eDNA Reveals the Associated Metazoan Diversity of Mediterranean Seagrass Sediments
Abstract
1. Introduction
2. Methods
2.1. Sediment Sampling
2.2. eDNA Analysis: Extraction, Amplification and Sequencing
2.3. Data Analysis
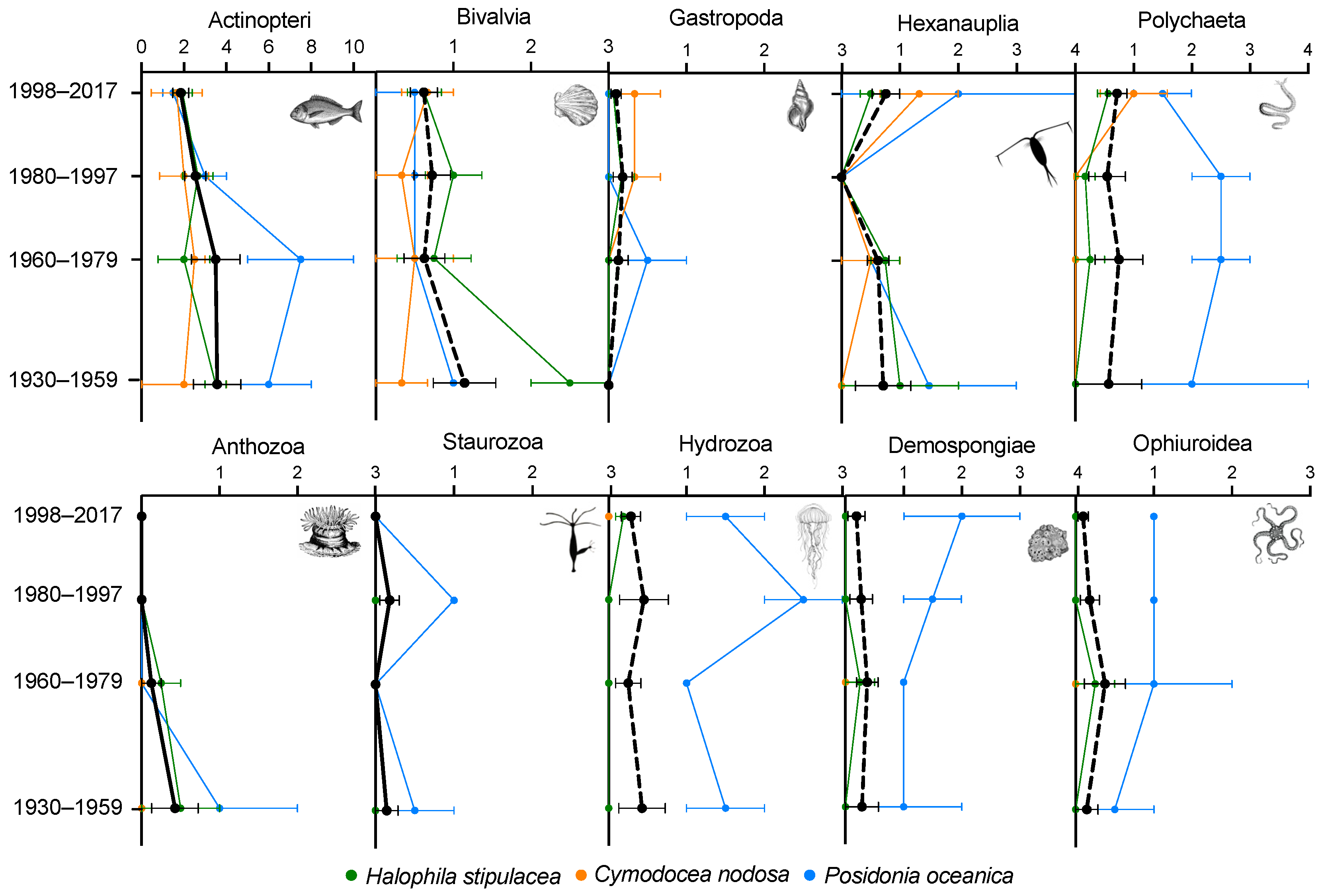
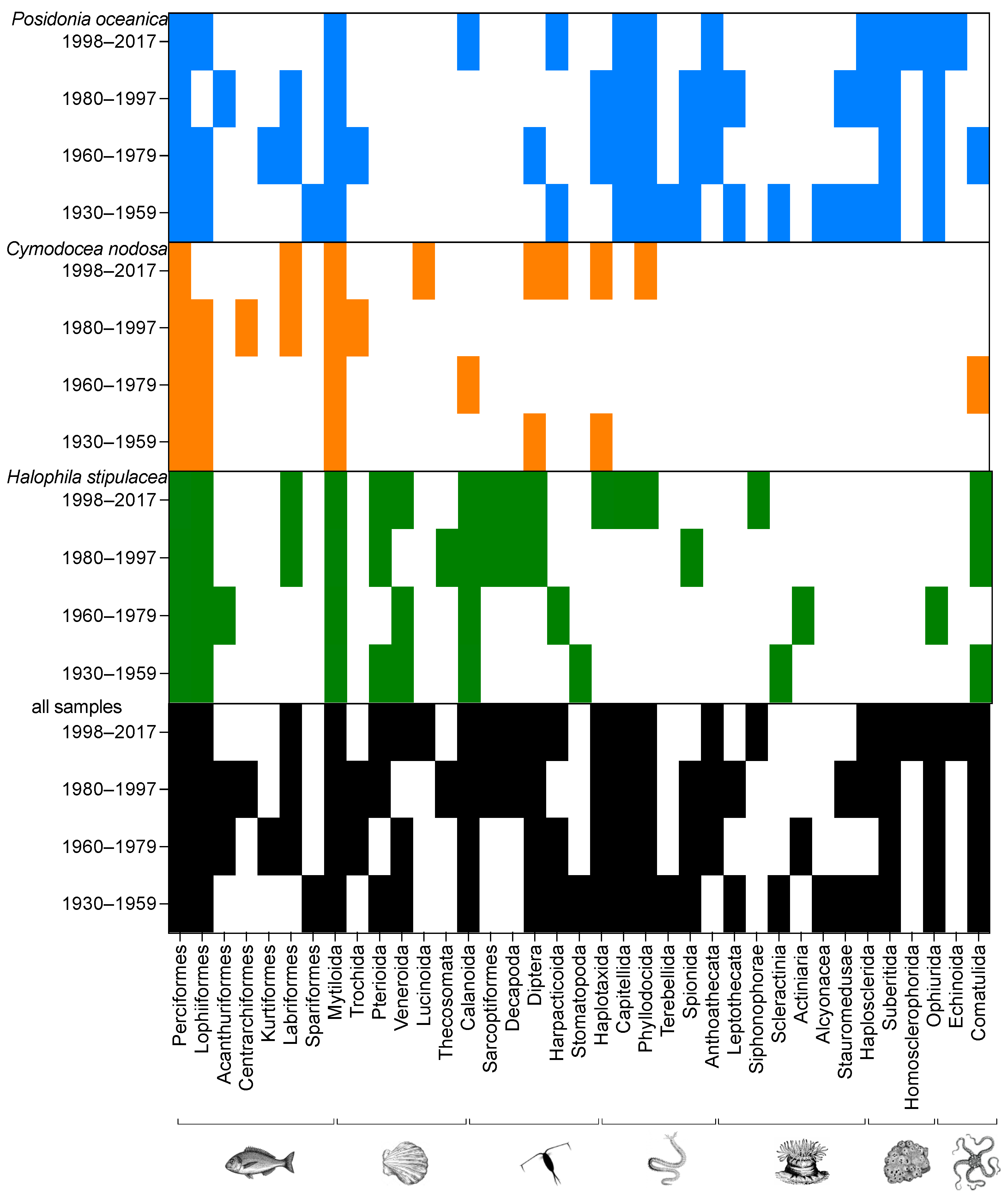
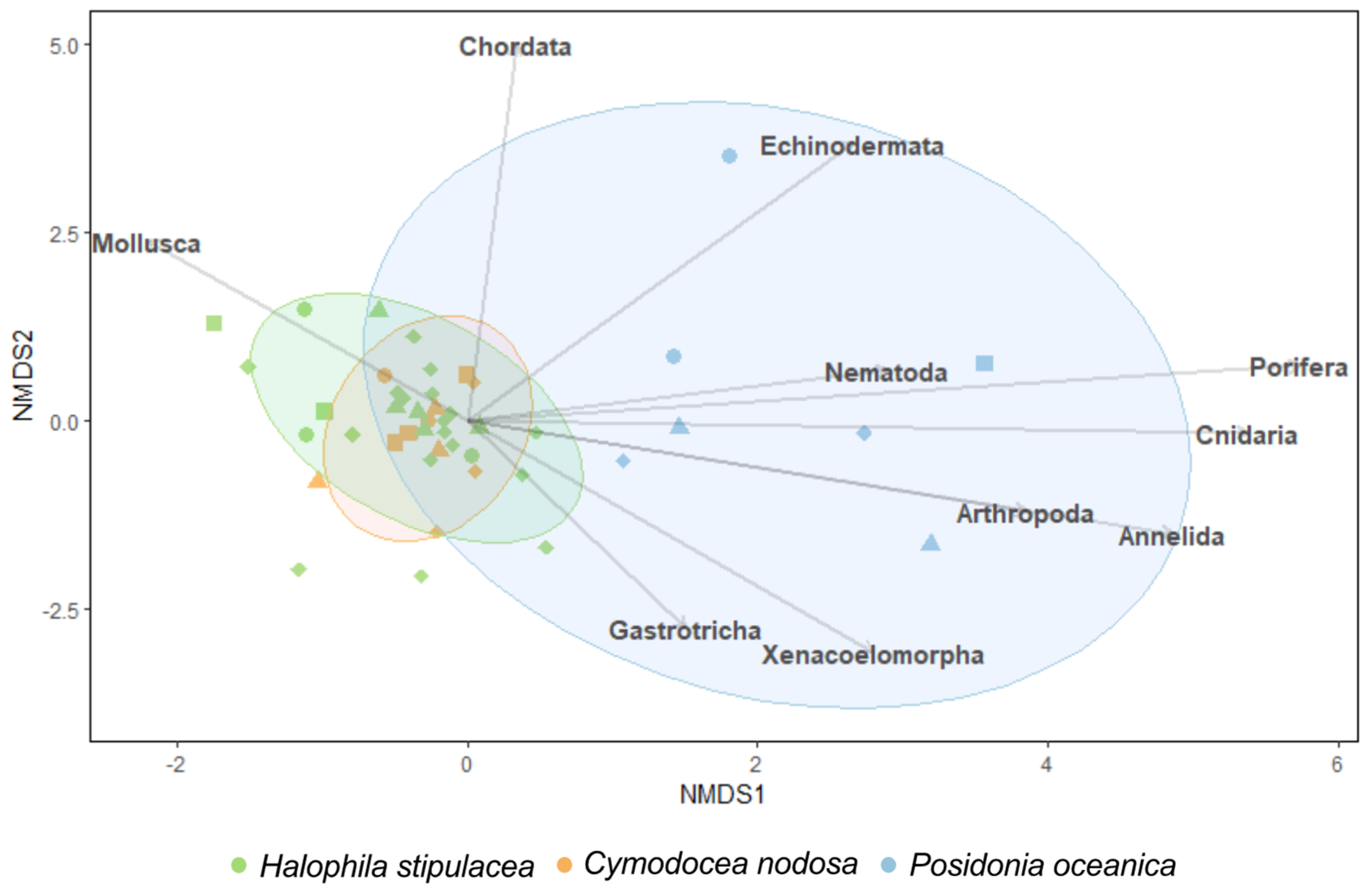
3. Results
4. Discussions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Steffen, W.; Grinevald, J.; Crutzen, P.; McNeill, J. The Anthropocene: Conceptual and historical perspectives. Philos. Trans. R. Soc. A 2011, 369, 842–867. [Google Scholar] [CrossRef] [PubMed]
- Millennium Ecosystem Assessment. Ecosystems and Human Well-Being: Biodiversity Synthesis; World Resources Institute: Washington, DC, USA, 2005. [Google Scholar]
- Travis, J.M.J. Climate change and habitat destruction: A deadly anthropogenic cocktail. Proc. R. Soc. B 2003, 270, 467–473. [Google Scholar] [CrossRef]
- Chen, I.C.; Hill, J.K.; Ohlemüller, R.; Roy, D.B.; Thomas, C.D. Rapid range shifts of species associated with high levels of climate warming. Science 2011, 333, 1024–1026. [Google Scholar] [CrossRef] [PubMed]
- Gottfried, M.; Pauli, H.; Futschik, A.; Akhalkatsi, M.; Barančok, P.; Alonso, J.L.B.; Coldea, G.; Dick, J.; Erschbamer, B.; Calzado, M.a.R.F.; et al. Continent-wide response of mountain vegetation to climate change. Nat. Clim. Chang. 2012, 2, 111–115. [Google Scholar] [CrossRef]
- Poloczanska, E.S.; Burrows, M.T.; Brown, C.J.; GarcíaMolinos, J.; Halpern, B.S.; Hoegh-Guldberg, O.; Kappel, C.V.; Moore, P.J.; Richardson, A.J.; Schoeman, D.S.; et al. Responses of marine organisms to climate change across oceans. Front. Mar. Sci. 2016, 3, 62. [Google Scholar] [CrossRef]
- Sunday, J.M.; Pecl, G.T.; Frusher, S.; Hobday, A.J.; Hill, N.; Holbrook, N.J.; Edgar, G.J.; Stuart-Smith, R.; Barrett, N.; Wernberg, T.; et al. Species traits and climate velocity explain geographic range shifts in an ocean-warming hotspot. Ecol. Lett. 2015, 18, 944–953. [Google Scholar] [CrossRef]
- Pecl, G.T.; Araújo, M.B.; Bell, J.D.; Blanchard, J.; Bonebrake, T.C.; Chen, I.C.; Clarck, T.A.; Colwell, R.K.; Danielsen, D.; Williams, S.E.; et al. Biodiversity redistribution under climate change: Impacts on ecosystems and human well-being. Science 2015, 355, eaai9214. [Google Scholar] [CrossRef]
- Barnosky, A.D.; Matzke, N.; Tomiya, S.; Wogan, G.O.; Swartz, B.; Quental, T.B.; Marshall, C.; McGuire, J.L.; Lindsey, E.L.; Maguire, K.C.; et al. Has the Earth’s sixth mass extinction already arrived? Nature 2011, 471, 51–57. [Google Scholar] [CrossRef]
