Litter, Root, and Mycorrhiza Input Affected Soil Microbial Community Structure in Schima superba Pure Forest in Subtropical China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Experimental Design and Sample Collection
2.3. Determination of Soil Chemical Properties and Enzyme Activities
2.4. Soil DNA Extraction, PCR Amplification, and Sequence Data Analysis
2.5. Statistical Analysis
3. Results
3.1. Effects of Different Treatments on Soil Chemical Properties and Enzyme Activities
3.2. Effects of Different Treatments on Soil Microbial OTUs and Diversity
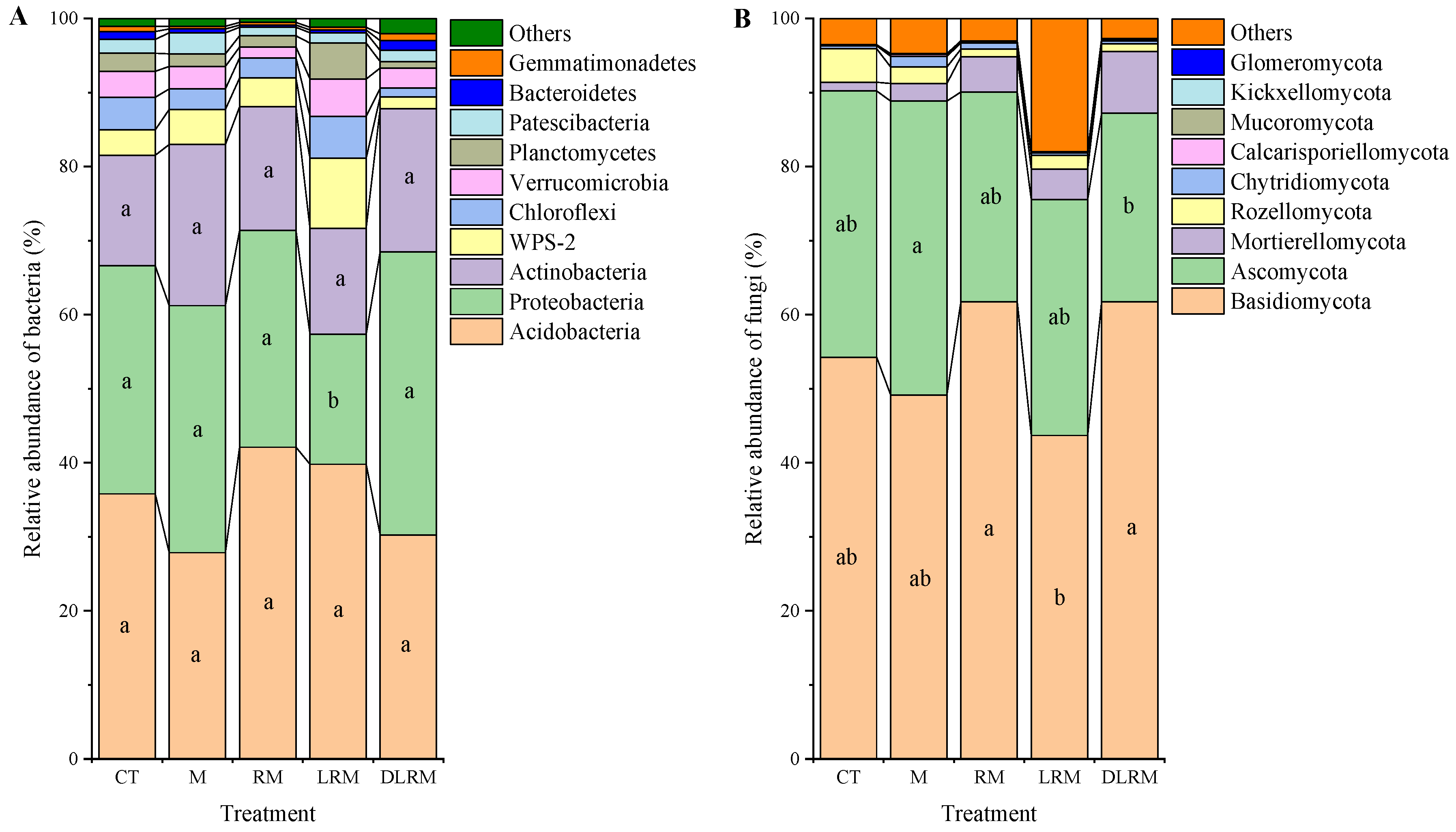
3.3. Effects of Different Treatments on Soil Microbial Community Composition and Structure
3.4. Effects of Soil Properties on Microbial Community Composition
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

| Type | Phylum | CT | M | RM | LRM | DLRM |
|---|---|---|---|---|---|---|
| Bacteria | Acidobacteria | 36 a | 28 a | 42 a | 40 a | 30 a |
| Proteobacteria | 31 a | 33 a | 29 a | 17.5 b | 38 a | |
| Actinobacteria | 15 a | 22 a | 17 a | 14 a | 19 a | |
| WPS−2 | 3.4 a | 4.7 a | 3.9 a | 9.5 a | 1.6 a | |
| Chloroflexi | 4.4 a | 2.8 a | 2.7 a | 5.7 a | 1.2 a | |
| Verrucomicrobia | 3.5 a | 3.1 a | 1.5 a | 5.0 a | 2.7 a | |
| Planctomycetes | 2.4 a | 1.7 a | 1.5 a | 4.9 a | 0.88 a | |
| Patescibacteria | 1.9 ab | 2.8 a | 1.2 b | 1.4 b | 1.5 b | |
| Bacteroidetes | 1.1 a | 0.55 b | 0.28 b | 0.41 b | 1.3 a | |
| Gemmatimonadetes | 0.70 ab | 0.35 b | 0.40 b | 0.33 b | 0.94 a | |
| Fungi | Basidiomycota | 54 ab | 49 ab | 62 a | 44 b | 62 a |
| Ascomycota | 36 ab | 40 a | 28 ab | 32 ab | 25 b | |
| Mortierellomycota | 1.2 d | 2.4 cd | 4.7 b | 4.1 bc | 8.4 a | |
| Rozellomycota | 4.5 a | 2.2 a | 1.0 a | 1.8 a | 1.0 a | |
| Chytridiomycota | 0.40 c | 1.4 a | 0.87 b | 0.35 c | 0.36 c | |
| Calcarisporiellomycota | 0.09 b | 0.26 a | 0.10 b | 0.06 b | 0.25 a | |
| Mucoromycota | 0.05 a | 0.05 a | 0.06 a | 0.11 a | 0.07 a | |
| Kickxellomycota | 0.01 b | 0.03 ab | 0.08 a | 0.01 b | 0.05 ab | |
| Glomeromycota | 0.004 b | 0.03 a | 0.005 b | 0.004 b | 0.002 b |
References
- Pankhurst, C.E.; Ophel-Keller, K.; Doube, B.M.; Gupta, V.V.S.R. Biodiversity of soil microbial communities in agricultural systems. Biodivers. Conserv. 1996, 5, 197–209. [Google Scholar] [CrossRef]
- Navarrete, A.A.; Tsai, S.M.; Mendes, L.W.; Faust, K.; de Hollander, M.; Cassman, N.A.; Raes, J.; van Veen, J.A.; Kuramae, E.E. Soil microbiome responses to the short-term effects of Amazonian deforestation. Mol. Ecol. 2015, 24, 2433–2448. [Google Scholar] [CrossRef] [PubMed]
- Averill, C.; Hawkes, C.V. Ectomycorrhizal fungi slow soil carbon cycling. Ecol. Lett. 2016, 19, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Van der Heijden, M.G.A.; Bardgett, R.D.; van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef]
- Luo, C.Y.; Zhang, B.X.; Liu, J.; Wang, X.X.; Han, F.P.; Zhou, J.H. Effects of different Ages of Robinia pseudoacacia plantations on soil physiochemical properties and microbial communities. Sustainability 2020, 12, 9161. [Google Scholar] [CrossRef]
- Kennedy, A.C.; Smith, K.L. Soil microbial diversity and the sustainability of agricultural soils. Plant Soil 1995, 170, 75–86. [Google Scholar] [CrossRef]
