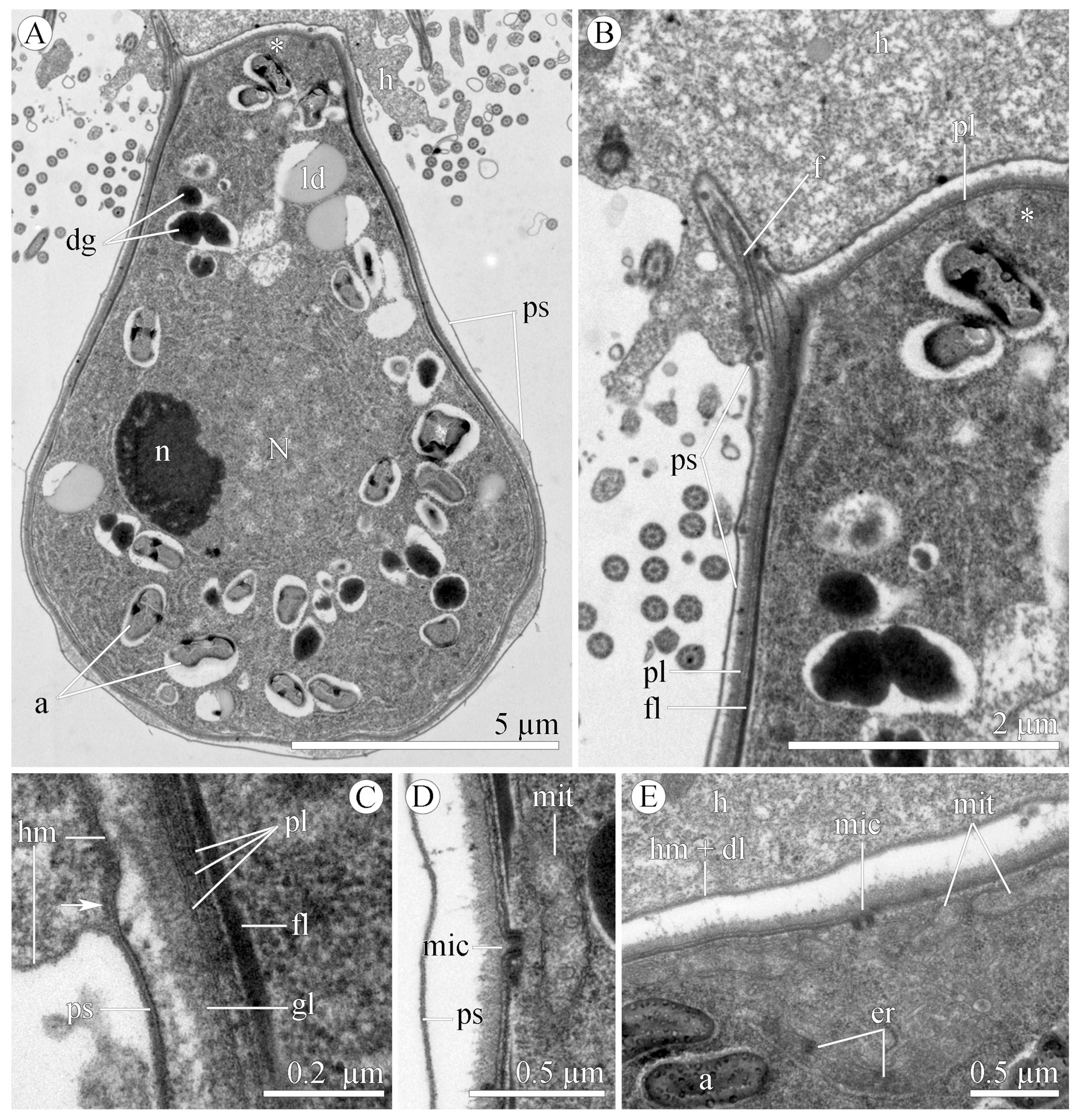
3.1.2. Morphology
3.2. E. planoratum sp. nov.
3.2.1. Occurrence
3.2.2. Morphology
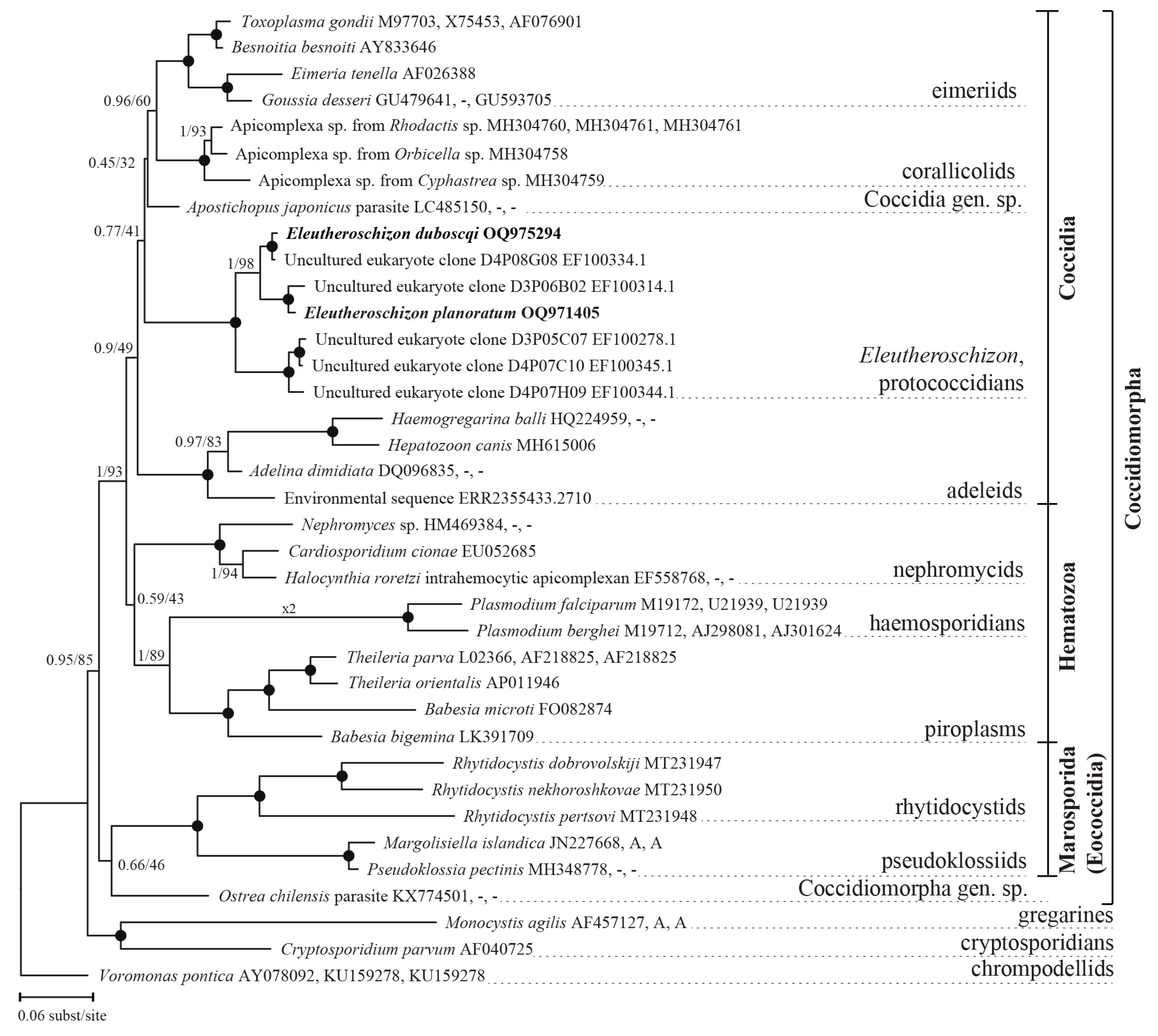
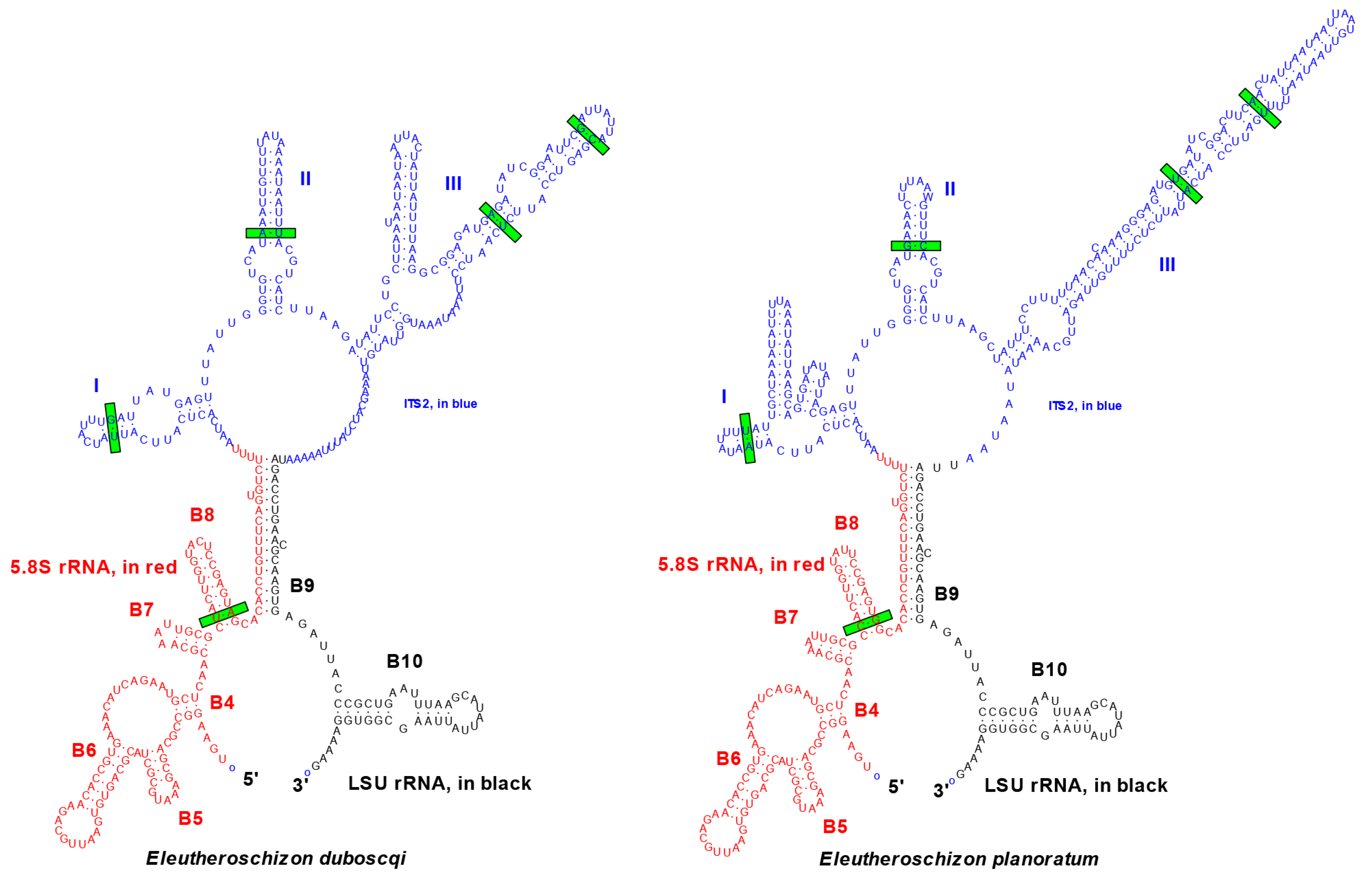
3.2.3. Phylogenetic Position of Eleutheroschizon spp.
4. Discussion
4.1. Justification of Species
4.2. Localization in a Closed Epicellular Niche
4.3. Parasitism in a Closed Epicellular Niche Evolved Convergently
4.4. Taxonomic Summary
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Janouškovec, J.; Paskerova, G.G.; Miroliubova, T.S.; Mikhailov, K.V.; Birley, T.; Aleoshin, V.V.; Simdyanov, T.G. Apicomplexan-like parasites are polyphyletic and widely but selectively dependent on cryptic plastid organelles. eLife 2019, 8, e49662. [Google Scholar] [CrossRef] [PubMed]
- Simdyanov, T.G.; Paskerova, G.G.; Valigurová, A.; Diakin, A.; Kováčiková, M.; Schrével, J.; Guillou, L.; Dobrovolskij, A.A.; Aleoshin, V.V. First ultrastructural and molecular phylogenetic evidence from the blastogregarines, an early branching lineage of plesiomorphic Apicomplexa. Protist 2018, 169, 697–726. [Google Scholar] [CrossRef] [PubMed]
- Kheisin, E.M. About the system of sporozoans (class Sporozoa, phylum Protozoa). Zool. Zhurnal 1956, 35, 1281–1298. (In Russian) [Google Scholar]
- Perkins, F.O.; Barta, J.R.; Clopton, R.E.; Peirce, M.A.; Upton, S.J. Phylum Apicomplexa Levine, 1970. In An Illustrated Guide to the Protozoa, 2nd ed.; Lee, J.J., Leedale, G.F., Bradbury, P., Eds.; Society of Protozoologists: Lawrence, KS, USA, 2000; Volume 11, pp. 190–369. [Google Scholar]
- Mathur, V.; Kwong, W.K.; Husnik, F.; Irwin, N.A.T.; Kristmundsson, A.; Gesta, C.; Freeman, M.; Keeling, P.J. Phylogenomics identifies a new major subgroup of apicomplexans, Marosporida class nov., with extreme apicoplast genome reduction. Genome Biol. Evol. 2020, 13, evaa244. [Google Scholar] [CrossRef]
- Mathur, V.; Wakeman, K.C.; Keeling, P.J. Parallel functional reduction in the mitochondria of apicomplexan parasites. Curr. Biol. 2021, 31, 2920–2928. [Google Scholar] [CrossRef]
- Mathur, V.; Salomaki, E.D.; Wakeman, K.C.; Na, I.; Kwong, W.; Kolisko, M.; Keeling, P.J. Reconstruction of Plastid Proteomes of Apicomplexans and Close Relatives Reveals the Major Evolutionary Outcomes of Cryptic Plastids. Mol. Biol. Evol. 2023, 40, msad002. [Google Scholar] [CrossRef]