- Eastwood, N.; Stubbings, W.A.; Abdallah, M.A.A.E.; Durance, I.; Paavola, J.; Dallimer, M.; Pantel, J.H.; Johnson, S.; Zhou, J.; Hosking, J.S.; et al. The Time Machine framework: Monitoring and prediction of biodiversity loss. Trends Ecol. Evol. 2021, 37, 138–146. [Google Scholar] [CrossRef]
- Vellend, M.; Brown, C.D.; Kharouba, H.M.; McCune, J.L.; Myers-Smith, I.H. Historical ecology: Using unconventional data sources to test for effects of global environmental change. Am. J. Bot. 2013, 100, 1294–1305. [Google Scholar] [CrossRef]
- Dornelas, M.; Gotelli, N.J.; McGill, B.; Shimadzu, H.; Moyes, F.; Sievers, C.; Magurran, A.E. Assemblage time series reveal biodiversity change but not systematic loss. Science 2014, 344, 296–299. [Google Scholar] [CrossRef] [PubMed]
- Mihoub, J.B.; Henle, K.; Titeux, N.; Brotons, L.; Brummitt, N.A.; Schmeller, D.S. Setting temporal baselines for biodiversity: The limits of available monitoring data for capturing the full impact of anthropogenic pressures. Sci. Rep. 2017, 7, 41591. [Google Scholar] [CrossRef] [PubMed]
- Hardesty, B.D.; Harari, J.; Isobe, A.; Lebreton, L.; Maximenko, N.; Potemra, J.; van Sebille, E.; Vethaak, A.D.; Wilcox, C. Using numerical model simulations to improve the understanding of micro-plastic distribution and pathways in the marine environment. Front. Mar. Sci. 2017, 4, 30. [Google Scholar] [CrossRef]
- Gillson, L. Biodiversity Conservation and Environmental Change: Using Palaeoecology to Manage Dynamic Landscapes in the Anthropocene; OUP Oxford: Oxford, UK, 2015. [Google Scholar]
- Troudet, J.; Grandcolas, P.; Blin, A.; Vignes-Lebbe, R.; Legendre, F. Taxonomic bias in biodiversity data and societal preferences. Sci. Rep. 2017, 7, 9132. [Google Scholar] [CrossRef]
- Mora, C.; Tittensor, D.P.; Myers, R.A. The completeness of taxonomic inventories for describing the global diversity and distribution of marine fishes. Proc. Royal Soc. B 2008, 275, 149–155. [Google Scholar] [CrossRef]
- Daan, N. The IBTS Database: A Plea for Quality Control; International Council for the Exploration of the Sea: Copenhagen, Denmark, 2001. [Google Scholar]
- Thomsen, P.F.; Willerslev, E. Environmental DNA–An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- Taberlet, P.; Bonin, A.; Zinger, L.; Coissac, E. Environmental DNA: For Biodiversity Research and Monitoring; Oxford University Press: Oxford, UK, 2018. [Google Scholar]
- Taberlet, P.; Coissac, E.; Pompanon, F.; Brochmann, C.; Willerslev, E. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 2012, 21, 2045–2050. [Google Scholar] [CrossRef]
