Effect of Cultivable Bacteria and Fungi on the Limestone Weathering Used in Historical Buildings
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteria and Fungi Extraction, Purification and Identification
2.2. Materials
2.3. Experimental Set up of Limestone Biodissolution
2.4. Solution Analyses
2.5. Solid Analyses
2.6. Statistical Analysis
3. Results
3.1. Identification of Bacterial and Fungal Strains
3.2. Bacterial and Fungal Control of Limestone Dissolution
3.3. Biocolonisation Assessment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mihajlovski, A.; Seyer, D.; Benamara, H.; Bousta, F.; Di Martino, P. An overview of techniques for the characterization and quantification of microbial colonization on stone monuments. Ann. Microbiol. 2015, 65, 1243–1255. [Google Scholar] [CrossRef]
- Paine, S.; Linggood, F.; Schimmer, F.; Thrupp, T. The Relationship of Micro-Organisms to the Decay of Stone. Philos. Trans. R Soc. London Ser. B. 1932, 222, 97–127. [Google Scholar]
- Villa, F.; Vasanthakumar, A.; Mitchell, R.; Cappitelli, F. RNA-based molecular survey of biodiversity of limestone tombstone microbiota in response to atmospheric sulphur pollution. Lett. Appl. Microbiol. 2015, 60, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Gutarowska, B.; Celikkol-Aydin, S.; Bonifay, V.; Otlewska, A.; Aydin, E.; Oldham, A.L.; Brauer, J.I.; Duncan, K.E.; Adamiak, J.; Sunner, J.; et al. Metabolomic and high-throughput sequencing analysis-modern approach for the assessment of biodeterioration of materials from historic buildings. Front. Microbiol. 2015, 6, 979. [Google Scholar] [CrossRef]
- Brewer, T.E.; Fierer, N. Tales from the tomb: The microbial ecology of exposed rock surfaces. Environ. Microbiol. 2018, 20, 958–970. [Google Scholar] [CrossRef]
- Gadd, G.M. Geomycology: Biogeochemical transformations of rocks, minerals, metals and radionuclides by fungi, bioweathering and bioremediation. Mycol. Res. 2007, 111, 3–49. [Google Scholar] [CrossRef] [PubMed]
- Popović, S.; Stupar, M.; Unković, N.; Subakov Simić, G.; Ljaljević Grbić, M. Diversity of Terrestrial Cyanobacteria Colonizing Selected Stone Monuments in Serbia. Stud. Conserv. 2018, 63, 292–302. [Google Scholar] [CrossRef]