- Brasil, L. Eleutheroschizon duboscqi, sporozoaire nouveau parasite de Scoloplos armiger O.F. Müller. Arch. Zool. Exp. Gen. 1906, 4, 17–22. [Google Scholar]
- Chatton, E.; Villeneuve, F. Le cycle évolutif de l’Eleutheroschizon duboscqui Brasil. Prevue expérimentale de l’absence de schizogonie chez la Siedleckia caulleryi Ch et Vill. C.R. Acad. Sci. D Nat. 1936, 203, 834–837. [Google Scholar]
- Valigurová, A.; Paskerova, G.G.; Diakin, A.; Kováčiková, M.; Simdyanov, T.G. Protococcidian Eleutheroschizon duboscqi, an unusual apicomplexan interconnecting gregarines and cryptosporidians. PLoS ONE 2015, 10, e0125063. [Google Scholar] [CrossRef] [Green Version]
- Valigurová, A.; Jirku, M.; Koudela, B.; Gelnar, M.; Modry, D.; Slapeta, J. Cryptosporidia: Epicellular parasites embraced by the host cell membrane. Int. J. Parasitol. 2008, 38, 913–922. [Google Scholar] [CrossRef]
- Kolářová, I.; Valigurová, A. Hide-and-seek: A game played between parasitic protists and their hosts. Microorganisms 2021, 9, 2434. [Google Scholar] [CrossRef]
- Paskerova, G.G.; Miroliubova, T.S.; Diakin, A.; Kováčiková, M.; Valigurová, A.; Guillou, L.; Aleoshin, V.V.; Simdyanov, T.G. Fine structure and molecular phylogenetic position of two marine gregarines, Selenidium pygospionis sp. n. and S. pherusae sp. n., with notes on the phylogeny of Archigregarinida (Apicomplexa). Protist 2018, 169, 826–852. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES science gateway for inference of large phylogenetic trees. In Gateway Computing Environments Workshop (GCE); IEEE: New Orleans, LA, USA, 2010; pp. 1–8. [Google Scholar] [CrossRef] [Green Version]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [Green Version]
- Felsenstein, J. Phylogenies and the Comparative Method. Am. Nat. 1985, 125, 1–15. Available online: http://www.jstor.org/stable/2461605 (accessed on 30 May 2023). [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Coleman, A.W. The significance of a coincidence between evolutionary landmarks found in mating affinity and a DNA sequence. Protist 2000, 151, 1–9. [Google Scholar] [CrossRef]