- Felske, A.; Wolterink, A.; van Lis, R.; de Vos, W.M.; Akkermans, A.D.L. Response of a soil bacterial community to grassland succession as monitored by 16S rRNA levels of the predominant ribotypes. Appl. Environ. Microbiol. 2000, 66, 3998–4003. [Google Scholar] [CrossRef] [Green Version]
- Bluhm, S.L.; Eitzinger, B.; Ferlian, O.; Bluhm, C.; Schröter, K.; Pena, R.; Maraun, M.; Scheu, S. Deprivation of root-derived resources affects microbial biomass but not community structure in litter and soil. PLoS ONE 2019, 14, e0214233. [Google Scholar] [CrossRef] [Green Version]
- Nevins, C.J.; Nakatsu, C.; Armstrong, S. Characterization of microbial community response to cover crop residue decomposition. Soil Biol. Biochem. 2018, 127, 39–49. [Google Scholar] [CrossRef]
- Xu, S.; Liu, L.; Sayer, E.J. Variability of aboveground litter inputs alters soil physicochemical and biological processes: A meta-analysis of litterfall-manipulation experiments. Biogeosciences 2013, 10, 5245–5272. [Google Scholar] [CrossRef]
- Brant, J.B.; Sulzman, E.W.; Myrold, D.D. Microbial community utilization of added carbon substrates in response to long-term carbon input manipulation. Soil Biol. Biochem. 2006, 38, 2219–2232. [Google Scholar] [CrossRef]
- Lu, X.R.; Yin, Y.; Feng, J.X.; Ma, H.L.; Gao, R.; Yin, Y.F. Effects of Chinese fir litter and its biochar amendment on soil microbial community structure. Acta Sci. Circumstantiae 2019, 39, 3090–3098. (In Chinese) [Google Scholar]
- Li, Y.; Zhou, C.F.; Qiu, Y.X.; Tigabu, M.; Ma, X.Q. Effects of biochar and litter on carbon and nitrogen mineralization and soil microbial community structure in a China fir plantation. J. For. Res. 2019, 30, 1913–1923. [Google Scholar] [CrossRef]
- Brant, J.B.; Myrold, D.D.; Sulzman, E.W. Root controls on soil microbial community structure in forest soils. Oecologia 2006, 148, 650–659. [Google Scholar] [CrossRef] [PubMed]
- Sayer, E.J. Using experimental manipulation to assess the roles of leaf litter in the functioning of forest ecosystems. Biol. Rev. 2006, 81, 1–31. [Google Scholar] [CrossRef]
- He, K.Y.; Shen, Y.W.; Feng, J.G.; Han, M.G.; Zhou, Y.Q.; Zhu, B. Effects of altered plant detritus input on soil respiration and its temperature sensitivity in a Pinus sylvestris var. mongolica Plantation. Acta Sci. Nat. Univ. Pekin. 2021, 57, 361–370. (In Chinese) [Google Scholar]
- Caesar-TonThat, A.J.; Espeland, E.; Sainju, U.M.; Lartey, R.T.; Gaskin, J.F. Effectsof Agaricus lilaceps fairy rings on soil aggregation and microbial communitystructure in relation to growth stimulation of western wheatgrass (Pascopyrumsmithii) in eastern Montana rangeland. Microb. Ecol. 2013, 66, 120–131. [Google Scholar] [CrossRef]
- Xu, J.; Liu, S.; Song, S.; Guo, H.; Tang, J.; Yong, J.W.; Ma, Y.; Chen, X. Arbuscular mycorrhizal fungi influence decomposition and the associated soil microbial community under different soil phosphorus availability. Soil Biol. Biochem. 2018, 120, 181–190. [Google Scholar] [CrossRef]
- Kaiser, C.; Fuchslueger, L.; Koranda, M.; Gorfer, M.; Stange, C.F.; Kitzler, B.; Rasche, F.; Strauss, J.; Sessitsch, A.; Zechmeister-Boltenstern, S.; et al. Plants control the seasonal dynamics of microbial N cycling in a beech forest soil by belowground Callocation. Pedobiologia 2011, 92, 1036–1051. [Google Scholar]