- Scheerer, S.; Ortega-Morales, O.; Gaylarde, C. Chapter 5 Microbial Deterioration of Stone Monuments—An Updated Overview. Adv. Appl. Microbiol. 2009, 66, 97–139. [Google Scholar]
- Krumbein, W.E. Zur Frage der biologischen Verwitterung: Einfluss der Mikroflora auf die Bausteinverwitterung und ihre Abhängigkeit von edapischen Faktoren. Z Allg. Mikrobiol. 1968, 8, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Trovão, J.; Portugal, A.; Soares, F.; Paiva, D.S.; Mesquita, N.; Coelho, C.; Pinheiro, A.C.; Catarino, L.; Gil, F.; Tiago, I. Fungal diversity and distribution across distinct biodeterioration phenomena in limestone walls of the old cathedral of Coimbra, UNESCO World Heritage Site. Int. Biodeterior. Biodegrad. 2019, 142, 91–102. [Google Scholar] [CrossRef]
- Balland, C.; Poszwa, A.; Leyval, C.; Mustin, C. Dissolution rates of phyllosilicates as a function of bacterial metabolic diversity. Geochim. Cosmochim. Acta 2010, 74, 5478–5493. [Google Scholar] [CrossRef]
- Bousserrhine, N.; Gasser, U.G.; Jeanroy, E.; Berthelin, J. Bacterial and chemical reductive dissolution of Mn-, Co-, Cr-, and Al-substituted goethites. Geomicrobiol. J. 1999, 16, 245–258. [Google Scholar] [CrossRef]
- Pinheiro, A.C.; Mesquita, N.; Trovão, J.; Soares, F.; Tiago, I.; Coelho, C.; de Carvalho, H.P.; Gil, F.; Catarino, L.; Piñar, G.; et al. Limestone biodeterioration: A review on the Portuguese cultural heritage scenario. J. Cult. Herit. 2018, 36, 275–285. [Google Scholar] [CrossRef]
- Abdel Ghany, T.M.; Omar, A.M.; Elwkeel, F.M.; Al Abboud, M.A.; Alawlaqi, M.M. Fungal deterioration of limestone false-door monument. Heliyon 2019, 5, e02673. [Google Scholar] [CrossRef] [PubMed]
- Friis, A.K.; Davis, T.A.; Figueira, M.M.; Paquette, J.; Mucci, A. Influence of Bacillus subtilis cell walls and EDTA on calcite dissolution rates and crystal surface features. Environ. Sci. Technol. 2003, 37, 2376–2382. [Google Scholar] [CrossRef]
- Jacobson, A.D.; Wu, L. Microbial dissolution of calcite at T = 28 °C and ambient pCO2. Geochim. Cosmochim. Acta 2009, 73, 2314–2331. [Google Scholar] [CrossRef]
- Li, W.; Yu, L.J.; Wu, Y.; Jia, L.P.; Yuan, D.X. Enhancement of Ca2+ release from limestone by microbial extracellular carbonic anhydrase. Bioresour. Technol. 2007, 98, 950–953. [Google Scholar] [CrossRef]