- Coleman, A.W. Is there a molecular key to the level of “biological species’’ in eukaryotes? A DNA guide. Mol. Phylogenet. Evol. 2009, 50, 197–203. [Google Scholar] [CrossRef]
- Müller, T.; Philippi, N.; Dandekar, T.; Schultz, J.; Wolf, M. Distinguishing species. RNA 2007, 13, 1469–1472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolf, M.; Chen, S.; Song, J.; Ankenbrand, M.; Müller, T. Compensatory base changes in ITS2 secondary structures correlate with the biological species concept despite intragenomic variability in ITS2 sequences—A proof of concept. PLoS ONE 2013, 8, e66726. [Google Scholar] [CrossRef] [PubMed]
- Awerinzew, S. Studien über parasitische Protozoen. Trav. Soc. Imper. Natur. St. Petersbourg 1908, 38, 1–139. (In Russian) [Google Scholar]
- Word Register of Marine Species. Polychaeta. Available online: https://www.marinespecies.org/aphia.php?p=taxdetails&id=883 (accessed on 30 May 2023).
- Miroliubova, T.S.; Simdyanov, T.G.; Mikhailov, K.V.; Aleoshin, V.V.; Janouškovec, J.; Belova, P.A.; Paskerova, G.G. Polyphyletic origin, intracellular invasion, and meiotic genes in the putatively asexual agamococcidians (Apicomplexa incertae sedis). Sci. Rep. 2020, 10, 15847. [Google Scholar] [CrossRef] [PubMed]
- Simdyanov, T.G.; Guillou, L.; Diakin, A.Y.; Mikhailov, K.V.; Schrével, J.; Aleoshin, V.V. A new view on the morphology and phylogeny of eugregarines suggested by the evidence from the gregarine Ancora sagittata (Leuckart, 1860) Labbé, 1899 (Apicomplexa: Eugregarinida). PeerJ 2017, 5, e3354. [Google Scholar] [CrossRef] [Green Version]
- Schultz, J.; Maisel, S.; Gerlach, D.; Müller, T.; Wolf, M. A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA 2005, 11, 361–364. [Google Scholar] [CrossRef] [Green Version]
- Huang, B.Q.; Chen, X.M.; LaRusso, N.F. Cryptosporidium parvum attachment to and internalization by human biliary epithelia in vitro: A morphologic study. J. Parasitol. 2004, 90, 212–221. [Google Scholar] [CrossRef]
- Lumb, R.; Smith, K.; O’Donoghue, P.J.; Lanser, J.A. Ultrastructure of the attachment of Cryptosporidium sporozoites to tissue culture cells. Parasitol. Res. 1988, 74, 531–536. [Google Scholar] [CrossRef]
