Microglena antarctica sp. nov. a New Antarctic Green Alga from Inexpressible Island (Terra Nova Bay, Ross Sea) Revealed through an Integrative Approach
Abstract
:1. Introduction
2. Materials and Methods
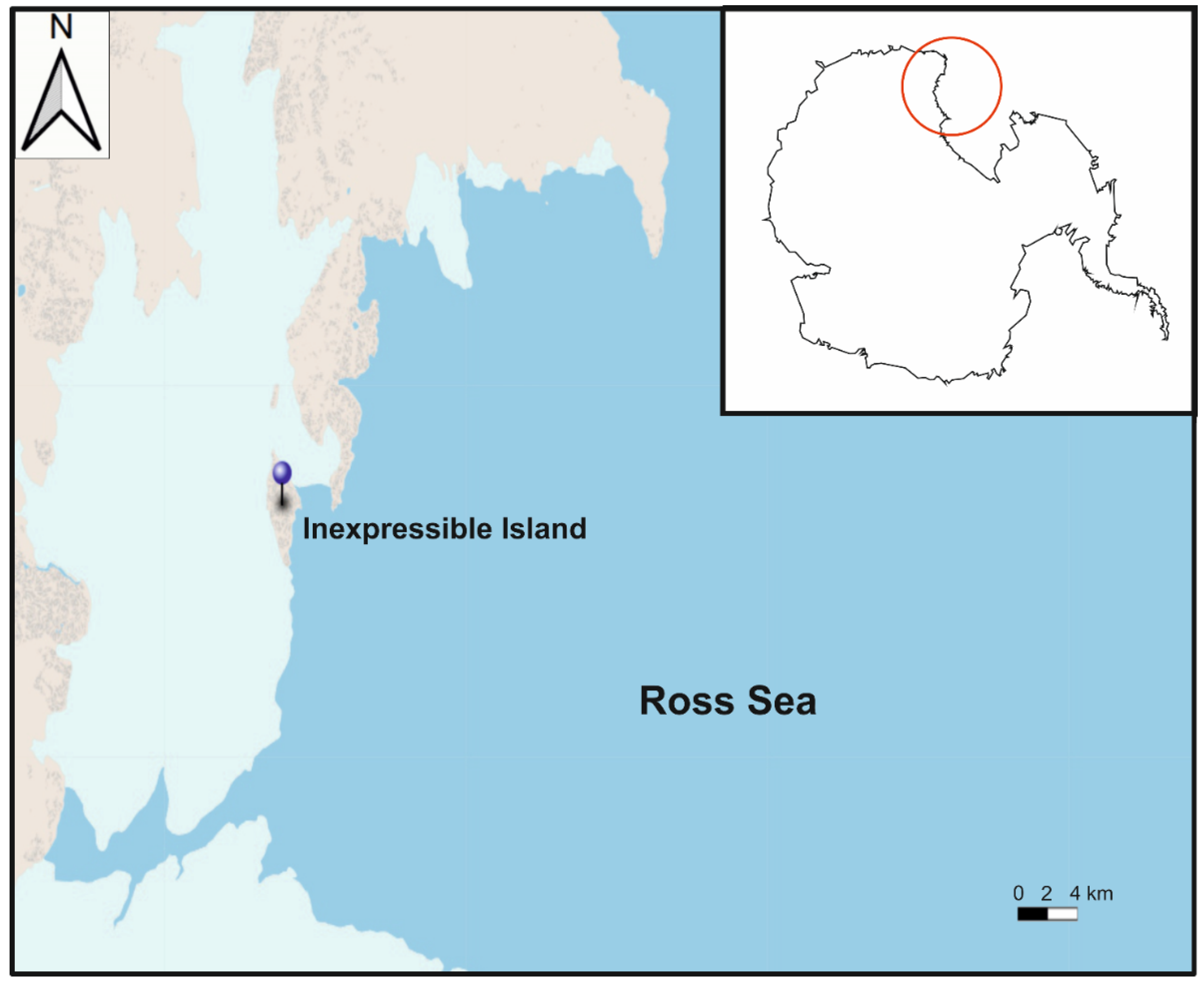
2.1. Isolation and Cultures
2.2. Molecular Analysis
2.2.1. DNA Extraction and Amplification of Selected Molecular Markers
2.2.2. Phylogenetic Analyses and Species Delimitation Methods
2.2.3. DNA Barcoding, CBCs/p-Distances
2.3. Morphological Analysis
3. Results
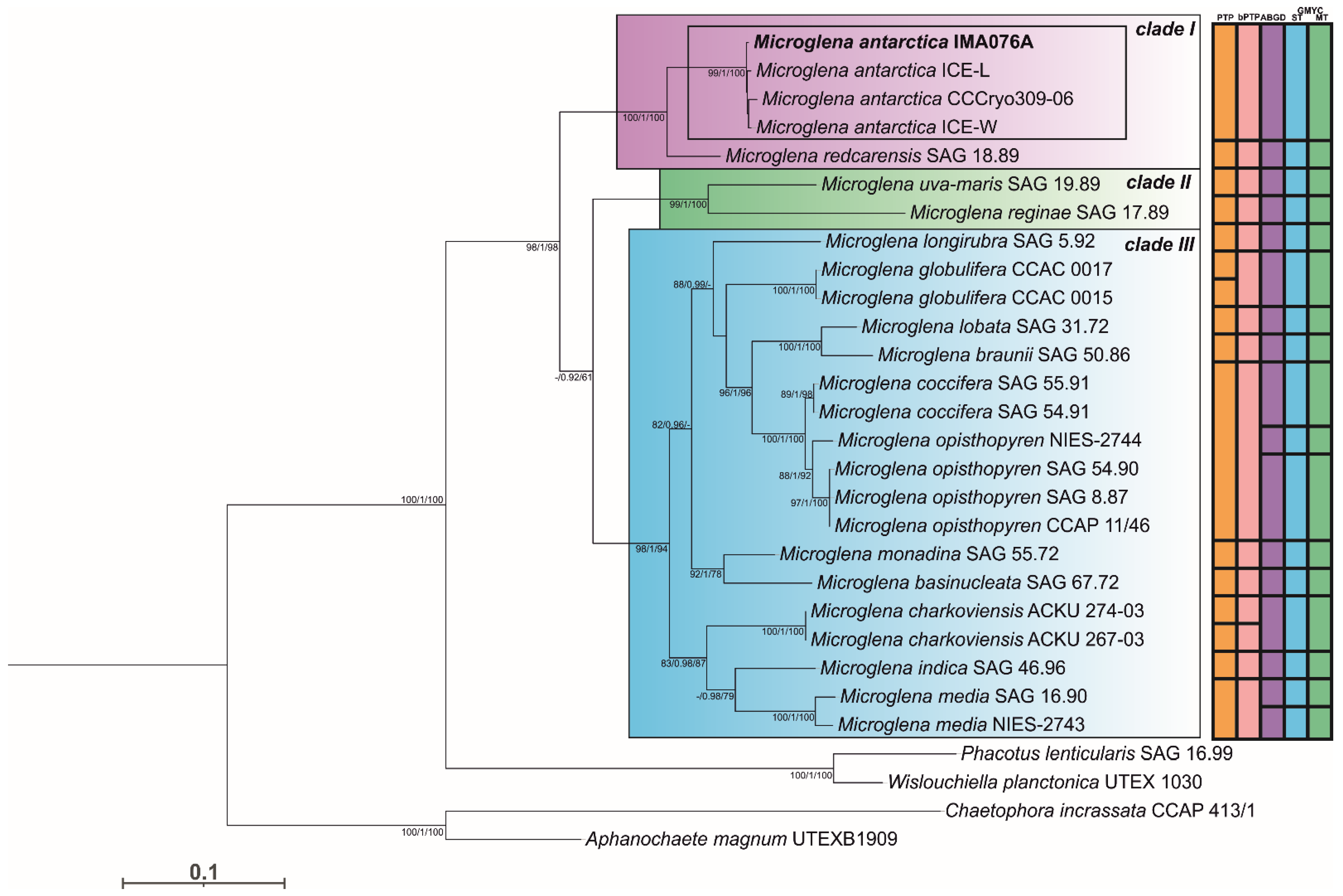
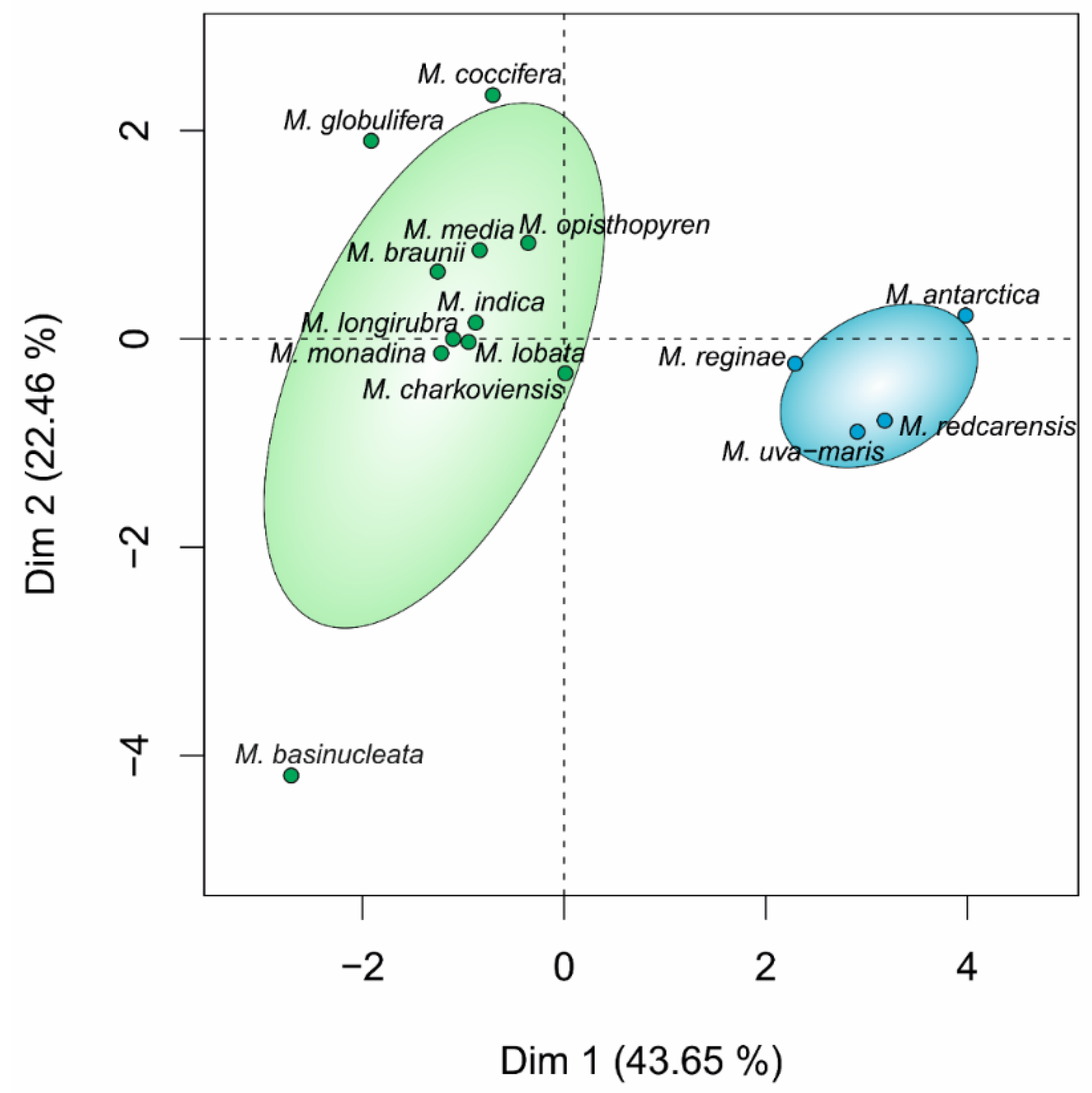
3.1. Phylogeny and Species Delimitation Methods
3.2. Hypothesis Testing
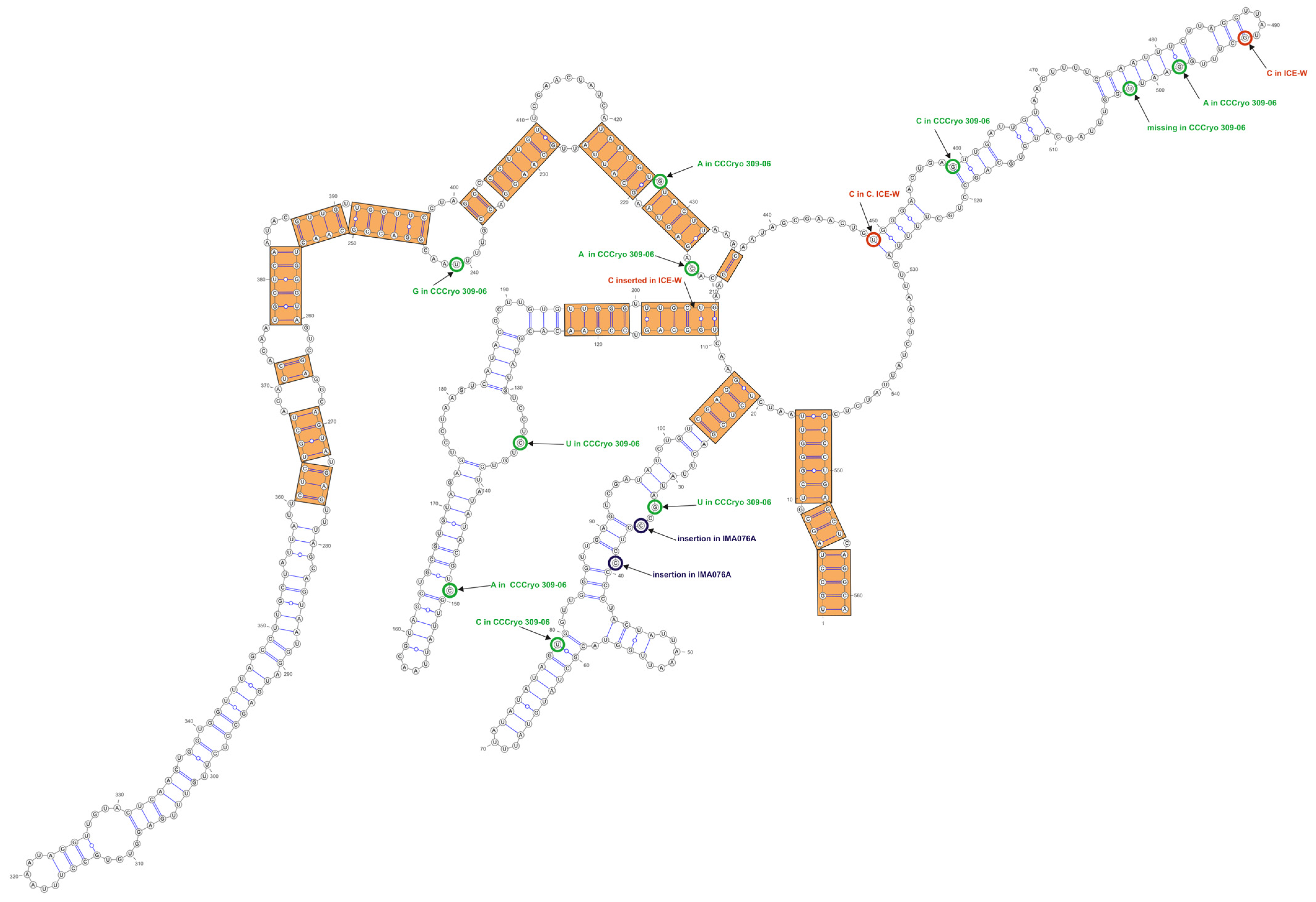
3.3. ITS-2 Secondary Structure, DNA Barcoding, CBCs/p-Distances
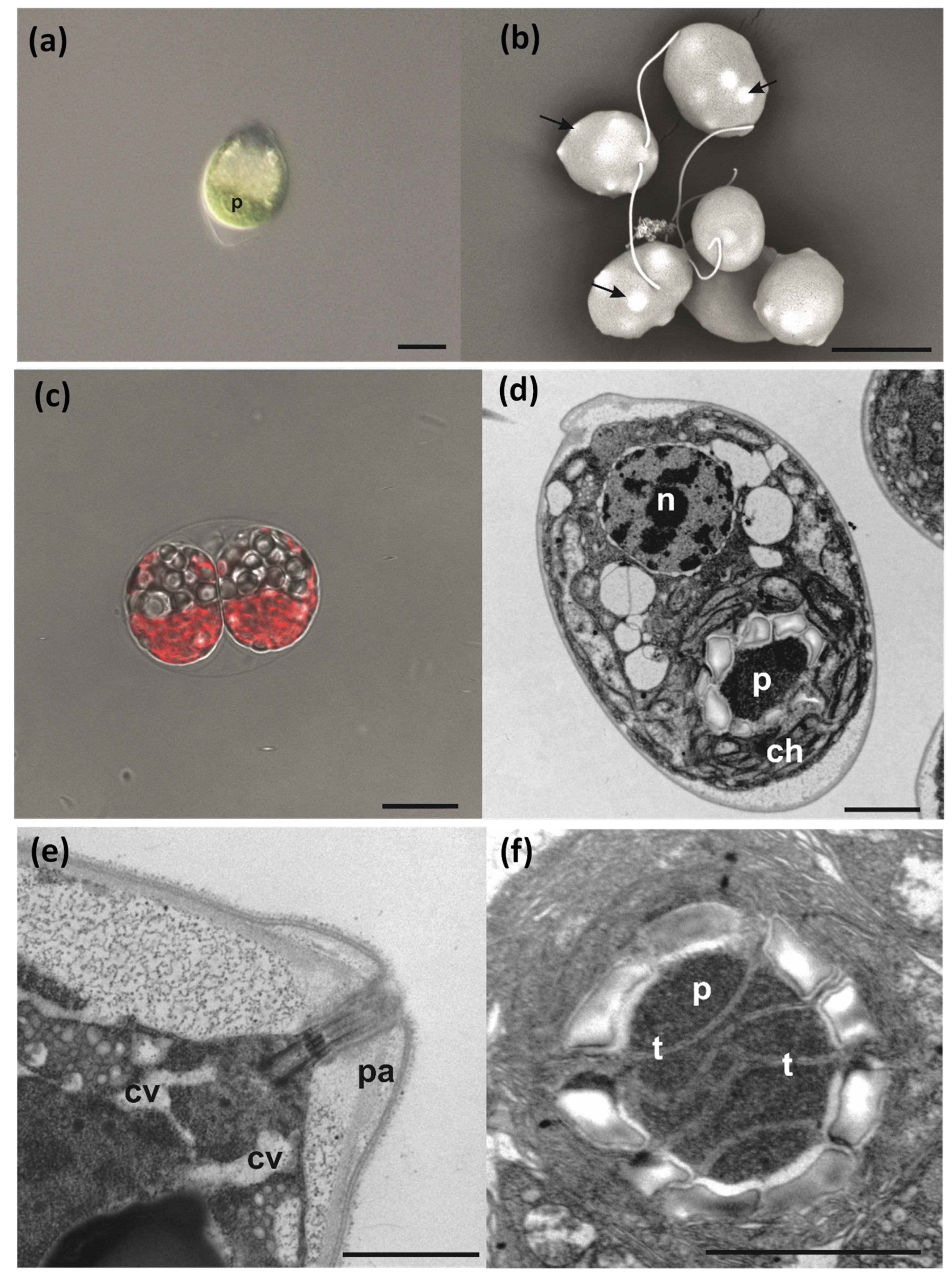
3.4. Cell Morphology and Ultrastructure
3.5. Taxonomy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wynn-Williams, D.D. Antarctic microbial diversity: The basis of polar ecosystem processes. Biodivers. Conserv. 1996, 5, 1271–1293. [Google Scholar] [CrossRef]
- Gálvez, F.E.; Saldarriaga-Córdoba, M.; Huovinen, P.; Silva, A.X.; Gómez, I. Revealing the Characteristics of the Antarctic Snow Alga Chlorominima collina gen. et sp. nov. Through Taxonomy, Physiology, and Transcriptomics. Front. Plant Sci. 2021, 12, 1050. [Google Scholar] [CrossRef]
- Kol, E.; Flint, E.A. Algae in green ice from the Balleny Islands, Antarctica. N. Zeal. J. Bot. 1968, 6, 249–261. [Google Scholar] [CrossRef]