- Lüttge, A.; Conrad, P.G. Direct Observation of Microbial Inhibition of Calcite Dissolution. Appl. Environ. Microbiol. 2004, 70, 1627–1632. [Google Scholar] [CrossRef]
- Balland-Bolou-Bi, C.; Saheb, M.; Bousserrhine, N.; Abbad-Andaloussi, S.; Alphonse, V.; Nowak, S.; Chabas, A.; Verney-Carron, A.; Desboeufs, K. Effect of Microorganisms Activities in A Polluted Area on the Alteration of Limestone Used in Historical Buildings. Sci. Art A Future Stone 2016, 1, 25–33. [Google Scholar]
- Rozenbaum, O.; Barbanson, L.; Muller, F.; Bruand, A. Significance of a combined approach for replacement stones in the heritage buildings’ conservation frame. Comptes Rendus Geosci. 2008, 340, 345–355. [Google Scholar] [CrossRef]
- Fronteau, G.; Moreau, C.; Thomachot-Schneider, C.; Barbin, V. Variability of some Lutetian building stones from the Paris Basin, from characterisation to conservation. Eng. Geol. 2010, 115, 158–166. [Google Scholar] [CrossRef]
- Angeli, M.; Bigas, J.P.-P.; Menendez, B.; Hebert, R.; David, C. Influence of capillary properties and evaporation on salt weathering of sedimentary rocks. In Heritage, Weathering and Conservation; Taylor&Francis/Balkema: Leiden, The Netherlands, 2006; pp. 253–259. [Google Scholar]
- Angeli, M.; Benavente, D.; Bigas, J.-P.; Menéndez, B.; Hébert, R.; David, C. Modification of the porous network by salt crystallization in experimentally weathered sedimentary stones. Mater. Struct. 2008, 41, 1091–1108. [Google Scholar] [CrossRef]
- Saheb, M.; Chabas, A.; Mertz, J.-D.; Colas, E.; Rozenbaum, O.; Sizun, J.-P.; Nowak, S.; Gentaz, L.; Verney-Carron, A. Weathering of limestone after several decades in an urban environment. Corros. Sci. 2016, 111, 742–752. [Google Scholar] [CrossRef]
- Van Hees, P.A.W.; Jones, D.L.; Jentschke, G.; Godbold, D.L. Organic acid concentrations in soil solution: Effects of young coniferous trees and ectomycorrhizal fungi. Soil Biol. Biochem. 2005, 37, 771–776. [Google Scholar] [CrossRef]
- Sazanova, K.V.; Zelenskaya, M.S.; Vlasov, A.D.; Bobir, S.Y.; Yakkonen, K.L.; Vlasov, D.Y. Microorganisms in Superficial Deposits on the Stone Monuments in Saint Petersburg. Microorganisms 2022, 10, 316. [Google Scholar] [CrossRef]