- Valigurová, A.; Hofmannová, L.; Koudela, B.; Vávra, J. An ultrastructural comparison of the attachment sites between Gregarina steini and Cryptosporidium muris. J. Eukaryot. Microbiol. 2007, 54, 495–510. [Google Scholar] [CrossRef]
- Umemiya, R.; Fukuda, M.; Fujisaki, K.; Matsui, T. Electron microscopic observation of the invasion process of Cryptosporidium parvum in severe combined immunodeficiency mice. J. Parasitol. 2005, 91, 1034–1039. [Google Scholar] [CrossRef]
- Valigurová, A.; Florent, I. Nutrient acquisition and attachment strategies in basal lineages: A tough nut to crack in the evolutionary puzzle of Apicomplexa. Microorganisms 2021, 9, 1430. [Google Scholar] [CrossRef]
- Bonnin, A.; Lapillonne, A.; Petrella, T.; Lopez, J.; Chaponnier, C.; Gabbiani, G.; Robine, S.; Dubremetz, J.F. Immunodetection of the microvillous cytoskeleton molecules villin and ezrin in the parasitophorous vacuole wall of Cryptosporidium parvum (Protozoa: Apicomplexa). Eur. J. Cell Biol. 1999, 78, 794–801. [Google Scholar] [CrossRef]
- Melicherová, J.; Hofmannová, L.; Valigurová, A. Response of cell lines to actual and simulated inoculation with Cryptosporidium proliferans. Eur. J. Protistol. 2018, 62, 101–121. [Google Scholar] [CrossRef]
- Barta, J.R.; Thompson, R.C.A. What is Cryptosporidium? Reappraising its biology and phylogenetic affinities. Trends Parasitol. 2006, 22, 463–468. [Google Scholar] [CrossRef]
- Bartošová-Sojková, P.; Oppenheim, R.D.; Soldati-Favre, D.; Lukeš, J. Epicellular apicomplexans: Parasites “on the way in”. PLoS Pathog. 2015, 11, e1005080. [Google Scholar] [CrossRef] [Green Version]
- Borowski, H.; Clode, P.L.; Thompson, R.C.A. Active invasion and/or encapsulation? A reappraisal of host-cell parasitism by Cryptosporidium. Trends Parasitol. 2008, 24, 509–516. [Google Scholar] [CrossRef]
- Elliott, D.A.; Clark, D.P. Cryptosporidium parvum induces host cell actin accumulation at the host-parasite interface. Infect. Immun. 2000, 68, 2315–2322. [Google Scholar] [CrossRef] [Green Version]
- Thompson, R.C.A.; Olson, M.E.; Zhu, G.; Enomoto, S.; Abrahamsen, M.S.; Hijjawi, N.S. Cryptosporidium and Cryptosporidiosis. Adv. Parasitol. 2005, 59, 77–158. [Google Scholar] [CrossRef]