- Balint, M.; Pfenninger, M.; Grossart, H.P.; Taberlet, P.; Vellend, M.; Leibold, M.A.; Englund, G.; Bowler, D. Environmental DNA time series in ecology. Trends Ecol. Evol. 2018, 33, 945–957. [Google Scholar] [CrossRef]
- Capo, E.; Giguet-Covex, C.; Rouillard, A.; Nota, K.; Heintzman, P.D.; Vuillemin, A.; Ariztegui, D.; Arnaud, F.; Belle, S.; Bertilsson, S.; et al. Lake sedimentary DNA research on past terrestrial and aquatic biodiversity: Overview and recommendations. Quaternary 2021, 4, 6. [Google Scholar] [CrossRef]
- Pedersen, M.W.; Ruter, A.; Schweger, C.; Friebe, H.; Staff, R.A.; Kjeldsen, K.K.; Mendoza, M.L.Z.; Beaudoin, A.B.; Zutter, C.; Larsen, N.K.; et al. Postglacial viability and colonization in North America’s ice-free corridor. Nature 2021, 537, 45–49. [Google Scholar] [CrossRef]
- Gomez Cabrera, M.C.; Young, J.M.; Roff, G.; Staples, T.; Ortiz, J.C.; Pandolfi, J.M.; Cooper, A. Broadening the taxonomic scope of coral reef palaeoecological studies using ancient DNA. Mol. Ecol. 2019, 28, 2636–2652. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Poulenard, J.; Sabatier, P.; Messager, E.; Gielly, L.; Leloup, A.; Etienne, D.; Bakke, J.; Malet, E.; Arnaud, F.; et al. DNA from lake sediments reveals long-term ecosystem changes after a biological invasion. Sci. Adv. 2018, 4, eaar4292. [Google Scholar] [CrossRef] [PubMed]
- Wesselmann, M.; Geraldi, N.R.; Duarte, C.M.; Garcia-Orellana, J.; Díaz-Rúa, R.; Arias-Ortiz, A.; Hendrisks, I.E.; Apostolaki, E.T.; Marbà, N. Seagrass (Halophila stipulacea) invasion enhances carbon sequestration in the Mediterranean Sea. Glob. Chang. Biol. 2021, 27, 2592–2607. [Google Scholar] [CrossRef] [PubMed]
- Costanza, R.; d’Arge, R.; De Groot, R.; Farber, S.; Grasso, M.; Hannon, B.; Limburg, K.; Naeem, S.; O’Neil, R.V.; Paruelo, J.; et al. The value of the world’s ecosystem services and natural capital. Nature 1998, 387, 253–260. [Google Scholar] [CrossRef]
- Duarte, C.M. Marine biodiversity and ecosystem services: An elusive link. J. Exp. Mar. Biol. Ecol. 2000, 250, 117–131. [Google Scholar] [CrossRef]
- Short, F.T.; Wyllie-Echeverria, S. Natural and human induced disturbance of seagrasses. Environment 1996, 23, 17–27. [Google Scholar] [CrossRef]
- Orth, R.J.; Carruthers, T.J.; Dennison, W.C.; Duarte, C.M.; Fourqurean, J.W.; Heck, K.L.; Hughes, A.R.; Kendrick, G.A.; Kenworthy, W.J.; Olyarnik, S.; et al. A global crisis for seagrass ecosystems. Bioscience 2006, 56, 987–996. [Google Scholar] [CrossRef]