- Tedersoo, L.; Nilsson, R.H.; Abarenkov, K.; Jairus, T.; Sadam, A.; Saar, I.; Bahram, M.; Bechem, E.; Chuyong, G.; Kõljalg, U. 454 Pyrosequencing and Sanger sequencing of tropical mycorrhizal fungi provide similar results but reveal substantial methodological biases. New Phytol. 2010, 188, 291–301. [Google Scholar] [CrossRef]
- Wei, X.; Li, Q.; Liu, Y.; Liu, S.; Guo, X.; Zhang, L.; Niu, D.; Zhang, W. Restoring ecosystem carbon sequestration through afforestation: A sub-tropic restoration case study. Forest Ecol. Manag. 2013, 300, 60–67. [Google Scholar] [CrossRef]
- Sparks, D.L.; Page, A.L.; Helmke, P.A.; Loeppert, R.H.; Soltanpour, P.N.; Tabatabai, M.A.; Johnston, C.T.; Sumner, M.E. Methods of Soil Analysis. Part 3: Chemical Methods; Soil Science Society of America Inc.: Madison, WI, USA, 1996. [Google Scholar]
- Hou, Q.; Wang, W.; Yang, Y.; Hu, J.; Bian, C.; Jin, L.; Li, G.; Xiong, X. Rhizosphere microbial diversity and community dynamics during potato cultivation. Eur. J. Soil Biol. 2020, 98, 103176. [Google Scholar] [CrossRef]
- Liu, B.; Wang, S.; Wang, J.; Zhang, X.; Shen, Z.; Shi, L.; Chen, Y. The great potential for phytoremediation of abandoned tailings pond using ectomycorrhizal Pinus sylvestris. Sci. Total Environ. 2020, 719, 137475. [Google Scholar] [CrossRef]
- Ren, Q.; Yuan, J.; Wang, J.; Liu, X.; Ma, S.; Zhou, L.; Miao, L.; Zhang, J. Water level has higher influence on soil organic carbon and microbial community in Poyang Lake wetland than vegetation type. Microorganisms 2022, 10, 131. [Google Scholar] [CrossRef]
- Nadelhoffer, K.J.; Boone, R.D.; Bowden, R.D.; Canary, J.D.; Kaye, J.; Micks, P.; Ricca, A.; Aitkenhead, J.A.; Lajtha, K.; McDowell, W.H. The DIRT experiment: Litter and root influences on forest soil organic matter stocks and function. In Forest Landscape Dynamics in New England: Ecosystem Structure and Function as a Consequence of 5000 Years of Change; Foster, D., Aber, J., Eds.; Oxford University Press: New York, NY, USA, 2004. [Google Scholar]
- Lemanski, K.; Scheu, S. Incorporation of 13C labelled glucose into soil microorganisms of grassland: Effects of fertilizer addition and plant functional group composition. Soil Biol. Biochem. 2014, 69, 38–45. [Google Scholar] [CrossRef]
- Rousk, J.; Hill, P.W.; Jones, D.L. Priming of the decomposition of ageing soil organic matter: Concentration dependence and microbial control. Funct. Ecol. 2015, 29, 285–296. [Google Scholar] [CrossRef] [Green Version]
- Elser, J.J.; Sterner, R.W.; Gorokhova, E.A.; Fagan, W.F.; Markow, T.A.; Cotner, J.B.; Harrison, J.F.; Hobbie, S.E.; Odell, G.M.; Weider, L.W. Biological stoichiometry from genes to ecosystems. Ecol. Lett. 2008, 3, 540–550. [Google Scholar] [CrossRef]
- DeAngelis, K.M.; Pold, G.; Topçuoğlu, B.D.; van Diepen, L.T.; Varney, R.M.; Blanchard, J.L.; Melillo, J.; Frey, S.D. Long-term forest soil warming alters microbial communities in temperate forest soils. Front. Microbiol. 2015, 6, 104. [Google Scholar] [CrossRef] [Green Version]
- Feng, W.T.; Zou, X.M.; Schaefer, D. Above- and belowground carbon inputs affect seasonal variations of soil microbial biomass in a subtropical monsoon forest of southwest China. Soil Biol. Biochem. 2009, 41, 978–983. [Google Scholar] [CrossRef]
