Distribution and Molecular Diversity of Paranoplocephala kalelai (Tenora, Haukisalmi & Henttonen, 1985) Tenora, Murai & Vaucher, 1986 in Voles (Rodentia: Myodes) in Eurasia
Abstract
1. Introduction
2. Materials and Methods
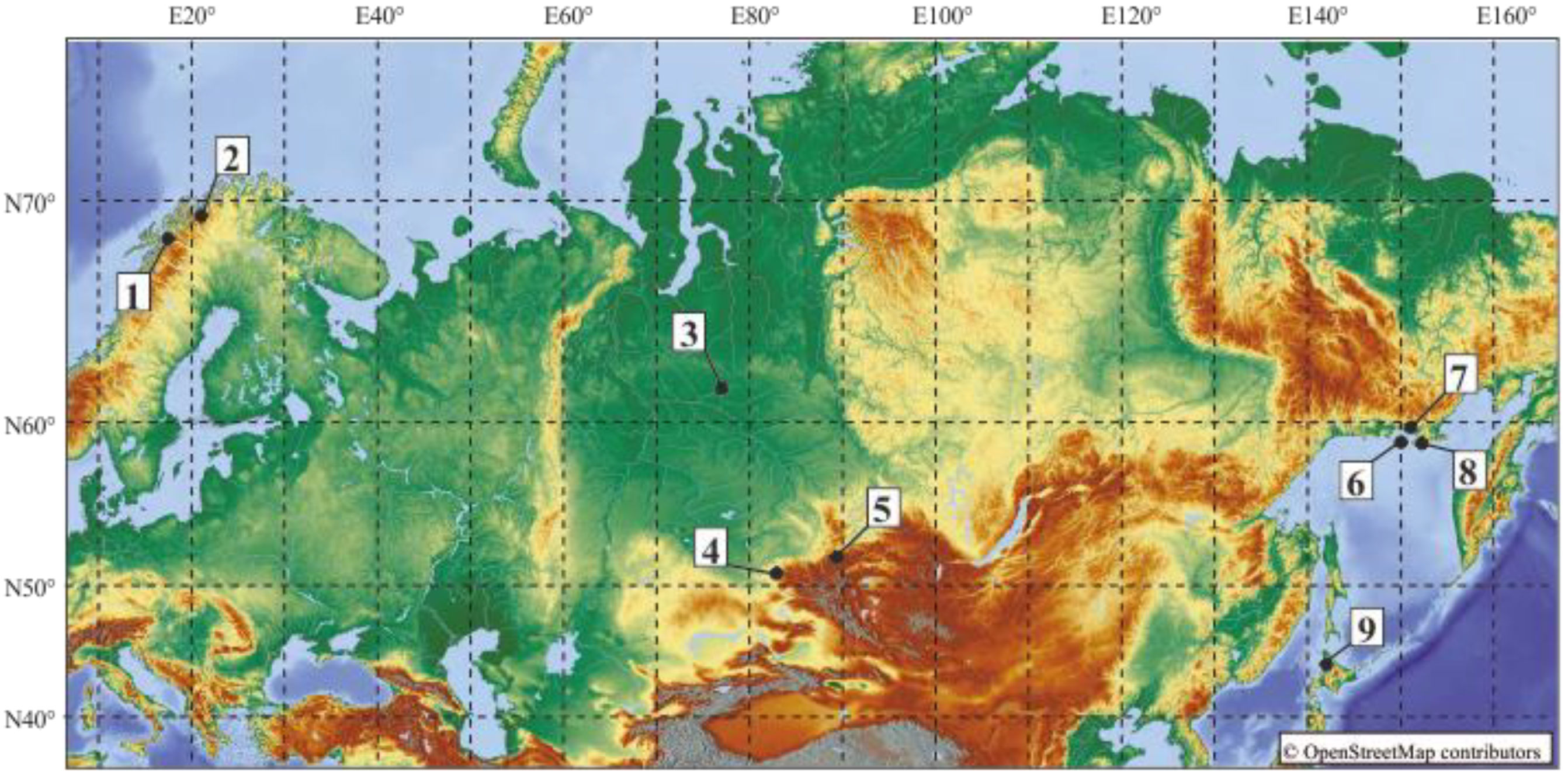
| Locality (Number) | GenBank acc. no. (cox1/nad1) | Final Host |
|---|---|---|
| Ola, Magadan Oblast, Russia (7) | ON533413/ON548169 | Myodes rufocanus |
| Zavyalov Island, Magadan Oblast, Russia (6) | ON533414/ON548171 | Myodes rufocanus |
| Zavyalov Island, Magadan Oblast, Russia (6) | ON533415/ON548172 | Myodes rufocanus |
| Ola, Magadan Oblast, Russia (7) | ON533416/ON548168 | Myodes rufocanus |
| Koni Peninsula, Magadan Oblast, Russia (8) | ON533417/ON548173 | Myodes rufocanus |
| Pozarym, Western Sayan, Russia (5) | ON533418/ON548175 | Myodes rutilus |
| Pozarym, Western Sayan, Russia (5) | ON533419/ON548176 | Myodes rufocanus |
| Raduzhny, KhMAO, Russia (3) | ON533420/ON548177 | Myodes rutilus |
| Ridder, Western Altai, Kazakhstan (4) | ON533421/--- | Myodes rufocanus |
| Ridder, Western Altai, Kazakhstan (4) | ---/ON548178 | Myodes rufocanus |
| Ola, Magadan Oblast, Russia (7) | ---/ON548174 | Myodes rufocanus |
| Ola, Magadan Oblast, Russia (7) | ---/ON548170 | Myodes rutilus |
| Kilpisjärvi, Finland (2) | AY181511/--- | Myodes rufocanus |
| Kilpisjärvi, Finland (2) | AY181512/--- | Myodes rufocanus |
| Kilpisjärvi, Finland (2) | EF583963/KJ778953 | Myodes rufocanus |
| Kilpisjärvi, Finland (2) | EF583962/--- | Myodes rufocanus |
| Kilpisjärvi, Finland (2) | EF583961/--- | Myodes rufocanus |
| Narvik, Norway (1) | AY181513/--- | Myodes rufocanus |
| Narvik, Norway (1) | AY189959/--- | Myodes rufocanus |
| Asahikawa, Hokkaido, Japan (9) | LC535262/--- | Myodes rufocanus |
| Asahikawa, Hokkaido, Japan (9) | LC535263/--- | Myodes rufocanus |
| Asahikawa, Hokkaido, Japan (9) | LC535264/--- | Myodes rufocanus |
| Asahikawa, Hokkaido, Japan (9) | LC535265/--- | Myodes rufocanus |
| Asahikawa, Hokkaido, Japan (9) | LC535266/--- | Myodes rufocanus |
| Asahikawa, Hokkaido, Japan (9) | LC535267/--- | Myodes rufocanus |