- Demchenko, E.; Mikhailyuk, T.; Coleman, A.W.; Pröschold, T. Generic and species concepts in Microglena (previously the Chlamydomonas monadina group) revised using an integrative approach. Eur. J. Phycol. 2012, 47, 264–290. [Google Scholar] [CrossRef]
- Hoham, R.W.; Remias, D. Snow and Glacial Algae: A Review1. J. Phycol. 2020, 56, 264–282. [Google Scholar] [CrossRef] [Green Version]
- Ehrenberg, C.G. Über die Entwickelung und Lebensdauer der Infusionsthiere; nebst ferneren Beiträgen zu einer Vergleichung ihrer organischen Systeme. Abh. Der Königlichen Akad. Wiss. Zu Berl. 1832, 288, 1–154, pls I.-IV. [Google Scholar]
- Ehrenberg, C.G. Dritter Beitrag zur Erkenntniss grosser Organisation in der Richtung des kleinsten Raumes. Abh. Der Königlichen Akad. Der Wiss. Zu Berl. 1834, 1833, 145–336, pls I-XIII. [Google Scholar]
- Buchheim, M.A.; Turmel, M.; Zimmer, E.A.; Chapman, R.L. Phylogeny of Chlamydomonas (Chlorophyta) Based on Cladistic Analysis of Nuclear 18s Rrna Sequence Data1. J. Phycol. 1990, 26, 689–699. [Google Scholar] [CrossRef]
- Buchheim, M.A.; Lemieux, C.; Otis, C.; Gutell, R.R.; Chapman, R.L.; Turmel, M. Phylogeny of the Chlamydomonadales (Chlorophyceae): A Comparison of Ribosomal RNA Gene Sequences from the Nucleus and the Chloroplast. Mol. Phylogenet. Evol. 1996, 5, 391–402. [Google Scholar] [CrossRef]
- Hepperle, D.; Nozaki, H.; Hohenberger, S.; Huss, V.A.R.; Morita, E.; Krienitz, L. Phylogenetic Position of the Phacotaceae Within the Chlamydophyceae as Revealed by Analysis of 18S rDNA and rbcL Sequences. J. Mol. Evol. 1998, 47, 420–430. [Google Scholar] [CrossRef]