- Wang, Q.K.; He, T.X.; Wang, S.L.; Wang, S.L.; Liu, L. Carbon input manipulation affects soil respiration and microbial community composition in a subtropical coniferous forest. Agric. Forest Meteorol. 2013, 178–179, 152–160. [Google Scholar] [CrossRef]
- Wan, X.H.; Huang, Z.Q.; He, Z.M.; Yu, Z.P.; Wang, M.H.; Liu, R.Q.; Zheng, L.J. Changes of above- and belowground carbon input affected soil microbial biomass and community composition in two tree species plantations in subtropical China. Acta Ecol. Sin. 2016, 36, 3582–3590. (In Chinese) [Google Scholar]
- Liu, J.; Liu, M.; Wu, M.; Jiang, C.; Chen, X.; Cai, Z.; Wang, B.; Zhang, J.; Zhang, T.; Li, Z. Soil pH rather than nutrients drive changes in microbial community following long-term fertilization in acidic Ultisols of southern China. J. Soil Sediments 2018, 18, 1853–1864. [Google Scholar] [CrossRef]
- Liu, G.Y.; Chen, L.L.; Shi, X.R.; Yuan, L.Y.; Lock, T.R.; Kallenbach, R.L. Changes in rhizosphere bacterial and fungal community composition with vegetation restoration in planted forests. Land Degrad. Dev. 2019, 30, 1147–1157. [Google Scholar] [CrossRef]
- Luo, Z.K.; Feng, W.T.; Luo, Y.Q.; Baldock, J.; Wang, E.L. Soil organic carbon dynamics jointly controlled by climate, carbon inputs, soil properties and soil carbon fractions. Glob. Chang. Biol. 2017, 23, 4430–4439. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Wang, L.; Hu, Y.; Tsang, Y.F.; Zhang, Y.; Wu, J.; Fu, X.; Sun, Y. Plant litter composition selects different soil microbial structures and in turn drives different litter decomposition pattern and soil carbon sequestration capability. Geoderma 2018, 319, 194–203. [Google Scholar] [CrossRef]
- Sang, R.; Li, S.; Huang, X.; Liu, W.; Liang, X.; Su, J. Effects of soil properties and plant diversity on soil microbial community composition and diversity during secondary succession. Forests 2021, 12, 805. [Google Scholar] [CrossRef]
- Hugenholtz, P.; Goebel, B.M.; Pace, N.R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 1998, 180, 4765–4774. [Google Scholar] [CrossRef] [Green Version]
- Barns, S.M.; Takala, S.L.; Kuske, C.R. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microb. 1999, 65, 1731–1737. [Google Scholar] [CrossRef] [Green Version]
- Bryant, D.A.; Costas, A.M.; Maresca, J.A.; Chew, A.G.; Klatt, C.G.; Bateson, M.M.; Tallon, L.J.; Hostetler, J.; Nelson, W.C.; Heidelberg, J.F.; et al. Candidatus Chloracidobacterium thermophilum: An aerobic phototrophic acidobacterium. Science 2007, 317, 523–526. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.P.; Gischkat, S.; Reiche, M.; Akob, D.M.; Hallberg, K.B.; Kusel, K. Ecophysiology of Fe-cycling bacteria in acidic sediments. Appl. Environ. Microb. 2010, 76, 8174–8183. [Google Scholar] [CrossRef] [Green Version]
- Pankratov, T.A.; Ivanova, A.O.; Dedysh, S.N.; Liesack, W. Bacterial populations and environmental factors controlling cellulose degradation in an acidic Sphagnum peat. Environ. Microbiol. 2011, 13, 1800–1814. [Google Scholar] [CrossRef]
- Sun, H.; Terhonena, E.; Koskinenb, K.; Paulinb, L.; Kasanena, R.; Asiegbu, F.O. Bacterial diversity and community structure along different peat soils in boreal forest. Appl. Soil Ecol. 2014, 74, 37–45. [Google Scholar] [CrossRef]