- Paskerova, G.G.; Miroliubova, T.S.; Valigurová, A.; Janouškovec, J.; Kováčiková, M.; Diakin, A.; Sokolova, Y.Y.; Mikhailov, K.V.; Aleoshin, V.V.; Simdyanov, T.G. Evidence from the resurrected family Polyrhabdinidae Kamm, Apicomplexa: Gregarinomorpha) supports the epimerite, an attachment organelle, as a major eugregarine innovation. PeerJ 2021, 9, e11912. [Google Scholar] [CrossRef]
- Koreny, L.; Mercado-Saavedra, B.N.; Klinger, C.M.; Barylyuk, K.; Butterworth, S.; Hirst, J.; Rivera-Cuevas, Y.; Zaccai, N.R.; Holzer, V.J.C.; Klingl, A.; et al. Stable endocytic structures navigate the complex pellicle of apicomplexan parasites. Nat. Commun. 2023, 14, 2167. [Google Scholar] [CrossRef]
- Piro, F.; Focaia, R.; Dou, Z.; Masci, S.; Smith, D.; Di Cristina, M. An uninvited seat at the dinner table: How apicomplexan parasites scavenge nutrients from the host. Microorganisms 2021, 9, 2592. [Google Scholar] [CrossRef] [PubMed]
- Wan, W.; Dong, H.; Lai, D.H.; Yang, J.; He, K.; Tang, X.; Liu, Q.; Hide, G.; Zhu, X.Q.; Sibley, L.D.; et al. The Toxoplasma micropore mediates endocytosis for selective nutrient salvage from host cell compartments. Nat. Commun. 2023, 14, 977. [Google Scholar] [CrossRef] [PubMed]
- Borowski, H.; Thompson, R.C.A.; Armstrong, T.; Clode, P.L. Morphological characterization of Cryptosporidium parvum life-cycle stages in an in vitro model system. Parasitology 2010, 137, 13–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frénal, K.; Dubremetz, J.F.; Lebrun, M.; Soldati-Favre, D. Gliding motility powers invasion and egress in Apicomplexa. Nat. Rev. Microbiol. 2017, 15, 645–660. [Google Scholar] [CrossRef] [Green Version]
- Stoeck, T.; Kasper, J.; Bunge, J.; Leslin, C.; Ilyin, V.; Epstein, S. Protistan Diversity in the Arctic: A Case of Paleoclimate Shaping Modern Biodiversity? PLoS ONE 2007, 2, e728. [Google Scholar] [CrossRef] [Green Version]
- Carreno, R.A.; Martin, D.S.; Barta, J.R. Cryptosporidium is more closely related to the gregarines than to coccidia as shown by phylogenetic analysis of apicomplexan parasites inferred using small-subunit ribosomal RNA gene sequences. Parasitol. Res. 1999, 85, 899–904. [Google Scholar] [CrossRef]
- Mathur, V.; Kolísko, M.; Hehenberger, E.; Irwin, N.A.T.; Leander, B.S.; Kristmundsson, Á.; Freeman, M.A.; Keeling, P.J. Multiple independent origins of apicomplexan-like parasites. Curr. Biol. 2019, 29, 2936–2941. [Google Scholar] [CrossRef]