- Wu, F.; Zhang, Y.; Gu, J.-D.; He, D.; Zhang, G.; Liu, X.; Guo, Q.; Cui, H.; Zhao, J.; Feng, H. Community assembly, potential functions and interactions between fungi and microalgae associated with biodeterioration of sandstone at the Beishiku Temple in Northwest China. Sci. Total. Environ. 2022, 835, 155372. [Google Scholar] [CrossRef] [PubMed]
- Subrahmanyam, G.; Vaghela, R.; Bhatt, N.P.; Archana, G. Carbonate-Dissolving Bacteria from ‘Miliolite’, a Bioclastic Limestone, from Gopnath, Gujarat, Western India. Microbes Environ. 2012, 27, 334–337. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, B.; Yang, X.; Ge, Q. Deterioration-associated microbiome of stone monuments: Structure, variation, and assembly. Appl. Environ. Microbiol. 2018, 84, e02680-17. [Google Scholar] [CrossRef]
- Gottschalk, G. Bacterial Metabolism; Springer: New-York, NY, USA, 1986; 359p. [Google Scholar]
- Negi, A.; Sarethy, I.P. Microbial Biodeterioration of Cultural Heritage: Events, Colonization, and Analyses. Microb. Ecol. 2019, 78, 1014–1029. [Google Scholar] [CrossRef]
- Welch, S.A.; Ullman, W.J. The effect of microbial glucose metabolism on bytownite feldspar dissolution rates between 5° and 35 °C. Geochim. Cosmochim. Acta 1999, 63, 3247–3259. [Google Scholar] [CrossRef]
- Huang, L.; Cao, J.; He, X.; Yang, H.; Li, X.; Shen, H. Simulation experiment on limestone dissolution by some low-molecular-weight organic acids. Earth Environ. 2006, 34, 44–50. [Google Scholar]
- Morse, J.W.; Mackenzie, F.T. Geochemistry of Sedimentary Carbonates, Developments in Sedimentology; Elsevier: Amsterdam, The Netherlands, 1990; Volume 48, p. 707. [Google Scholar]
- Franke, W.A.; Teschner-Steinhardt, R. An experimental approach to the sequence of the stability of rock-forming minerals towards chemical weathering. Catena 1994, 21, 279–290. [Google Scholar] [CrossRef]
- Roussel, E.; André, M. Quantitative assessment of pre- and post-restoration weathering rates of limestone Mayan temples (Uxmal, Yucatán). Geogr. Fis. Din. Quat. 2013, 36, 169–179. [Google Scholar]
- Swoboda-Colberg, N.G.; Drever, J.I. Mineral dissolution rates in plot-scale field and laboratory experiments. Chem. Geol. 1993, 105, 51–69. [Google Scholar] [CrossRef]
- Fiol, L.; Fornos, J.J.; Gines, A. Effects of biokarstic processes on the development of sollutional Rillenkarren in limestone rocks. Earth Surf. Process Landf. 1996, 21, 447–452. [Google Scholar] [CrossRef]