- Nakada, T.; Takahashi, S.; Tomita, M. Microglena redcarensis sp. nov. (Volvocales, Chlorophyceae), a brackish water chlamydomonad with contractile vacuoles. Phycol. Res. 2018, 66, 310–317. [Google Scholar] [CrossRef]
- Nakada, T.; Tomita, M. Light Microscopy and Phylogenetic Analyses of Chlamydomonas Species (Volvocales, Chlorophyceae). II. Molecular Phylogeny, Secondary Structure of ITS-2, Cell Morphology, and Nomenclature of Microglena opisthopyren, and M. media, comb. nov. Acta Phytotaxon. Geobot. 2014, 65, 67–73. [Google Scholar] [CrossRef]
- Coleman, A.W. The Significance of a Coincidence between Evolutionary Landmarks Found in Mating Affinity and a DNA Sequence. Protist 2000, 151, 1–9. [Google Scholar] [CrossRef]
- Darienko, T.; Gustavs, L.; Eggert, A.; Wolf, W.; Pröschold, T. Evaluating the Species Boundaries of Green Microalgae (Coccomyxa, Trebouxiophyceae, Chlorophyta) Using Integrative Taxonomy and DNA Barcoding with Further Implications for the Species Identification in Environmental Samples. PLoS ONE 2015, 10, e0127838. [Google Scholar] [CrossRef]
- Guillard, R.R.L. Culture of Phytoplankton for Feeding Marine Invertebrates. Cult. Mar. Invertebr. Anim. 1975, 29–60. [Google Scholar] [CrossRef]
- Hall, J.D.; Fučíková, K.; Lo, C.; Lewis, L.A.; Karol, K.G. An assessment of proposed DNA barcodes in freshwater green algae. Cryptogam. Algol. 2010, 31, 529–555. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guid. Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Dawson, S.C.; Pace, N.R. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc. Natl. Acad. Sci. USA 2002, 99, 8324–8329. [Google Scholar] [CrossRef] [Green Version]