- McCaig, A.E.; Glover, L.A.; Prosser, J.I. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl. Environ. Microbiol. 1999, 65, 1721–1730. [Google Scholar] [CrossRef] [Green Version]
- Navarrete, I.A.; Tsutsuki, K. Land-use impact on soil carbon, nitrogen, neutral sugar composition and related chemical properties in a degraded Ultisol in Leyte, Philippines. Soil Sci. Plant Nutr. 2008, 54, 321–331. [Google Scholar] [CrossRef]
- Eilers, K.G.; Lauber, C.L.; Knight, R.; Fierer, N. Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biol. Biochem. 2010, 42, 896–903. [Google Scholar] [CrossRef]
- Wang, H.; Liu, S.; Zhang, X.; Mao, Q.; Li, X.; You, Y.; Wang, J.; Zheng, M.; Zhang, W.; Lu, X.; et al. Nitrogen addition reduces soil bacterial richness, while phosphorus addition alters community composition in an old-growth N-rich tropical forest in southern China. Soil Biol. Biochem. 2018, 127, 22–30. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungis. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [Green Version]
- Lopes, A.R.; Manaia, C.M.; Nunes, O.C. Bacterial community variations in an alfalfa-rice rotation system revealed by 16S rRNA gene 454-pyrosequencing. FEMS Microbiol. Ecol. 2014, 87, 650–663. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Oliva, F.; Sveshtarova, B.; Oliva, M. Seasonal effects on soil organic carbon dynamics in a tropical deciduous forest ecosystem in westernMexico. J. Trop. Ecol. 2003, 19, 179–188. [Google Scholar] [CrossRef]
- Beimforde, C.; Feldberg, K.; Nylinder, S.; Rikkinen, J.; Tuovila, H.; Dörfelt, H.; Gube, M.; Jackson, D.J.; Reitner, J.; Seyfullah, L.; et al. Estimating the phanerozoic history of the ascomycota lineages: Combining fossil and molecular data. Mol. Phylogenet. Evol. 2014, 78, 386–398. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, Z.; Li, Y.; Wang, G.; Liu, J.; Liu, J.; Liu, X.; Jin, J. Microbial association with the dynamics of particulate organic carbon in response to the amendment of elevated CO2-derived wheat residue into a Mollisol. Sci. Total Environ. 2017, 607–608, 972–981. [Google Scholar] [CrossRef] [PubMed]
- Chau, J.F.; Bagtzoglou, A.C.; Willig, M.R. The effect of soil texture on richness and diversity of bacterial communities. Environ. Forensics 2011, 12, 333–341. [Google Scholar] [CrossRef]
- Naether, A.; Foesel, B.U.; Naegele, V.; Wüst, P.K.; Weinert, J.; Bonkowski, M.; Alt, F.; Oelmann, Y.; Polle, A.; Lohaus, G.; et al. Environmental factors affect acidobacterial communities below the subgroup level in grassland and forest soils. Appl. Environ. Microbiol. 2012, 78, 7398–7406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Liu, N.; Zhang, Y. Soil aggregates regulate the impact of soil bacterial and fungal communities on soil respiration. Geoderma 2019, 337, 444–452. [Google Scholar] [CrossRef]
- Liu, J.; Sui, Y.; Yu, Z.; Shi, Y.; Chu, H.; Jin, J.; Liu, X.; Wang, G. High throughput sequencing analysis of biogeographical distribution of bacterial communities in the black soils of northeast China. Soil Biol. Biochem. 2014, 70, 113–122. [Google Scholar] [CrossRef]
- Zhong, Y.; Yan, W.; Shangguan, Z. Impact of long-term N additions upon coupling between soil microbial community structure and activity, and nutrient-use efficiencies. Soil Biol. Biochem. 2015, 91, 151–159. [Google Scholar] [CrossRef]