- Salomaki, E.D.; Terpis, K.X.; Rueckert, S.; Kotyk, M.; Varadínová, Z.K.; Čepička, I.; Lane, C.E.; Kolisko, M. Gregarine single-cell transcriptomics reveals differential mitochondrial remodeling and adaptation in apicomplexans. BMC Biol. 2021, 19, 77. [Google Scholar] [CrossRef]
| Species/ Characteristics | E. duboscqi Brasil, 1906, Type Species | E. duboscqi; Emended Description | E. murmanicum Awerintzew, 1908 | E. planoratum sp. nov. |
|---|---|---|---|---|
| Host | Scoloplos armiger (Müller, 1776), Protoaricia oerstedii (Claparède, 1864) (former Theostoma oerstedi (Claparède, 1864)) | Scoloplos armiger (Müller, 1776) | Ophelia limacina (Rathke, 1843) | Naineris quadricuspida (Fabricius, 1780) |
| Locality | Luc-sur-Mer, English Channel, East Atlantic (S. armiger); L’Étang de Thau, Mediterranean Sea (P. oerstedii) | Velikaja Salma strait (WSBS) and Bolshoy Goreliy (ERS), Kandalaksha Bay, White Sea | Strait Ekaterininskaya Gavan’, Kola Bay, Barents Sea | Vichennaya Luda Isl, Keret Archipelago, Kandalaksha Bay, White Sea |
| Localization in the host | at the end of the first third of the intestine, often in the ventral groove | throughout the midgut; attached to the intestinal epithelium or free in the intestine cavity | attached to the intestinal epithelium or free in the intestine cavity | throughout the mid- and hindgut; usually attached to the intestinal epithelium |
| Infection: extensity (percentage of infected hosts), intensity (number of parasites per host) | about 5%, abundant | about 11% (16 out of 146) at WSBS and 16% (4 out of 25) at ERS, few tens | not in each host, abundant | about 83% (100 out of 121), several to several dozen cells |
| Cell shape | elongated bell, widest part facing to the host epithelium | helmet or elongated bell, straight or slightly curved, widest part facing to the host intestine | wide low cone, widest part facing to the host intestine | barrel-shaped, straight or slightly curved, flattened anterior end facing to the host intestine |
| Cell size (width x height (av. ± SE, dataset), µm) | up to 30 in height | 6.5–15 × 8.5–27 × (11 ± 1.7 × 15 ± 3.3, n = 18) | up to 50–60 × 25–38 | 6–24 × 7–36 (15 ± 3.2 × 24 ± 4.5, n = 100) |