- Marbà, N.; Duarte, C.M. Mediterranean warming triggers seagrass (Posidonia oceanica) shoot mortality. Glob. Chang. Biol. 2010, 16, 2366–2375. [Google Scholar] [CrossRef]
- Arias-Ortiz, A.; Serrano, O.; Masqué, P.; Lavery, P.S.; Mueller, U.; Kendrick, G.A.; Rozaimi, M.; Esteban, A.; Fourqurean, W.J.; Marba, N.; et al. A marine heatwave drives massive losses from the world’s largest seagrass carbon stocks. Nat. Clim. Chang. 2018, 8, 338–344. [Google Scholar] [CrossRef]
- Hemminga, M.A.; Duarte, C.M. Seagrass Ecology; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Hendriks, I.E.; Olsen, Y.S.; Ramajo, L.; Basso, L.; Steckbauer, A.; Moore, T.S.; Howard, J.; Duarte, C.M. Photosynthetic activity buffers ocean acidification in seagrass meadows. Biogeosciences 2014, 11, 333–346. [Google Scholar] [CrossRef]
- Béthoux, J.P.; Copin-Montégut, G. Biological fixation of atmospheric nitrogen in the Mediterranean Sea. Limnol. Oceanogr. 1986, 31, 1353–1358. [Google Scholar] [CrossRef]
- Pasqualini, V.; Pergent-Martini, C.; Clabaut, P.; Pergent, G. Mapping of Posidonia oceanica using Aerial Photographs and Side Scan Sonar: Application off the Island of Corsica (France). Estuar. Coast. Shelf Sci. 1998, 47, 359–367. [Google Scholar] [CrossRef]
- Montefalcone, M.; Morri, C.; Peirano, A.; Albertelli, G.; Bianchi, C.N. Substitution and phase shift within the Posidonia oceanica seagrass meadows of NW Mediterranean Sea. Estuar. Coast. Shelf Sci. 2007, 75, 63–71. [Google Scholar] [CrossRef]
- Winters, G.; Beer, S.; Willette, D.A.; Viana, I.G.; Chiquillo, K.L.; Beca-Carretero, P.; Villamayor, B.; Azcarate-Garcia, T.; Shem-Tov, R.; Mwabvu, B.; et al. The tropical seagrass Halophila stipulacea: Reviewing what we know from its native and invasive habitats, alongside identifying knowledge gaps. Front. Mar. Sci. 2020, 7, 300. [Google Scholar] [CrossRef]
- Gambi, M.C.; Barbieri, F.; Bianchi, C.N. New record of the alien seagrass Halophila stipulacea (Hydrocharitaceae) in the western Mediterranean: A further clue to changing Mediterranean Sea biogeography. Mar. Biodivers. Rec. 2009, 2, E84. [Google Scholar] [CrossRef]
- Orfanidis, S.; Alvito, A.; Azzurro, E.; Badreddine, A.; Soussi, J.B.; Chamorro, M.; Crochetta, F.; Dalyan, C.; Fortic, A.; Galanti, L.; et al. New Alien Mediterranean Biodiversity Records (March 2021) Mediterr. Mar. Sci. 2021, 22, 180–198. [Google Scholar]
- Thibaut, T.; Blanfuné, A.; Boudouresque, C.F.; Holon, F.; Agel, N.; Descamps, P.; Deter, J.; Pavy, T.; Delaruelle, G.; Verlaque, M. Distribution of the seagrass Halophila stipulacea: A big jump to the northwestern Mediterranean Sea. Aquat. Bot. 2022, 176, 103465. [Google Scholar] [CrossRef]