- Zeng, Q.; Liu, Y.; An, S. Impact of litter quantity on the soil bacteria community during the decomposition of Quercus wutaishanica litter. PeerJ 2017, 5, e3777. [Google Scholar] [CrossRef]
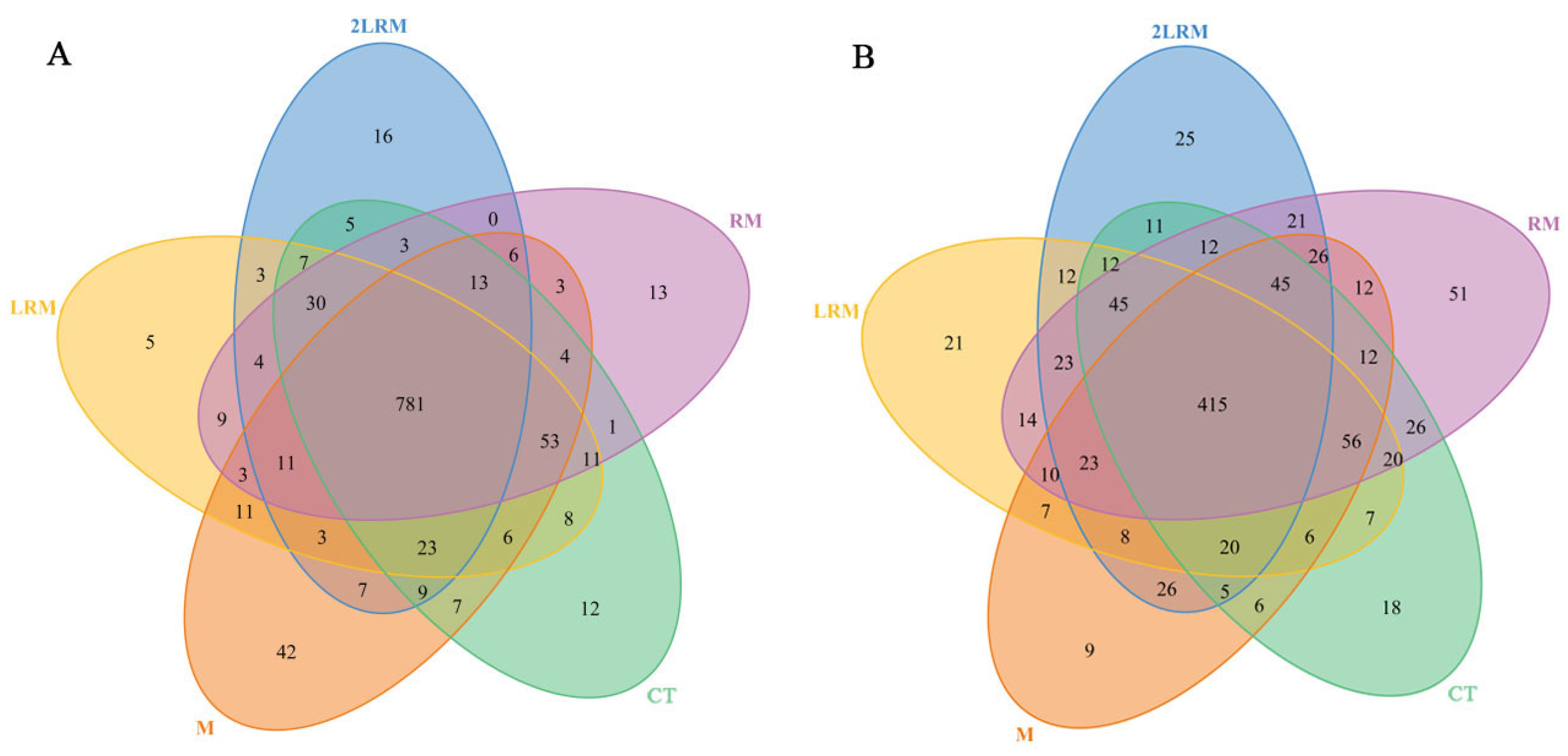

| Treament | Abbreviation | Description |
|---|---|---|
| Control treatment | CT | Removing aboveground litter + removing root + removing mycorrhiza |
| Mycorrhiza | M | Removing aboveground litter + removing root + retaining mycorrhiza |
| Root + mycorrhiza | RM | Removing aboveground litter + retaining root + retaining mycorrhiza |
| Litter + root + mycorrhiza | LRM | Retaining aboveground litter + retaining root + retaining mycorrhiza |
| Double litter+ root + mycorrhiza | DLRM | Doubling aboveground litter + retaining root + retaining mycorrhiza |
| Index | CT | M | RM | LRM | DLRM |
|---|---|---|---|---|---|
| Chemical properties | |||||
| pH | 4.1 ± 0.1 a | 4.2 ± 0.2 a | 4.1 ± 0.02 a | 4.0 ± 0.1 a | 4.2 ± 0.04 a |
| SOC/(g kg−1) | 29 ± 3 b | 30 ± 3 b | 30 ± 3 b | 33 ± 2 b | 45 ± 6 a |
| TN/(g kg−1) | 2.1 ± 0.2 b | 2.2 ± 0.2 ab | 2.2 ± 0.2 ab | 2.3 ± 0.1 ab | 2.8 ± 0.3 a |
| AP/(mg kg−1) | 0.03 ± 0.003 a | 0.03 ± 0.002 a | 0.04 ± 0.008 a | 0.03 ± 0.003 a | 0.04 ± 0.005 a |
| AK/(mg kg−1) | 96 ± 6 a | 116 ± 7 a | 77 ± 19 a | 96 ± 13 a | 105 ± 10 a |
| Enzymatic activity | |||||
| SC/(Ug−1) | 7.1 ± 1.0 a | 11 ± 3 a | 16 ± 5 a | 10 ± 1 a | 16 ± 2 a |
| UE/(Ug−1) | 658 ± 98 a | 767 ± 77 a | 712 ± 105 a | 687 ± 41 a | 787 ± 55 a |
| ACP/(Ug−1) | 48,620 ± 1460 a | 45,954 ± 340 a | 45,324 ± 1360 a | 48,984 ± 3409 a | 44,593 ± 3002 a |
| AI/(Ug−1) | 36 ± 9 ab | 38 ± 17 ab | 34 ± 2 ab | 25 ± 5 a | 24 ± 7 b |
| PPO/(Ug−1) | 49 ± 5 a | 49 ± 6 a | 32 ± 5 b | 19 ± 2 b | 105 ± 10 b |
| Type | Treatment | Population/ (106 · g−1 Dry Soil) | OTUs | Richness | Diversity | ||
|---|---|---|---|---|---|---|---|
| ACE | Chao1 | Simpson | Shannon | ||||
| Bacteria | CT | 1982 ± 568 a | 852 ± 22 a | 920 ± 20 a | 946 ± 25 a | 0.99 ± 0.001 a | 7.9 ± 0.05 a |
| M | 892 ± 109 b | 790 ± 40 a | 858 ± 44 a | 864 ± 50 a | 0.99 ± 0.001 a | 7.8 ± 0.11 ab | |
| RM | 413 ± 239 b | 799 ± 9 a | 880 ± 3 a | 898 ± 8 a | 0.99 ± 0.001 a | 7.6 ± 0.01 ab | |
| LRM | 652 ± 338 b | 789 ± 36 a | 880 ± 30 a | 889 ± 27 a | 0.98 ± 0.008 a | 7.3 ± 0.38 b | |
| DLRM | 875 ± 171 b | 782 ± 29 a | 852 ± 34 a | 858 ± 37 a | 0.99 ± 0.001 a | 7.7 ± 0.05 ab | |
| Fungi | CT | 39 ± 7 a | 501 ± 17 b | 590 ± 13 b | 580 ± 14 b | 0.91 ± 0.023 a | 5.2 ± 0.31 b |
| M | 59 ± 7 a | 686 ± 32 a | 791 ± 32 a | 804 ± 35 a | 0.97 ± 0.026 a | 6.7 ± 0.39 a | |
| RM | 54 ± 36 a | 664 ± 49 a | 760 ± 62 a | 768 ± 66 a | 0.92 ± 0.019 a | 6.0 ± 0.26 ab | |
| LRM | 77 ± 57 a | 470 ± 24 b | 566 ± 14 b | 579 ± 24 b | 0.91 ± 0.054 a | 5.2 ± 0.81 b | |
| DLRM | 111 ± 20 a | 624 ± 38 a | 738 ± 37 a | 743 ± 38 a | 0.94 ± 0.010 a | 5.9 ± 0.17 ab | |
| Type | Class | CT | M | RM | LRM | DLRM |
|---|---|---|---|---|---|---|
| Bacteria | Acidobacteriia | 36 a | 28 a | 42 a | 39 a | 30 a |
| Alphaproteobacteria | 24 a | 25 a | 22 a | 11 b | 28 a | |
| Actinobacteria | 6.2 a | 6.7 a | 7.3 a | 7.4 a | 6.3 a | |
| Gammaproteobacteria | 5.9 a | 7.4 a | 5.7 a | 6.1 a | 8.3 a | |