- Edgcomb, V.P.; Kysela, D.T.; Teske, A.; Gomez, A.; de, V.; Sogin, M.L.; Edgcomb, V.P.; Kysela, D.T.; Teske, A.; Gomez, A.d.V.; et al. Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. PNAS 2002, 99, 7658–7662. [Google Scholar] [CrossRef] [Green Version]
- Stoeck, T.; Schwarz, M.V.J.; Boenigk, J.; Schweikert, M.; von der Heyden, S.; Behnke, A. Cellular identity of an 18S rRNA gene sequence clade within the class Kinetoplastea: The novel genus Actuariola gen. nov. (Neobodonida) with description of the type species Actuariola framvarensis sp. nov. Int. J. Syst. Evol. Microbiol. 2005, 55, 2623–2635. [Google Scholar] [CrossRef]
- Giovannoni, S.J.; DeLong, E.F.; Olsen, G.J.; Pace, N.R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J. Bacteriol. 1988, 170, 720–726. [Google Scholar] [CrossRef] [Green Version]
- Medlin, L.; Elwood, H.J.; Stickel, S.; Sogin, M.L. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 1988, 71, 491–499. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Markham, N.R.; Zuker, M. UNAFold. Methods Mol. Biol. 2008, 453, 3–31. [Google Scholar] [CrossRef]
- Seibel, P.N.; Müller, T.; Dandekar, T.; Schultz, J.; Wolf, M. 4SALE–A tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinform. 2006, 7, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [Green Version]