- Wesselmann, M.; Chefaoui, R.; Marbà, N.; Serrao, E.A.; Duarte, C.M. Warming threatens to propel the range expansion of the exotic seagrass Halophila stipulacea. Front. Mar. Sci. 2021, 8, 759676. [Google Scholar] [CrossRef]
- Benoit, G.; Comeau, A. A Sustainable Future for the Mediterranean, The Blue Plan’s Environment & Development Outlook; Earthscan: London, UK, 2005. [Google Scholar]
- Zenetos, A.; Galanidi, M. Mediterranean non indigenous species at the start of the 2020s: Recent changes. Mar. Biodivers. Rec. 2020, 13, 10. [Google Scholar] [CrossRef]
- Geraldi, N.R.; Díaz-Rúa, R.; Shea, L.A.; Duarte, C.M. Performance of extraction methods for extracellular DNA from sediments across marine habitats. Environ. DNA 2020, 2, 91–98. [Google Scholar] [CrossRef]
- Ushio, M.; Murakami, H.; Masuda, R.; Sado, T.; Miya, M.; Sakurai, S.; Yamanaka, H.; Minamoto, T.; Kondoh, M. Quantitative monitoring of multispecies fish environmental DNA using high-throughput sequencing. Metabarcoding Metagenom. 2017, 2, e23297. [Google Scholar]
- Seebens, H.; Blackburn, T.M.; Dyer, E.E.; Genovesi, P.; Hulme, P.E.; Jeschke, J.M.; Pagad, S.; Pysek, P.; Winter, M.; Arianoutsou, M.; et al. No saturation in the accumulation of alien species worldwide. Nat. Commun. 2017, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Salat, J.; Pascual, J. The oceanographic and meteorological station at L’Estartit (NW Mediterranean). In Tracking Long-Term Hydrological Change in the Mediterranean Sea; Proceedings of the CIESM Workshop Series; CIESM: Monaco, 2002; Volume 16. [Google Scholar]
- Salat, J.; Pascual, J. Principales Tendencias Climatológicas en el Mediterráneo Noroccidental, a Partir de Más de 30 Años de Observaciones Oceanográficas y Meteorológicas en la Costa Catalana. In Clima, Socie—Dad y Medio Ambiente; Cuadrat Prats, J.M., Saz Sánchez, M.A., Vicente Serrano, S.M., Lanjeri, S., de Luis Arrillaga, M., González-Hidalgo, J.C., Eds.; Publicaciones de la Sociedad Española de Climatología: Zaragoza, Spain, 2006. [Google Scholar]
- Vargas-Yáñez, M.; García, M.J.; Salat, J.; García-Martínez, M.C.; Pascual, J.; Moya, F. Warming trends and decadal variability in the Western Mediterranean shelf. Glob. Planet. Chang. 2008, 63, 177–184. [Google Scholar] [CrossRef]
- Bates, D.; Maechler, M.; Bolker, B. lme4: Linear Mixed-Effects Models Using Eigen and S4. (R Package Version 1.1-6). Available online: http://CRAN.R-project.org/package=lme4 (accessed on 1 September 2021).