- Li, W.; Zhou, P.P.; Jia, L.P.; Yu, L.J.; Li, X.L.; Zhu, M. Limestone Dissolution Induced by Fungal Mycelia, Acidic Materials, and Carbonic Anhydrase from Fungi. Mycopathologia 2009, 167, 37–46. [Google Scholar] [CrossRef]
- Little, B.J.; Wagner, P.A.; Lewandowski, Z. Spatial relationships between bacteria and mineral surfaces. Rev. Mineral. 1997, 35, 123–159. [Google Scholar]
- Barker, W.W.; Banfield, J.F. Zones of chemical and physical interaction at interfaces between microbial communities and minerals: A model. Geomicrobiol. J. 1998, 15, 223–244. [Google Scholar] [CrossRef]
- Sulu-Gambari, F. Bacterially-Induced Dissolution of Calcite: The Role of Bacteria in Limestone Weathering. Ph.D. Thesis, McGill University, Montréal, QC, Canada, 2011. [Google Scholar]
- Rampazzi, L.; Andreotti, A.; Bonaduce, I.; Colombini, M.; Colombo, C.; Toniolo, L. Analytical investigation of calcium oxalate films on marble monuments. Talanta 2004, 63, 967–977. [Google Scholar] [CrossRef]
- Rosado, T.; Gil, M.; Mirão, J.; Candeias, A.; Caldeira, A.T. Oxalate biofilm formation in mural paintings due to microorganisms—A comprehensive study. Int. Biodeterior. Biodegrad. 2013, 85, 1–7. [Google Scholar] [CrossRef]
- Rosado, T.; Silva, M.; Galvão, A.; Mirão, J.; Candeias, A.; Caldeira, A.T. A first insight on the biodegradation of limestone: The case of the World Heritage Convent of Christ. Appl. Phys. A 2016, 122, 1012. [Google Scholar] [CrossRef]
- Mitchell, R.; Gu, J.D. Changes in the biofilm microflora of limestone caused by atmospheric pollutants. Int. Biodeterior. Biodegrad. 2000, 46, 299–303. [Google Scholar] [CrossRef]
- Herrera, A.; Cockell, C.S.; Self, S.; Blaxter, M.; Reitner, J.; Arp, G.; Dröse, W.; Thorsteinsson, T.; Tindle, A.G. Bacterial Colonization and Weathering of Terrestrial Obsidian in Iceland. Geomicrobiol. J. 2008, 25, 25–37. [Google Scholar] [CrossRef]
- Lepleux, C.; Turpault, M.P.; Oger, P.; Frey-Klett, P.; Uroz, S. Correlation of the abundance of betaproteobacteria on mineral surfaces with mineral weathering in forest soils. Appl. Environ. Microbiol. 2012, 78, 7114–7119. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, B.; He, Z.; Yang, X. Distribution and diversity of bacteria and fungi colonization in stone monuments analyzed by high-throughput sequencing. PLoS ONE 2016, 11, e0163287. [Google Scholar] [CrossRef]