| Thermoleophilia | 5.4 bc | 9.8 a | 4.6 bc | 2.3 c | 7.3 ab | |
| Acidimicrobiia | 3.4 a | 5.3 a | 4.8 a | 4.6 a | 5.7 a | |
| uncultured_bacterium_p_WPS−2 | 3.4 a | 4.7 a | 3.9 a | 9.5 a | 1.6 a | |
| Verrucomicrobiae | 3.5 a | 3.1 a | 1.5 a | 5.0 a | 2.7 a | |
| Planctomycetacia | 2.1 a | 1.3 a | 1.4 a | 4.5 a | 0.7 a | |
| Ktedonobacteria | 3.0 a | 1.3 ab | 1.2 ab | 2.5 ab | 0.47 b | |
| Saccharimonadia | 1.8 ab | 2.8 a | 1.1 b | 1.3 b | 1.3 b | |
| Deltaproteobacteria | 1.1 b | 1.1 b | 1.1 b | 0.83 b | 1.8 a | |
| AD3 | 0.78 a | 0.81 a | 0.79 a | 2.0 a | 0.38 a | |
| Bacteroidia | 1.0 a | 0.42 b | 0.22 b | 0.37 b | 1.0 a | |
| Gemmatimonadetes | 0.70 ab | 0.35 b | 0.40 b | 0.33 b | 0.94 a | |
| JG30-KF-CM66 | 0.39 a | 0.32 a | 0.40 a | 0.63 a | 0.22 a | |
| Thermoplasmata | 0.33 b | 0.18 b | 0.04 b | 0.16 b | 1.1 a | |
| Fimbriimonadia | 0.17 b | 0.32 a | 0.14 b | 0.11 b | 0.42 a | |
| Phycisphaerae | 0.27 a | 0.32 a | 0.10 a | 0.32 a | 0.13 a | |
| Subgroup_6 | 0.18 a | 0.16 a | 0.12 a | 0.50 a | 0.11 a | |
| Fungi | Agaricomycetes | 45 a | 41 a | 58 a | 36 a | 56 a |
| Sordariomycetes | 15 a | 16 a | 7.8 b | 7.8 b | 5.3 b | |
| Leotiomycetes | 13 a | 12 a | 9.7 a | 7.1 a | 3.7 a | |
| Mortierellomycetes | 1.2 d | 2.4 cd | 4.7 b | 4.1 bc | 8.4 a | |
| Eurotiomycetes | 3.2 a | 4.1 a | 3.7 a | 3.7 a | 3.2 a | |
| Dothideomycetes | 1.4 ab | 1.3 ab | 1.8 a | 0.65 b | 1.5 ab | |
| Rozellomycotina_cls_Incertae_sedis | 3.6 a | 0.55 a | 0.24 a | 1.2 a | 0.46 a | |
| Chytridiomycetes | 0.22 b | 1.0 a | 0.20 b | 0.20 b | 0.13 b | |
| Archaeorhizomycetes | 0.16 a | 0.52 a | 0.27 a | 0.15 a | 0.44 a | |
| GS13 | 0.05 b | 0.18 b | 0.51 a | 0.04 b | 0.11 b | |
| Calcarisporiellomycetes | 0.09 b | 0.26 a | 0.10 b | 0.06 b | 0.25 a | |
| Umbelopsidomycetes | 0.05 a | 0.05 a | 0.06 a | 0.11 a | 0.07 a | |
| Tremellomycetes | 0.04 a | 0.05 a | 0.02 bc | 0.02 c | 0.04 ab | |
| Pezizomycetes | 0.004 c | 0.07 a | 0.03 bc | 0.002 c | 0.04 b | |
| Orbiliomycetes | 0.03 a | 0.03 a | 0.02 a | 0.01 a | 0.03 a | |
| Rhizophydiomycetes | 0.01 a | 0.01 a | 0.04 a | 0.01 a | 0.03 a | |
| Microbotryomycetes | 0.01 b | 0.01 b | 0.02 ab | 0.01 b | 0.04 a | |
| Cystobasidiomycetes | 0.005 a | 0.01 a | 0.02 a | 0.01 a | 0.01 a | |
| Saccharomycetes | 0.01 a | 0.01 a | 0.004 a | 0.01 a | 0.01 a |
| Class | Alpha Diversity | Soil Chemical Properties and Enzymatic Activity | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACE | Chao1 | Simpson | Shannon | pH | SOC | TN | AP | AK | SC | UE | ACP | AI | PPO | |
| Bacteria | ||||||||||||||
| Acidobacteriia | 0.047 | 0.098 | −0.372 | −0.337 | −0.439 | −0.004 | 0.085 | 0.285 | 0.077 | 0.044 | 0.063 | 0.015 | −0.209 | 0.054 |
| Alphaproteobacteria | 0.033 | 0.052 | 0.519 * | 0.483 | 0.025 | 0.085 | 0.160 | 0.240 | 0.056 | 0.154 | 0.115 | −0.011 | −0.572 * | 0.279 |
| Actinobacteria | 0.078 | 0.045 | 0.633 * | 0.536 * | 0.419 | −0.178 | −0.188 | −0.184 | −0.402 | 0.140 | −0.133 | 0.065 | 0.104 | 0.044 |
| Gammaproteobacteria | −0.038 | −0.095 | 0.279 | 0.246 | 0.048 | 0.213 | 0.349 | 0.263 | 0.300 | −0.140 | 0.007 | 0.012 | −0.504 | 0.053 |
| Thermoleophilia | −0.300 | −0.311 | 0.501 | 0.442 | 0.169 | 0.168 | 0.231 | 0.213 | 0.351 | −0.070 | 0.291 | −0.218 | −0.519 * | 0.235 |