- Buia, M.C.; Gambi, M.C.; Zupo, V. Structure and functioning of Mediterranean seagrass ecosystems: An overview. Biol. Mar. Mediterr. 2000, 7, 167–190. [Google Scholar]
- Pranovi, F.; Da Ponte, F.; Torricelli, P. Historical changes in the structure and functioning of the benthic community in the lagoon of Venice. Estuar. Coast. Shelf Sci. 2008, 76, 753–764. [Google Scholar] [CrossRef]
- Bertolino, M.; Betti, F.; Bo, M.; Cattaneo-Vietti, R.; Pansini, M.; Romero, J.; Bavestrello, G. Changes and stability of a Mediterranean hard bottom benthic community over 25 years. J. Mar. Biol. Assoc. UK 2016, 96, 341–350. [Google Scholar] [CrossRef]
- Piroddi, C.; Coll, M.; Liquete, C.; Macias, D.; Greer, K.; Buszowski, J.; Steenbeek, J.; Danovaro, R.; Christensen, V. Historical changes of the Mediterranean Sea ecosystem: Modelling the role and impact of primary productivity and fisheries changes over time. Sci. Rep. 2017, 7, 44491. [Google Scholar] [CrossRef]
- Verdura, J.; Linares, C.; Ballesteros, E.; Coma, R.; Uriz, M.J.; Bensoussan, N.; Cebrian, E. Biodiversity loss in a Mediterranean ecosystem due to an extreme warming event unveils the role of an engineering gorgonian species. Sci. Rep. 2019, 9, 5911. [Google Scholar] [CrossRef]
- Claudet, J.; Fraschetti, S. Human-driven impacts on marine habitats: A regional meta-analysis in the Mediterranean Sea. Biol. Conserv. 2010, 143, 2195–2206. [Google Scholar] [CrossRef]
- Gardner, T.A.; Côté, I.M.; Gill, J.A.; Grant, A.; Watkinson, A.R. Long-term region-wide declines in Caribbean corals. Science 2003, 301, 958–960. [Google Scholar] [CrossRef]
- Coma, R.; Ribes, M.; Serrano, E.; Jiménez, E.; Salat, J.; Pascual, J. Global warming-enhanced stratification and mass mortality events in the Mediterranean. Proc. Natl. Acad. Sci. USA 2009, 106, 6176–6181. [Google Scholar] [CrossRef] [PubMed]
- Ferretti, F.; Osio, G.C.; Jenkins, C.J.; Rosenberg, A.A.; Lotze, H.K. Long-term change in a meso-predator community in response to prolonged and heterogeneous human impact. Sci. Rep. 2013, 3, 1057. [Google Scholar] [CrossRef] [PubMed]
- Veneroni, B.; Fernandes, P.G. Fishers’ knowledge detects ecological decay in the Mediterranean Sea. Ambio 2021, 50, 1159–1171. [Google Scholar] [CrossRef] [PubMed]
- Seytre, C.; Francour, P. A long-term survey of Posidonia oceanica fish assemblages in a Mediterranean Marine Protected Area: Emphasis on stability and no-take area effectiveness. Mar. Freshw. Res. 2013, 65, 244–254. [Google Scholar] [CrossRef]
- Azzurro, E.; La Mesa, G.; Fanelli, E. The rocky-reef fish assemblages of Malta and Lampedusa islands (Strait of Sicily, Mediterranean Sea): A visual census study in a changing biogeographical sector. J. Mar. Biolog. Assoc. UK 2013, 93, 2015–2026. [Google Scholar] [CrossRef]
- Harmelin-Vivien, M.L.; Bitar, G.; Harmelin, J.G.; Monestiez, P. The littoral fish community of the Lebanese rocky coast (eastern Mediterranean Sea) with emphasis on Red Sea immigrants. Biol. Invasions 2005, 7, 625–637. [Google Scholar] [CrossRef]
- Kalogirou, S.; Corsini-Foka, M.; Sioulas, A.; Wennhage, H.; Pihl, L. Diversity, structure and function of fish assemblages associated with Posidonia oceanica beds in an area of the eastern Mediterranean Sea and the role of non-indigenous species. J. Fish Biol. 2010, 77, 2338–2357. [Google Scholar] [CrossRef]
- Froese, R.; Pauly, D. Fishbase. 2015. Available online: http://www.fishbase.org (accessed on 30 November 2021).