- Frey, B.; Rieder, S.R.; Brunner, I.; Plötze, M.; Koetzsch, S.; Lapanje, A.; Brandl, H.; Furrer, G. Weathering-Associated Bacteria from the Damma Glacier Forefield: Physiological Capabilities and Impact on Granite Dissolution. Appl. Environ. Microbiol. 2010, 76, 4788–4796. [Google Scholar] [CrossRef] [PubMed]
- Hutchens, E. Microbial selectivity on mineral surfaces: Possible implications for weathering processes. Fungal Biol. Rev. 2009, 23, 115–121. [Google Scholar] [CrossRef]
- Guillitte, O. Bioreceptivity: A new concept for building ecology studies. Sci. Total Environ. 1995, 167, 215–220. [Google Scholar] [CrossRef]
- Miller, A.; Sanmartín, P.; Pereira-Pardo, L.; Dionísio, A.; Saiz-Jimenez, C.; Macedo, M.; Prieto, B. Bioreceptivity of building stones: A review. Sci. Total. Environ. 2012, 426, 1–12. [Google Scholar] [CrossRef]
- Ortega-Morales, O.; Montero-Muñoz, J.L.; Baptista Neto, J.A.; Beech, I.B.; Sunner, J.; Gaylarde, C. Deterioration and microbial colonization of cultural heritage stone buildings in polluted and unpolluted tropical and subtropical climates: A meta-analysis. Int. Biodeterior. Biodegrad. 2019, 143, 104734. [Google Scholar] [CrossRef]
- Zanardini, E.; Abbruscato, P.; Ghedini, N.; Realini, M.; Sorlini, C. Influence of atmospheric pollutants on the biodeterioration of stone. Int. Biodeterior. Biodegrad. 2000, 45, 35–42. [Google Scholar] [CrossRef]
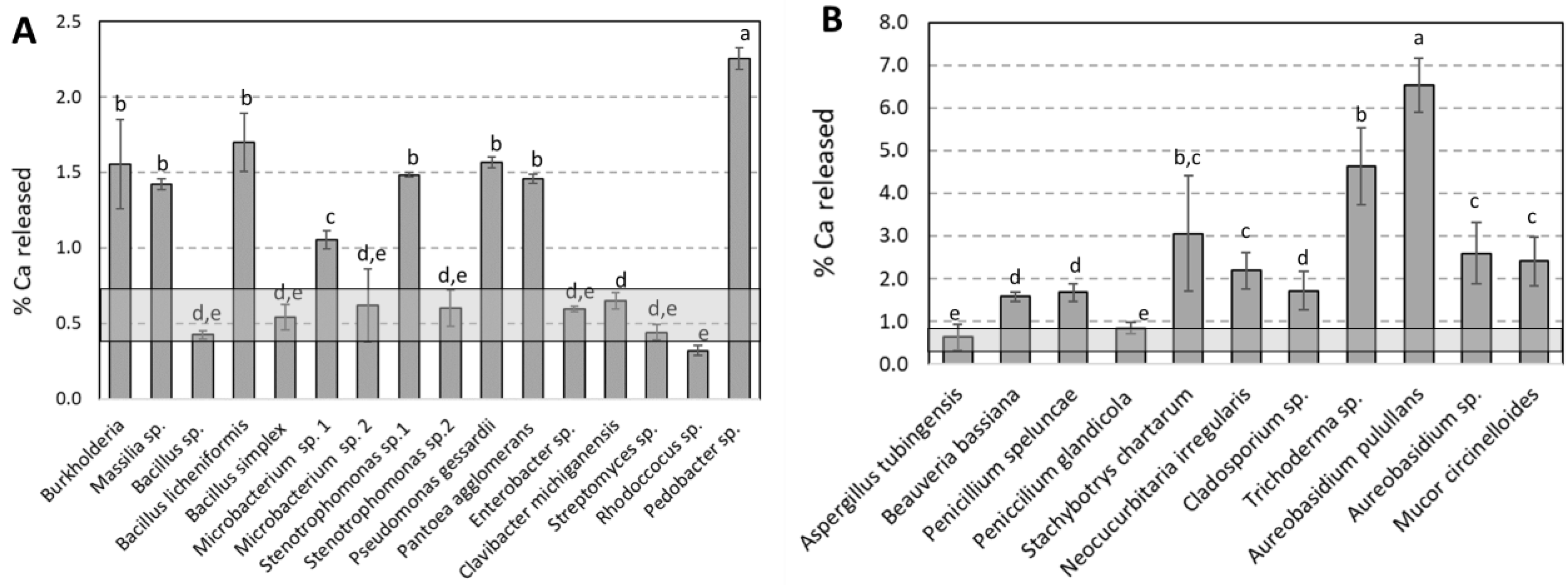
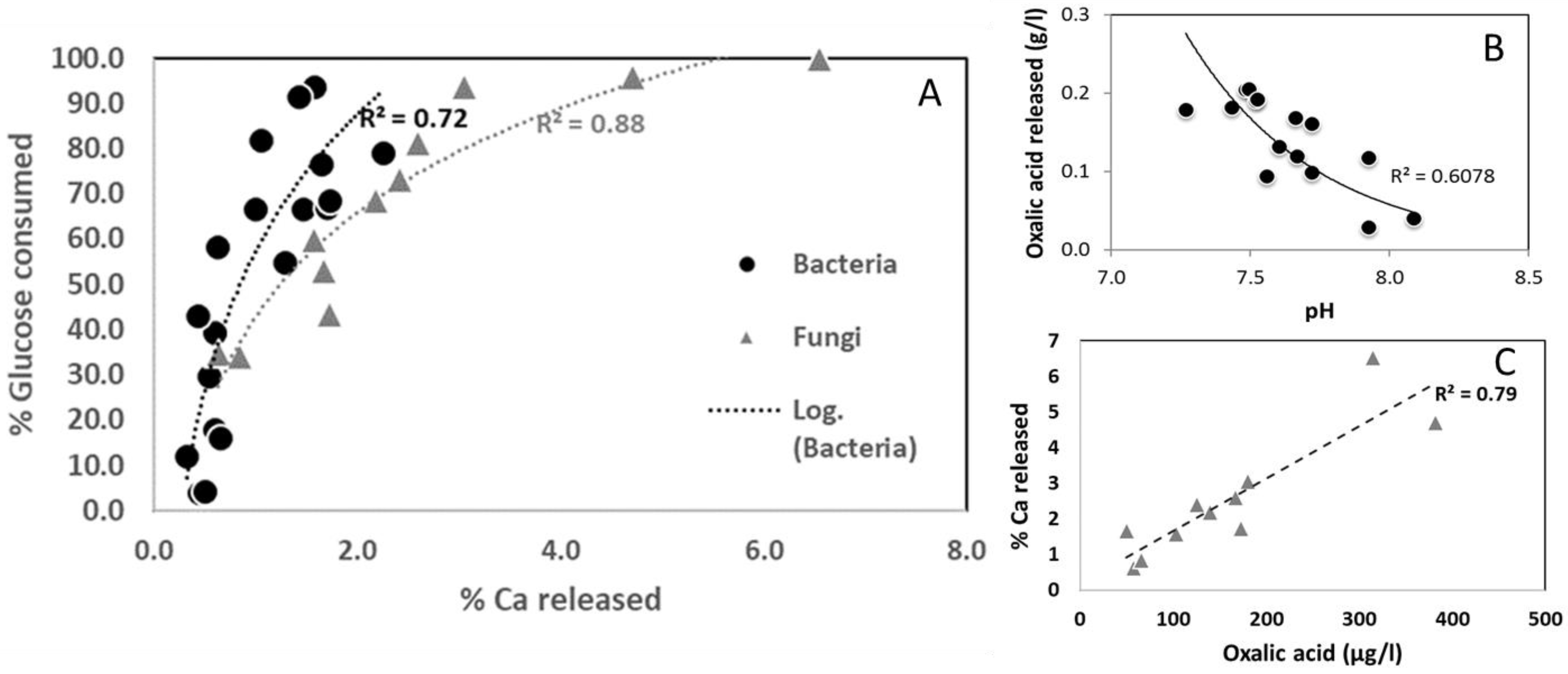
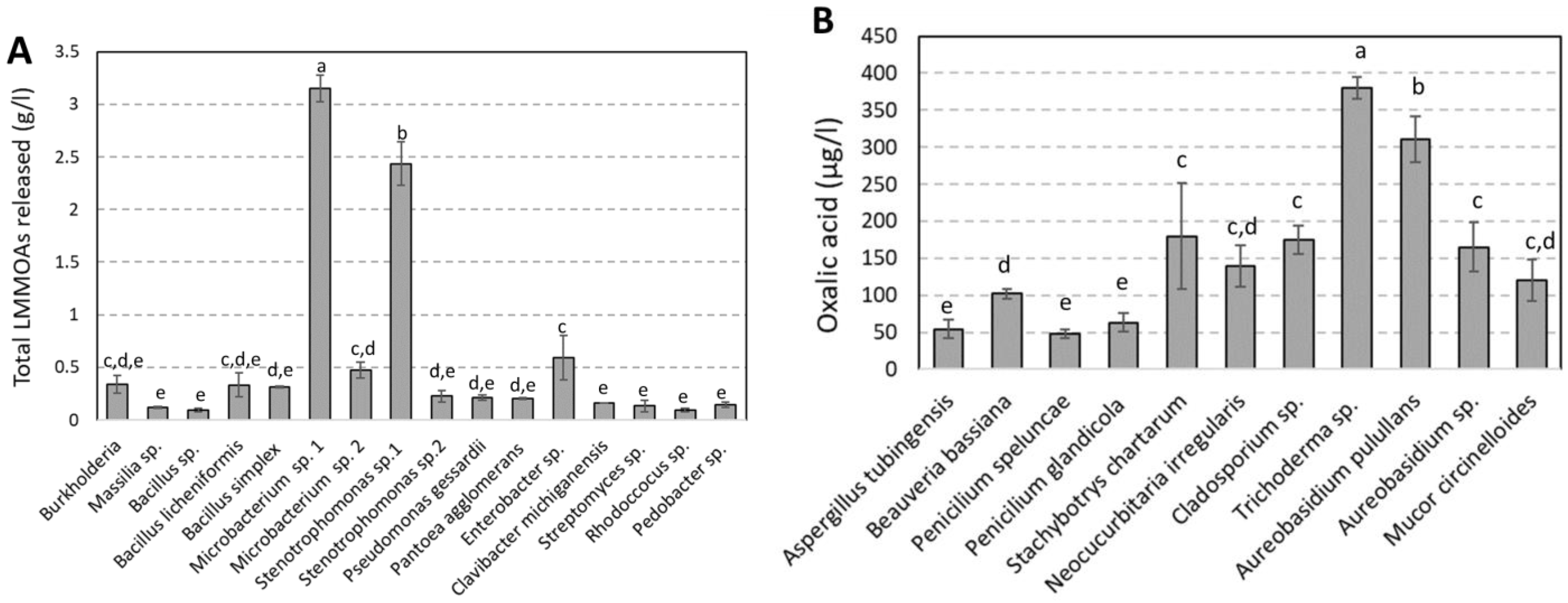
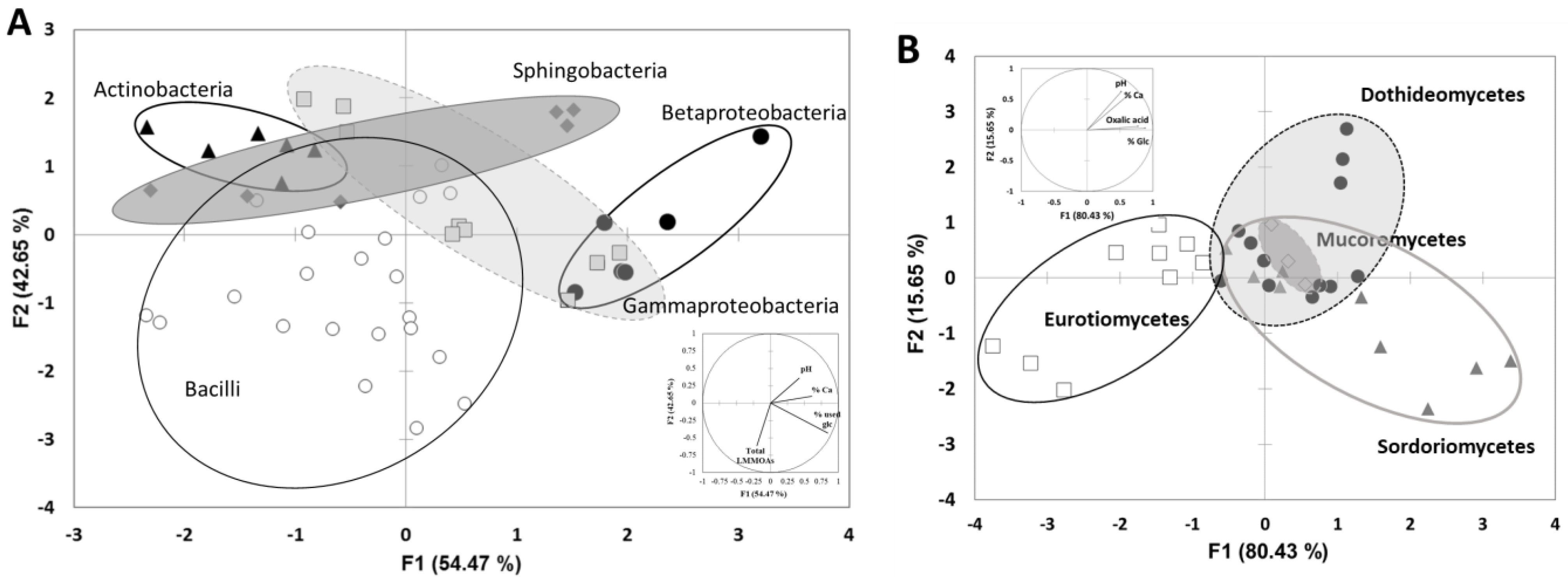
| PHYLUM | CLASS | ORDER | FAMILLY | Closest Match According to 16S rRNA Gene Sequence | Number of Colony Identified | Accession Number | % Similarity |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Oxalobacteriaceae | Burkholderia | 1 | AY839565.1 | 97 |
| Massilia sp. | 2 | FR865957.1 | 99 | ||||
| Bacilli | Bacilliaceae | Bacillus simplex | 2 | KJ586283.1 | 99 | ||
| Bacillus licheniformis | 1 | MF581456.1 | 98 | ||||
| Bacillus muralis | 2 | KM036074.1 | 96 | ||||
| Bacillus sp. | 1 | HM566651.1 | 99 | ||||
| Paenibacilliaiceae | Paenibacillus sp. | 2 | KY446062.1 | 98 | |||
| Microbacterium sp. 1 | 1 | KR085857.1 | 97 | ||||