| Acidimicrobiia | −0.261 | −0.296 | 0.564 * | 0.421 | 0.418 | 0.156 | 0.104 | −0.008 | −0.179 | 0.256 | 0.149 | −0.226 | −0.037 | −0.206 |
| uncultured_bacterium_p_WPS−2 | 0.071 | 0.057 | 0.027 | 0.027 | 0.070 | −0.42 | −0.414 | −0.397 | −0.215 | −0.082 | −0.175 | 0.100 | 0.556 * | 0.184 |
| Verrucomicrobiae | 0.356 | 0.295 | 0.13 | 0.250 | 0.262 | 0.018 | −0.069 | −0.242 | 0.096 | −0.261 | −0.165 | 0.212 | 0.212 | 0.105 |
| Planctomycetacia | 0.313 | 0.292 | 0.019 | 0.085 | 0.130 | −0.344 | −0.396 | −0.422 | −0.261 | −0.152 | −0.301 | 0.304 | 0.525 * | 0.154 |
| Ktedonobacteria | 0.589 * | 0.591 * | 0.143 | 0.300 | 0.004 | −0.446 | −0.465 | −0.391 | −0.235 | −0.297 | −0.310 | 0.284 | 0.455 | 0.452 |
| Saccharimonadia | 0.061 | 0.007 | 0.672 ** | 0.676 ** | 0.319 | −0.219 | −0.174 | −0.263 | 0.124 | −0.250 | −0.014 | 0.073 | 0.039 | 0.510 |
| Deltaproteobacteria | −0.074 | −0.064 | 0.257 | 0.214 | 0.069 | 0.402 | 0.416 | 0.339 | 0.113 | 0.238 | 0.038 | −0.138 | −0.703 ** | −0.067 |
| Fungi | ||||||||||||||
| Agaricomycetes | 0.262 | 0.238 | 0.147 | 0.173 | 0.151 | 0.139 | 0.118 | 0.299 | −0.085 | 0.277 | 0.116 | −0.324 | −0.453 | −0.300 |
| Sordariomycetes | 0.040 | −0.007 | 0.613 * | 0.561 * | −0.185 | −0.465 | −0.335 | −0.202 | 0.209 | −0.425 | −0.187 | 0.227 | −0.150 | 0.762 ** |
| Leotiomycetes | 0.144 | 0.115 | 0.193 | 0.298 | −0.152 | −0.423 | −0.304 | −0.071 | −0.031 | −0.138 | 0.036 | 0.287 | 0.217 | 0.578 * |
| Mortierellomycetes | 0.363 | 0.371 | 0.389 | 0.383 | −0.119 | 0.632 * | 0.578 * | 0.422 | 0.013 | 0.576 * | 0.203 | −0.338 | −0.255 | −0.662 ** |
| Eurotiomycetes | 0.133 | 0.105 | 0.611 * | 0.666 ** | −0.138 | −0.190 | 0.025 | 0.143 | −0.026 | −0.028 | −0.008 | 0.360 | −0.160 | 0.252 |
| Dothideomycetes | 0.441 | 0.397 | 0.563 * | 0.589 * | 0.004 | −0.033 | −0.033 | 0.052 | −0.299 | 0.114 | −0.267 | −0.056 | −0.600 * | 0.294 |
| Rozellomycotina_cls_Incertae_sedis | −0.294 | −0.273 | −0.112 | −0.254 | −0.127 | −0.172 | −0.153 | −0.323 | −0.049 | −0.288 | −0.549 * | −0.084 | −0.088 | 0.331 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, L.; Huang, R.; Wang, J.; Huang, G.; Guan, H.; Lin, L.; Yang, M.; Li, Y.; Zou, X. Litter, Root, and Mycorrhiza Input Affected Soil Microbial Community Structure in Schima superba Pure Forest in Subtropical China. Diversity 2023, 15, 82. https://doi.org/10.3390/d15010082
Zhu L, Huang R, Wang J, Huang G, Guan H, Lin L, Yang M, Li Y, Zou X. Litter, Root, and Mycorrhiza Input Affected Soil Microbial Community Structure in Schima superba Pure Forest in Subtropical China. Diversity. 2023; 15(1):82. https://doi.org/10.3390/d15010082
Chicago/Turabian StyleZhu, Liqin, Rongzhen Huang, Jinping Wang, Guomin Huang, Hongzhi Guan, Lijing Lin, Mengjia Yang, Yanyan Li, and Xianhua Zou. 2023. "Litter, Root, and Mycorrhiza Input Affected Soil Microbial Community Structure in Schima superba Pure Forest in Subtropical China" Diversity 15, no. 1: 82. https://doi.org/10.3390/d15010082
APA StyleZhu, L., Huang, R., Wang, J., Huang, G., Guan, H., Lin, L., Yang, M., Li, Y., & Zou, X. (2023). Litter, Root, and Mycorrhiza Input Affected Soil Microbial Community Structure in Schima superba Pure Forest in Subtropical China. Diversity, 15(1), 82. https://doi.org/10.3390/d15010082