3. Results and Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haukisalmi, V.; Henttonen, H.; Tenora, F. Parasitism by helminths in the grey-sided vole (Clethrionomys rufocanus) in northern Finland: Influence of density, habitat and sex of the host. J. Wildl. Dis. 1987, 23, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Yushkov, V.F. The fauna of the European northeast of Russia. In Helminths of Mammals; Izd. “Nauka”: St. Petersburg, Russian, 1995; Volume 3, p. 202. (In Russian) [Google Scholar]
- Makarikov, A.; Dokuchaev, N.; Konyaev, S. Cestodes of Rodents of the Northern Priokhotye; Bulletin of the North-Eastern Scientific Center FEB RAS: Magadan, Russia, 2016; pp. 52–61. (In Russian) [Google Scholar]
- Haukisalmi, V.; Wickström, L.; Henttonen, H.; Hantula, J.; Gubányi, A. Molecular and morphological evidence for multiple species within Paranoplocephala omphalodes (Cestoda, Anoplocephalidae) in Microtus voles (Arvicolinae). Zool. Scr. 2004, 33, 277–290. [Google Scholar] [CrossRef]
- Haukisalmi, V.; Hardman, L.M.; Niemimaa, J.; Henttonen, H. Taxonomy and genetic divergence of Paranoplocephala kalelai (Tenora, Haukisalmi & Henttonen, 1985) (Cestoda: Anoplocephalidae) in the grey-sided vole Myodes rufocanus in northern Fennoscandia. Acta Parasitol. 2007, 52, 335–341. [Google Scholar] [CrossRef]
- Wilson, D.E.; Reeder, D.M. (Eds.) Mammal Species of the World: A Taxonomic and Geographic Reference, 3rd ed.; Johns Hopkins University Press: Baltimore, MD, USA, 2005. [Google Scholar]
- Sasaki, M.; Anders, J.L.; Nakao, M. Cestode fauna of murid and cricetid rodents in Hokkaido, Japan, with assignment of DNA barcodes. Species Divers. 2021, 26, 255–272. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Haukisalmi, V.; Hardman, L.; Hoberg, E.P.; Henttonen, H. Phylogenetic relationships and taxonomic revision of Paranoplocephala Lühe, 1910 sensu lato (Cestoda, Cyclophyllidea, Anoplocephalidae). Zootaxa 2014, 3873, 371–415. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Abramson, N.; Petrova, T.; Dokuchaev, N.; Obolenskaya, E.; Lissovsky, A. Phylogeography of the gray red-backed vole Craseomys rufocanus (Rodentia: Cricetidae) across the distribution range inferred from nonrecombining molecular markers. Russ. J. Theriol. 2012, 11, 137–156. [Google Scholar] [CrossRef]