| Microbacterium sp. 2 | 4 | KM035942.1 | 99 | ||||
| Stenotrophomonas maltophilia | 1 | LN867305.1 | 99 | ||||
| Stenotrophomonas sp.1 | 1 | KC464789.1 | 99 | ||||
| Stenotrophomonas sp.2 | 2 | KX588618.1 | 99 | ||||
| Gammaproteobacteria | Pseudomonales | Pseudomonadaceae | Pseudomonas gessardii | 1 | KT184489.1 | 99 | |
| Pseudomonas sp. | 1 | KR006341.1 | 98 | ||||
| Enterobacteriales | Enterobacteriaceae | Pantoea agglomerans | 9 | KT075163.1 | 99 | ||
| Pantoea vagans | 1 | KY127412.1 | 98 | ||||
| Enterobacter sp. | 1 | KR189819.1 | 98 | ||||
| Erwinia billingiae | 1 | HQ256807.1 | 97 | ||||
| Actinobacteria | Actinobacteria | Actinomycetales | Brevibacteriaceae | Brevibacterium frigoritolerans | 1 | HQ202870.1 | 99 |
| Micrococcales | Micrococcaceae | Arthrobacter sp. 1 | 1 | KC019208.1 | 99 | ||
| Arthrobacter sp. 2 | 2 | HQ202815.1 | 99 | ||||
| Microbacteriales | Microbacteriaceae | Clavibacter michiganensis | 1 | NR_133729.1 | 98 | ||
| Curtobacterium sp. | 1 | KR906476.1 | 98 | ||||
| Streptomycetales | Streptomycetaceae | Streptomyces sp. | 3 | GU211901.1 | 98 | ||
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Sphingobacteriaceae | Pedobacter sp. | 1 | HF548384.1 | 98 |
| Rhodoccocus sp. | 1 | JF923558.1 | 97 | ||||
| PHYLUM | CLASS | ORDER | FAMILLY | Closest Match According to ITS Sequence | Number of Colony Identified | Accesion Number | % Similarity |
| Ascomycota | Sordariomycetes | Hypocreales | Cordycipitaceae | Beauveria bassiana | 1 | NR_111594.1 | 98 |
| Stachybotryaceaea | Stachybotrys chartarum | 1 | NR_145083.1 | 99 | |||
| Hypocreacea | Trichoderma sp. | 2 | NR_077207.1 | 100 | |||
| Eurotiomycetes | Eurotiales | Aspergillaceae | Penicillium speluncae | 1 | NR_172035.1 | 94 | |
| Penicillium glandicola | 4 | NR_119395.1 | 100 | ||||
| Penicillium citrinum | 1 | NR_121224.1 | 99 | ||||
| Aspergillus tubingensis | 1 | NR_131293.1 | 99 | ||||
| Dothideomycetes | Pleosporales | Cucurbitariacea | Neocucurbitaria irregularis | 1 | NR_160337.1 | 97 | |
| Cladosporiales | Cladosporiaceae | Cladosporium sp. | 3 | NR_119730.1 | 99 | ||
| Dothideales | Saccotheciaceae | Aureobasidium melanogenum | 1 | NR_159598.1 | 98 | ||
| Aureobasidium sp. | 1 | NR_159598.1 | 71 | ||||