- Kimura, M. Journal of Molecular Evolution A Simple Method for Estimating Evolutionary Rates of Base Substitutions Through Comparative Studies of Nucleotide Sequences. J. Mol. Evol 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. Estimation of evolutionary distances between homologous nucleotide sequences. Proc. Natl. Acad. Sci. USA 1981, 78, 454–458. [Google Scholar] [CrossRef] [Green Version]
- Anisimova, M.; Gascuel, O. Approximate Likelihood-Ratio Test for Branches: A Fast, Accurate, and Powerful Alternative. Syst. Biol. 2006, 55, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Kishino, H.; Miyata, T.; Hasegawa, M. Maximum likelihood inference of protein phylogeny and the origin of chloroplasts. J. Mol. Evol. 1990, 31, 151–160. [Google Scholar] [CrossRef]
- Kishino, H.; Hasegawa, M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J. Mol. Evol. 1989, 29, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Shimodaira, H.; Hasegawa, M. Multiple Comparisons of Log-Likelihoods with Applications to Phylogenetic Inference. Mol. Biol. Evol. 1999, 16, 1114–1116. [Google Scholar] [CrossRef] [Green Version]
- Strimmer, K.; Rambaut, A. Inferring confidence sets of possibly misspecified gene trees. Proc. R. Soc. London. Ser. B Biol. Sci. 2002, 269, 137–142. [Google Scholar] [CrossRef]
- Shimodaira, H. An Approximately Unbiased Test of Phylogenetic Tree Selection. Syst. Biol. 2002, 51, 492–508. [Google Scholar] [CrossRef] [Green Version]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujisawa, T.; Barraclough, T.G. Delimiting