| Number of grooves (av., (min-max, dataset)) | -- | usually 12 (10–13, n = 21) | -- | usually 12 (9–12, n = 15) |
| Anterior end (attachment base) | wide, with 2 circles of rounded lobes (“denticles” in origin.) | roundish, × 4.4–12.7 (9.7 ± 2.2, n = 11) µm in diameter, convex, with 1–2 circles of rounded lobes (up to about 20 in total) and 1 peripheral ring of fascicles of long filaments alternating with short hook-shaped filaments | wide, convex, with 1 peripheral ring of 12–20 conical lobes (“denticles” in origin.) | oval, 9–19 (14.5 ± 2.6, n = 5) µm in max. diameter, flat, with wavy contour and 1 peripheral ring of fascicles of filaments |
| Posterior end; “tail” (distal part of the parasitophorous sac) | rounded; with 1 “tail”, conical, pointed, sometimes hooked or ended in a very small ball | rounded, sometimes with a depression at the apex; with 1 (rarely 2–3 or absent) “tail” (conical, pointed, sometimes hooked or ended in a very small ball) | rounded; without “tail” | rounded, sometimes with a depression at the apex; with or without 1 “tail” (conical, pointed, sometimes hooked) |
| Nucleus: number, shape, size (width x length (av. ± SE, dataset), µm), position in parasite cell | -- | 1 large, roundish, 3.9–8.1 × 3.0–9.0 (5.8 ± 1.3 × 5.7 ± 1.3, n = 10), located in the widest cell part (macrogamonts); several small, spherical, 1.2–1.7 × 1.2–1.6 (×1.4 ± 0.1, n = 20), evenly distributed throughout the cytoplasm (microgamonts) | 1 large, oval, centrally located (macrogamonts); multiple small nuclei at the cell periphery and 1 irregular oval nuclear residue (microgamonts) | 1 large, spherical, 4–11 × 4–11 (×7 ± 1.2, n = 57), located in the widest cell part (macrogamonts); several small, evenly distributed throughout the cytoplasm (microgamonts) |
| Nucleolus: quantity, shape, size (widthwise x lengthwise, av. ± SE, dataset), µm), position in nucleus | -- | 1 large, spherical, 1.5–3.0 × 1.4–2.8 (2.4 ± 0.5 × 2.3 ± 0.6, n = 5), eccentrically located (macrogamonts); several fragmented (microgamonts) | 1 large, spherical, centrically located (macrogamonts) | 1 large, oval, 2.8–4.4 × 1.6–3.5 (3.5 ± 0.5 × 2.4 ± 0.4, n = 10), eccentrically located (macrogamonts) |
| Merogony | absent | absent | absent | absent |