- Crespo, J.; Rey, J.C.; Garcia, A. Primera cita de Acanthurus monrovae Steindachner, 1876 y de Diodon eydouxii Brissout de Barneville, 1846 para la ictioma europea. Misc. Zool. 1987, 11, 271–275. [Google Scholar]
- Guidetti, P.; Magnali, L.; Navone, A. First record of the acanthurid fish Zebrasoma xanthurum (Blyth 1987, 1852) in the Mediterranean Sea, with some considerations on the risk associated with aquarium trade. Mediterr. Mar. Sci. 2016, 17, 147–151. [Google Scholar] [CrossRef]
- Langeneck, J.; Marcelli, M.; Simak, H.C. Unexpected alien species in Cyprus waters: Acanthurus coeruleus (Actinopterygii: Acanthuridae). Mar. Biodivers. Rec. 2012, 5, E116. [Google Scholar] [CrossRef]
- Langeneck, J.; Boyer, M.; De Cecco, P.G.; Luciani, C.; Marcelli, M.; Vacchi, M. First record of Acanthurus chirurgus (Perciformes: Acanthuridae) in the Mediterranean Sea, with some distributional notes on Mediterranean Acanthuridae. Mediterr. Mar. Sci. 2015, 16, 427–431. [Google Scholar] [CrossRef][Green Version]
- Marcelli, M.; Dayan, A.R.; Langeneck, J. Finding Dory: First record of Paracanthurus hepatus (Perciformes: Acanthuridae) in the Mediterranean Sea. Mar. Biodivers. 2017, 47, 599–602. [Google Scholar] [CrossRef]
- Weitzmann, B.; Mercader, L.; Azzurro, E. First sighting of Zebrasoma flavescens (Teleostei: Acanthuridae) and Balistoides conspicillum (Teleostei: Balistidae) in the Mediterranean Sea: Two likely aquarium releases. Mediterr. Mar. Sci. 2015, 16, 147–150. [Google Scholar] [CrossRef][Green Version]
- Raitsos, D.E.; Beaugrand, G.; Georgopoulos, D.; Zenetos, A.; Pancucci-Papadopoulou, A.M.; Theocharis, A.; Papathanassiou, E. Global climate change amplifies the entry of tropical species into the Eastern Mediterranean Sea. Limnol. Oceanograph. 2010, 55, 1478–1484. [Google Scholar] [CrossRef]
- Sardain, A.; Sardain, E.; Leung, B. Global forecasts of shipping traffic and biological invasions to 2050. Nat. Sustain. 2019, 2, 274–282. [Google Scholar] [CrossRef]
- Pérès, J.M.; Picard, J. Nouveau Manuel de Bionomie Benthique de la Mer Méditerranée. Recl. Trav. Stn. Mar. D’endoume 1964, 31, 1–137. [Google Scholar]
- Bianchi, C.N.; Bedulli, D.; Morri, C.; Occhipinti Ambrogi, A. L’herbier de Posidonies: Écosysteme ou Carrefour Eco-Ethologique. In Proceedings of the International Workshop on Posidonia Beds; Boudouresque, C.F., Meinesz, A., Fresi, E., Gravez, V., Eds.; GIS Posidonie: Marseille, France, 1989; Volume 2, pp. 257–272. [Google Scholar]
- Gambi, M.C.; Conti, G.; Bremec, C.S. Polychate distribution, diversity and seasonality related to seagrass cover in shallow bottoms of the Tyrrenian Sea (Italy). Sci. Mar. 1998, 62, 1–17. [Google Scholar]
- Sardà, R. Polychaete communities related to plant covering in the mediolittoral and infralittoral zones of the Balearic Islands (Western Mediterranean). Mar. Ecol. 1991, 12, 341–360. [Google Scholar] [CrossRef]
- Cancemi, G. Fenologia e Produzione Primaria di Cymodocea nodosa (Ucria) Aschers. In Un Prato Superficiale Dell’Kola d’Ischia (Golfo di Napoli). Ph.D. Thesis, Universita di Napoli, Naples, Italy, 1991. [Google Scholar]
- Mazzella, L.; Scipione, M.B.; Gambi, M.C.; Buia, M.C.; Lorenti, M.; Zupo, V.; Cancemi, G. The Mediterranean seagrass Posidonia oceanica and Cymodocea nodosa. A comparative overview. In A comparative overview. In Proceedings of the First International Conference on the Mediterranean Coastal Environment, MEDCOAST, Antalya, Turkey, 2–5 November 1993; Volume 93, pp. 103–116. [Google Scholar]
- Kuo, J.; Den Hartog, C. Seagrass taxonomy and identification key. Glob. Seagrass Res. Methods 2001, 33, 31–58. [Google Scholar]
- Duarte, C.M. Temporal biomass variability and production/biomass relationships of seagrass communities. Mar. Ecol. Prog. Ser. 1989, 51, 269–276. [Google Scholar] [CrossRef]
- Pansini, M.; Pronzato, R. Distribution and Ecology of Epiphytic Porifera in two Posidonia oceanica (L.) DELILE Meadows of the Ligurian and Tyrrhenian Sea. Mar. Ecol. 1985, 6, 1–11. [Google Scholar] [CrossRef]
- Hillman, K.; McComb, A.J.; Walker, D.I. The distribution, biomass and primary production of the seagrass Halophila ovalis in the Swan/Canning Estuary, Western Australia. Aquat. Bot. 1995, 51, 1–54. [Google Scholar] [CrossRef]
- Ben Brahim, M.; Mabrouk, L.; Hamza, A.; Jribi, I. Comparison of spatial scale variability of shoot density and epiphytic leaf assemblages of Halophila stipulacea and Cymodocea nodosa on the Eastern Coast of Tunisia. Pl. Biosyst. 2020, 154, 413–426. [Google Scholar] [CrossRef]
- De Troch, M.; Fiers, F.; Vincx, M. Alpha and beta diversity of harpacticoid copepods in a tropical seagrass bed: The relation between diversity and species’ range size distribution. Mar. Ecol. Prog. Ser. 2001, 215, 225–236. [Google Scholar] [CrossRef]