Species Using Single-Locus Data and the Generalized Mixed Yule Coalescent Approach: A Revised Method and Evaluation on Simulated Data Sets. Syst. Biol. 2013, 62, 707–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pons, J.; Barraclough, T.G.; Gomez-Zurita, J.; Cardoso, A.; Duran, D.P.; Hazell, S.; Kamoun, S.; Sumlin, W.D.; Vogler, A.P. Sequence-Based Species Delimitation for the DNA Taxonomy of Undescribed Insects. Syst. Biol. 2006, 55, 595–609. [Google Scholar] [CrossRef] [Green Version]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.-H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [Green Version]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siebert, S.; Backofen, R. MARNA: Multiple alignment and consensus structure prediction of RNAs based on sequence structure comparisons. Bioinformatics 2005, 21, 3352–3359. [Google Scholar] [CrossRef] [Green Version]
- Coleman, A.W. Is there a molecular key to the level of “biological species” in eukaryotes? A DNA guide. Mol. Phylogenet. Evol. 2009, 50, 197–203. [Google Scholar] [CrossRef]
- Wolf, M.; Friedrich, J.; Dandekar, T.; Müller, T. CBCAnalyzer: Inferring Phylogenies Based on Compensatory Base Changes in RNA Secondary Structures. Silico Biol. 2005, 5, 291–294. [Google Scholar]
- Swofford, D.L. PAUP *. Phylogenetic Analysis Using Parsimony (* and Other Methods); Version 4.0b10; Sinauer: Sunderland, MA, USA, 2002. [Google Scholar]
- The R Foundation R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 10 April 2022).