| Mucoromycota | Mucoromycetes | Mucorales | Mucoraceae | Mucor circinelloides | 1 | NR_126116.1 | 64 |
| Modalities | Chemical Weathering Rates (mol.m−2.s−1) | References |
|---|---|---|
| Laboratory experiments pH 4 | 2.25 × 10−6 | Franke and Teschner-Steinhardt (1994) [35] |
| Laboratory experiments pH 5.5 | 9.17 × 10−8 | |
| Field experiments pH 4.5 | 1.59 × 10−6 | Swoboda-Colberg and Drever (1993) [37] |
| Field experiments pH 6 | 7.02 × 10−9 | |
| Field experiments pH 6.5 | 2.25 × 10−9 | |
| Field experiments pH 7.9 | 1.10 × 10−10 | |
| Field experiments | 1.32 × 10−9 | Roussel and André (2013) [36] |
| Laboratory experiments Bare rock with cyanobacteria | 9.86 × 10−10 | Fiol et al. (1996) [38] |
| Lichen-covered rock | 6.18 × 10−10 | |
| Laboratory experiments by fungal mycelia | 1.54 × 10−4 | Li et al. (2009) [39] |
| Laboratory experiments by bacterial strains: Burkholderia sp.; Pantoea agglomerans; Pseudomonas gessardii; Bacillus simplex | 3.79 × 10−5 | Our study |
| Laboratory experiments by fungal strain: Aureobasidium pulullans, Trichoderma sp., Stachybotrys chartarum | 9.92 × 10−5 | Our study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balland-Bolou-Bi, C.; Saheb, M.; Alphonse, V.; Livet, A.; Reboah, P.; Abbad-Andaloussi, S.; Verney-Carron, A. Effect of Cultivable Bacteria and Fungi on the Limestone Weathering Used in Historical Buildings. Diversity 2023, 15, 587. https://doi.org/10.3390/d15050587
Balland-Bolou-Bi C, Saheb M, Alphonse V, Livet A, Reboah P, Abbad-Andaloussi S, Verney-Carron A. Effect of Cultivable Bacteria and Fungi on the Limestone Weathering Used in Historical Buildings. Diversity. 2023; 15(5):587. https://doi.org/10.3390/d15050587
Chicago/Turabian StyleBalland-Bolou-Bi, Clarisse, Mandana Saheb, Vanessa Alphonse, Alexandre Livet, Paloma Reboah, Samir Abbad-Andaloussi, and Aurélie Verney-Carron. 2023. "Effect of Cultivable Bacteria and Fungi on the Limestone Weathering Used in Historical Buildings" Diversity 15, no. 5: 587. https://doi.org/10.3390/d15050587
APA StyleBalland-Bolou-Bi, C., Saheb, M., Alphonse, V., Livet, A., Reboah, P., Abbad-Andaloussi, S., & Verney-Carron, A. (2023). Effect of Cultivable Bacteria and Fungi on the Limestone Weathering Used in Historical Buildings. Diversity, 15(5), 587. https://doi.org/10.3390/d15050587