- Lehmkuhl, F.; Klinge, M.; Stauch, G. The Extent and Timing of Late Pleistocene Glaciations in the Altai and Neighbouring Mountain Systems. In Developments in Quaternary Sciences; Elsevier: Amsterdam, The Netherlands, 2011; pp. 967–979. [Google Scholar] [CrossRef]
- Rudoy, A.N. Glacier-dammed lakes and geological work of glacial superfloods in the Late Pleistocene, Southern Siberia, Altai Mountains. Quat. Int. 2002, 87, 119–140. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krivopalov, A.; Vlasenko, P.; Abramov, S.; Akimova, L.; Barkhatova, A.; Dokuchaev, N.; Gromov, A.; Konyaev, S.; Lopatina, N.; Vlasov, E.; et al. Distribution and Molecular Diversity of Paranoplocephala kalelai (Tenora, Haukisalmi & Henttonen, 1985) Tenora, Murai & Vaucher, 1986 in Voles (Rodentia: Myodes) in Eurasia. Diversity 2022, 14, 472. https://doi.org/10.3390/d14060472
Krivopalov A, Vlasenko P, Abramov S, Akimova L, Barkhatova A, Dokuchaev N, Gromov A, Konyaev S, Lopatina N, Vlasov E, et al. Distribution and Molecular Diversity of Paranoplocephala kalelai (Tenora, Haukisalmi & Henttonen, 1985) Tenora, Murai & Vaucher, 1986 in Voles (Rodentia: Myodes) in Eurasia. Diversity. 2022; 14(6):472. https://doi.org/10.3390/d14060472
Chicago/Turabian StyleKrivopalov, Anton, Pavel Vlasenko, Sergey Abramov, Lyudmila Akimova, Alina Barkhatova, Nikolai Dokuchaev, Anton Gromov, Sergey Konyaev, Natalia Lopatina, Egor Vlasov, and et al. 2022. "Distribution and Molecular Diversity of Paranoplocephala kalelai (Tenora, Haukisalmi & Henttonen, 1985) Tenora, Murai & Vaucher, 1986 in Voles (Rodentia: Myodes) in Eurasia" Diversity 14, no. 6: 472. https://doi.org/10.3390/d14060472
APA StyleKrivopalov, A., Vlasenko, P., Abramov, S., Akimova, L., Barkhatova, A., Dokuchaev, N., Gromov, A., Konyaev, S., Lopatina, N., Vlasov, E., & Zakharov, E. (2022). Distribution and Molecular Diversity of Paranoplocephala kalelai (Tenora, Haukisalmi & Henttonen, 1985) Tenora, Murai & Vaucher, 1986 in Voles (Rodentia: Myodes) in Eurasia. Diversity, 14(6), 472. https://doi.org/10.3390/d14060472