- Lemieux, C.; Vincent, A.T.; Labarre, A.; Otis, C.; Turmel, M. Chloroplast phylogenomic analysis of chlorophyte green algae identifies a novel lineage sister to the Sphaeropleales (Chlorophyceae) Phylogenetics and phylogeography. BMC Evol. Biol. 2015, 15, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Ettl, H. Chlorophyta I, Phytomonadina. In Süβwasserflora von Mitteleuropa; Gustaw Fischer Verlag: Jena, Germany, 1983; Volume 9. [Google Scholar]
- Nakada, T.; Misawa, K.; Nozaki, H. Molecular systematics of Volvocales (Chlorophyceae, Chlorophyta) based on exhaustive 18S rRNA phylogenetic analyses. Mol. Phylogenet. Evol. 2008, 48, 281–291. [Google Scholar] [CrossRef]
- Sugasawa, M.; Matsuzaki, R.; Arakaki, Y.; Nozaki, H. Morphology and phylogenetic position of a rare four-celled green alga, Pascherina tetras (Volvocales, Chlorophyceae), based on cultured material. Phycologia 2015, 54, 342–348. [Google Scholar] [CrossRef]
- An, M.; Mou, S.; Zhang, X.; Zheng, Z.; Ye, N.; Wang, D.; Zhang, W.; Miao, J. Expression of fatty acid desaturase genes and fatty acid accumulation in Chlamydomonas sp. ICE-L under salt stress. Bioresour. Technol. 2013, 149, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, J.; Jahn, R.; Gemeinholzer, B. Barcoding diatoms: Evaluation of the V4 subregion on the 18S rRNA gene, including new primers and protocols. Org. Divers. Evol. 2011, 11, 173–192. [Google Scholar] [CrossRef]

| Tree | logl | deltal | bp-RELL | p-KH | p-SH | p-WKH | p-WSH | c-ELW | p-AU |
|---|---|---|---|---|---|---|---|---|---|
| BT | −9255.53 | 0 | 0.625+ | 0.645+ | 1+ | 0.645+ | 0.881+ | 0.62+ | 0.648+ |
| Clade I+II | −9257.77 | 2.2402 | 0.349+ | 0.354+ | 0.803+ | 0.354+ | 0.782+ | 0.348+ | 0.483+ |
| Clade I+III | −9320.01 | 64.485 | 0− | 0.0001− | 0.0002− | 0.0001− | 0.0001− | 0.0002 | 0.0398− |
| Clade II+R | −9266.84 | 11.313 | 0.0263− | 0.12+ | 0.321+ | 0.0578+ | 0.108+ | 0.0326− | 0.0355− |
| Species - Strains | Barcode | 5.8S-LSU | Helix I | Helix II | Helix III |
|---|---|---|---|---|---|
| M. antarctica IMA076/CCCryo 309-06/ICE-L/ICE-W | A | 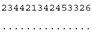 |  |  |  |
| M. redcarensis SAG 18. 89 | B | 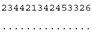 |  |  |  |
| M. reginae SAG 17.89 | C | 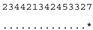 |  |  | 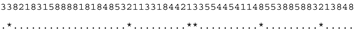 |
| M. uva-maris SAG 19.89 | D | 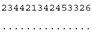 |  |  |  |
| M. lobata SAG 31.72 | E | 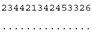 |  |  |  |
| M. braunii SAG 50.86 | F | 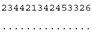 | 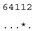 | 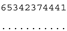 |  |
| M. globulifera CCAC 0015/CCAC 0017 | G | 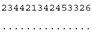 |  |  |  |
| M. longirubra SAG 5.92 | H | 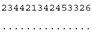 |  |  |  |
| M. coccifera SAG 54.91/SAG 55.91 | I | 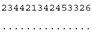 |  |  |  |
| M. opisthopyren SAG 8.87/SAG 54.90/CCAP 11/46 ★ | J1 | 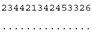 |  |  |  |
| M. opisthopyren NIES-2744 ★ | J2 | 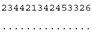 |  |  |  |
| M. basinucleata SAG 67.72 | K |  |  | 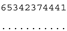 |  |
| M. monadina SAG 55.72 | L | 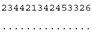 |  |  |  |
| M. charkoviensis ACKU267-03/ACKU274-03 | M | 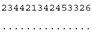 |  |  |  |
| M. indica SAG 46.96 | N | 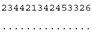 |  |  | 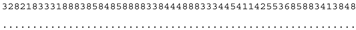 |
| M. media SAG 16.90 ● | O1 | 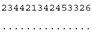 | 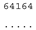 |  |  |