| Sporozoites (width × length, µm) | 2 × 5, pointed; the nucleus located in the apical end | -- | similar to E. duboscqi | -- |
| Oocyst | several fan-shaped clusters of sporozoites connected at one end to a residual body | -- | -- | -- |
| Motility | immotile | immotile | -- | immotile |
| Characteristic features | -- | formation of a parasitophorous sac from fused outgrowths of the infected enterocyte | paired association of one- or multinuclear (2–3 nuclei) cells by expanded parts, lobes absent (syzygy?) | formation of a parasitophorous sac from fused outgrowths of the infected enterocyte |
| rDNA sequences | -- | SSU, 5.8S, LSU rDNA, ITS1, ITS2 | -- | SSU, 5.8S, LSU rDNA, ITS1, ITS2 |
| References | [8,9] | [10], this study | [25] | this study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paskerova, G.G.; Miroliubova, T.S.; Valigurová, A.; Aleoshin, V.V.; Simdyanov, T.G. Morphological and Phylogenetic Study of Protococcidians Sheds Light on the Evolution of Epicellular Parasitism in Sporozoa (Apicomplexa), with the Description of Eleutheroschizon planoratum sp. nov. Diversity 2023, 15, 863. https://doi.org/10.3390/d15070863
Paskerova GG, Miroliubova TS, Valigurová A, Aleoshin VV, Simdyanov TG. Morphological and Phylogenetic Study of Protococcidians Sheds Light on the Evolution of Epicellular Parasitism in Sporozoa (Apicomplexa), with the Description of Eleutheroschizon planoratum sp. nov. Diversity. 2023; 15(7):863. https://doi.org/10.3390/d15070863
Chicago/Turabian StylePaskerova, Gita G., Tatiana S. Miroliubova, Andrea Valigurová, Vladimir V. Aleoshin, and Timur G. Simdyanov. 2023. "Morphological and Phylogenetic Study of Protococcidians Sheds Light on the Evolution of Epicellular Parasitism in Sporozoa (Apicomplexa), with the Description of Eleutheroschizon planoratum sp. nov" Diversity 15, no. 7: 863. https://doi.org/10.3390/d15070863
APA StylePaskerova, G. G., Miroliubova, T. S., Valigurová, A., Aleoshin, V. V., & Simdyanov, T. G. (2023). Morphological and Phylogenetic Study of Protococcidians Sheds Light on the Evolution of Epicellular Parasitism in Sporozoa (Apicomplexa), with the Description of Eleutheroschizon planoratum sp. nov. Diversity, 15(7), 863. https://doi.org/10.3390/d15070863