- Marbà, N.; Duarte, C.M. Rhizome elongation and seagrass clonal growth. Mar. Ecol. Prog. Ser. 1998, 174, 269–280. [Google Scholar] [CrossRef]
- Díaz-Almela, E.; Marbà, N.; Duarte, C.M. Consequences of Mediterranean warming events in seagrass (Posidonia oceanica) flowering records. Glob. Chang. Biol. 2007, 13, 224–235. [Google Scholar] [CrossRef]
- Wesselmann, M.; Anton, A.; Duarte, C.M.; Hendriks, I.E.; Agustí, S.; Savva, I.; Apostolaki, E.T.; Marbà, N. Tropical seagrass Halophila stipulacea shifts thermal tolerance during Mediterranean invasion. Proc. R. Soc. B 2020, 287, 20193001. [Google Scholar] [CrossRef]
- Savva, I.; Bennett, S.; Roca, G.; Jordà, G.; Marbà, N. Thermal tolerance of Mediterranean marine macrophytes: Vulnerability to global warming. Ecol. Evol. 2018, 8, 12032–12043. [Google Scholar] [CrossRef]
- Jordà, G.; Marbà, N.; Duarte, C.M. Mediterranean seagrass vulnerable to regional climate warming. Nat. Clim. Chang. 2012, 2, 821–824. [Google Scholar] [CrossRef]
- Chefaoui, R.M.; Varela-Álvarez, E. Niche conservatism and spread of seaweed invasive lineages with different residence time in the Mediterranean Sea. Biol. Invasions 2018, 20, 423–435. [Google Scholar] [CrossRef]
- Bennett, S.; Alcoverro, T.; Kletou, D.; Antoniou, C.; Boada, J.; Buñuel, X.; Cucala, L.; Jorda, G.; Kleitou, P.; Roca, G.; et al. Resilience of seagrass populations to thermal stress does not reflect regional differences in ocean climate. New Phytol. 2022, 233, 1657–1666. [Google Scholar] [CrossRef] [PubMed]
- Stat, M.; Huggett, M.J.; Bernasconi, R.; DiBattista, J.D.; Newman, S.J.; Harvey, E.S.; Bunce, M.; Berry, T.E. Ecosystem biomonitoring with eDNA: Metabarcoding across the tree of life in a tropical marine environment. Sci. Rep. 2017, 7, 12240. [Google Scholar] [CrossRef]
- Yang, C.; Wang, X.; Miller, J.A.; de Blécourt, M.; Ji, Y.; Yang, C.; Harrison, R.D.; Yu, D.W. Using metabarcoding to ask if easily collected soil and leaf-litter samples can be used as a general biodiversity indicator. Ecol. Indic. 2014, 46, 379–389. [Google Scholar] [CrossRef]
- Corinaldesi, C.; Barucca, M.; Luna, G.M.; Dell’Anno, A. Preservation, origin and genetic imprint of extracellular DNA in permanently anoxic deep-sea sediments. Mol. Ecol. 2011, 20, 642–654. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, M.W.; Overballe-Petersen, S.; Ermini, L.; Sarkissian, C.D.; Haile, J.; Hellstrom, M.; Spens, J.; Thomsen, P.F.; Bohmann, K.; Cappellini, E.; et al. Ancient and modern environmental DNA. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20130383. [Google Scholar] [CrossRef]
- Krehenwinkel, H.; Wolf, M.; Lim, J.Y.; Rominger, A.J.; Simison, W.B.; Gillespie, R.G. Estimating and mitigating amplification bias in qualitative and quantitative arthropod metabarcoding. Sci. Rep. 2017, 7, 17668. [Google Scholar]
- Kelly, R.P.; Gallego, R.; Jacobs-Palmer, E. The effect of tides on nearshore environmental DNA. PeerJ 2018, 6, e4521. [Google Scholar] [CrossRef]
- Parducci, L.; Bennett, K.D.; Ficetola, G.F.; Alsos, I.G.; Suyama, Y.; Wood, J.R.; Pedersen, M.W. Ancient plant DNA in lake sediments. New Phytol. 2017, 214, 924–942. [Google Scholar] [CrossRef]
- Garcés-Pastor, S.; Wangensteen, O.S.; Pérez-Haase, A.; Pèlachs, A.; Pérez-Obiol, R.; Cañellas-Boltà, N.; Mariani, S.; Vegas-Vilarrúbia, T. DNA metabarcoding reveals modern and past eukaryotic communities in a high-mountain peat bog system. J. Paleolimnol. 2019, 62, 425–441. [Google Scholar] [CrossRef]
- Yates, M.C.; Fraser, D.J.; Derry, A.M. Meta-analysis supports further refinement of eDNA for monitoring aquatic species-specific abundance in nature. Environ. DNA 2019, 1, 5–13. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wesselmann, M.; Geraldi, N.R.; Marbà, N.; Hendriks, I.E.; Díaz-Rúa, R.; Duarte, C.M. eDNA Reveals the Associated Metazoan Diversity of Mediterranean Seagrass Sediments. Diversity 2022, 14, 549. https://doi.org/10.3390/d14070549
Wesselmann M, Geraldi NR, Marbà N, Hendriks IE, Díaz-Rúa R, Duarte CM. eDNA Reveals the Associated Metazoan Diversity of Mediterranean Seagrass Sediments. Diversity. 2022; 14(7):549. https://doi.org/10.3390/d14070549
Chicago/Turabian StyleWesselmann, Marlene, Nathan R. Geraldi, Núria Marbà, Iris E. Hendriks, Rubén Díaz-Rúa, and Carlos M. Duarte. 2022. "eDNA Reveals the Associated Metazoan Diversity of Mediterranean Seagrass Sediments" Diversity 14, no. 7: 549. https://doi.org/10.3390/d14070549
APA StyleWesselmann, M., Geraldi, N. R., Marbà, N., Hendriks, I. E., Díaz-Rúa, R., & Duarte, C. M. (2022). eDNA Reveals the Associated Metazoan Diversity of Mediterranean Seagrass Sediments. Diversity, 14(7), 549. https://doi.org/10.3390/d14070549