| M. media NIES-2743 ● | O2 | 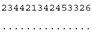 | 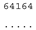 |  |  |
| Species | Chloroplast Morphotype | Pyrenoid-Shape | Pyrenoid Fragmentation | Pyrenoid-Starch Granules | Basal Thickening of the Chloroplast of Mature Cells | Nucleus-Position | Cell Shape | Ecology |
|---|---|---|---|---|---|---|---|---|
| M. uva-maris | (I) | ellipsoid | absent | several and large | present | central | ellipsoid to widely ellipsoidal | marine |
| M. redcarensis | (I) | ellipsoid | absent | several and large | present | central | ellipsoid to spherical | marine |
| M. reginae | (I) | ellipsoid | absent | many and small | present | central | ellipsoid to oval to spherical | marine |
| M. antarctica | (I) | ellipsoid to spherical | absent | several and large | present | central-anterior | ellipsoid to oval | marine |
| M. globulifera | (III) | halfring | present | many and small | absent | central | ellipsoidal to spherical | freshwater |
| M. braunii | (II) | halfring | sometimes fragmented | many and small | absent | central | ellipsoidal to widely ellipsoidal to spherical | freshwater |
| M. lobata | (II) | halfring | absent | many and small | absent | central | ellipsoid to widely ellipsoidal | freshwater |
| M. longirubra | (II) | halfring | absent | many and small | absent | central | ellipsoid to widely ellipsoidal | freshwater |
| M. charkoviensis | (I) | ellipsoid | absent | many and small | absent | central | ellipsoid to widely ellipsoidal | freshwater |
| M. monadina | (II) | halfring | absent | many and small | absent | central | ellipsoid to widely ellipsoid | freshwater |
| M. basinucleata | (II) | halfring | absent | no starch | absent | basal | ellipsoid to widely ellipsoid | freshwater |
| M. coccifera | (III) | ellipsoid to spherical | present | many and small | present | central | widely ellipsoid | freshwater |
| M. indica | (II) | halfring | absent | many and small | absent | central | widely ellipsoid to spherical | freshwater |
| M. opisthopyren | (II) | halfring to ellipsoid | sometimes fragmented | many and small | present | central | ellipsoid to widely ellipsoid | freshwater |
| M media | (II) | ellipsoid | sometimes fragmented | many and small | absent | central | ellipsoid to widely ellipsoid | freshwater |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trentin, R.; Negrisolo, E.; Moschin, E.; Veronese, D.; Cecchetto, M.; Moro, I. Microglena antarctica sp. nov. a New Antarctic Green Alga from Inexpressible Island (Terra Nova Bay, Ross Sea) Revealed through an Integrative Approach. Diversity 2022, 14, 337. https://doi.org/10.3390/d14050337
Trentin R, Negrisolo E, Moschin E, Veronese D, Cecchetto M, Moro I. Microglena antarctica sp. nov. a New Antarctic Green Alga from Inexpressible Island (Terra Nova Bay, Ross Sea) Revealed through an Integrative Approach. Diversity. 2022; 14(5):337. https://doi.org/10.3390/d14050337
Chicago/Turabian StyleTrentin, Riccardo, Enrico Negrisolo, Emanuela Moschin, Davide Veronese, Matteo Cecchetto, and Isabella Moro. 2022. "Microglena antarctica sp. nov. a New Antarctic Green Alga from Inexpressible Island (Terra Nova Bay, Ross Sea) Revealed through an Integrative Approach" Diversity 14, no. 5: 337. https://doi.org/10.3390/d14050337
APA StyleTrentin, R., Negrisolo, E., Moschin, E., Veronese, D., Cecchetto, M., & Moro, I. (2022). Microglena antarctica sp. nov. a New Antarctic Green Alga from Inexpressible Island (Terra Nova Bay, Ross Sea) Revealed through an Integrative Approach. Diversity, 14(5), 337. https://doi.org/10.3390/d14050337






