Phenotypic Polymorphism in Two Endosymbiotic Bacteria of the Ciliate Paramecium: Pseudolyticum multiflagellatum and “Ca. Megaira venefica”
Abstract
:1. Introduction
2. Materials and Methods
2.1. Host Isolation, Cultivation and Identification
2.2. DNA Extraction, PCR, Cloning and Sequencing
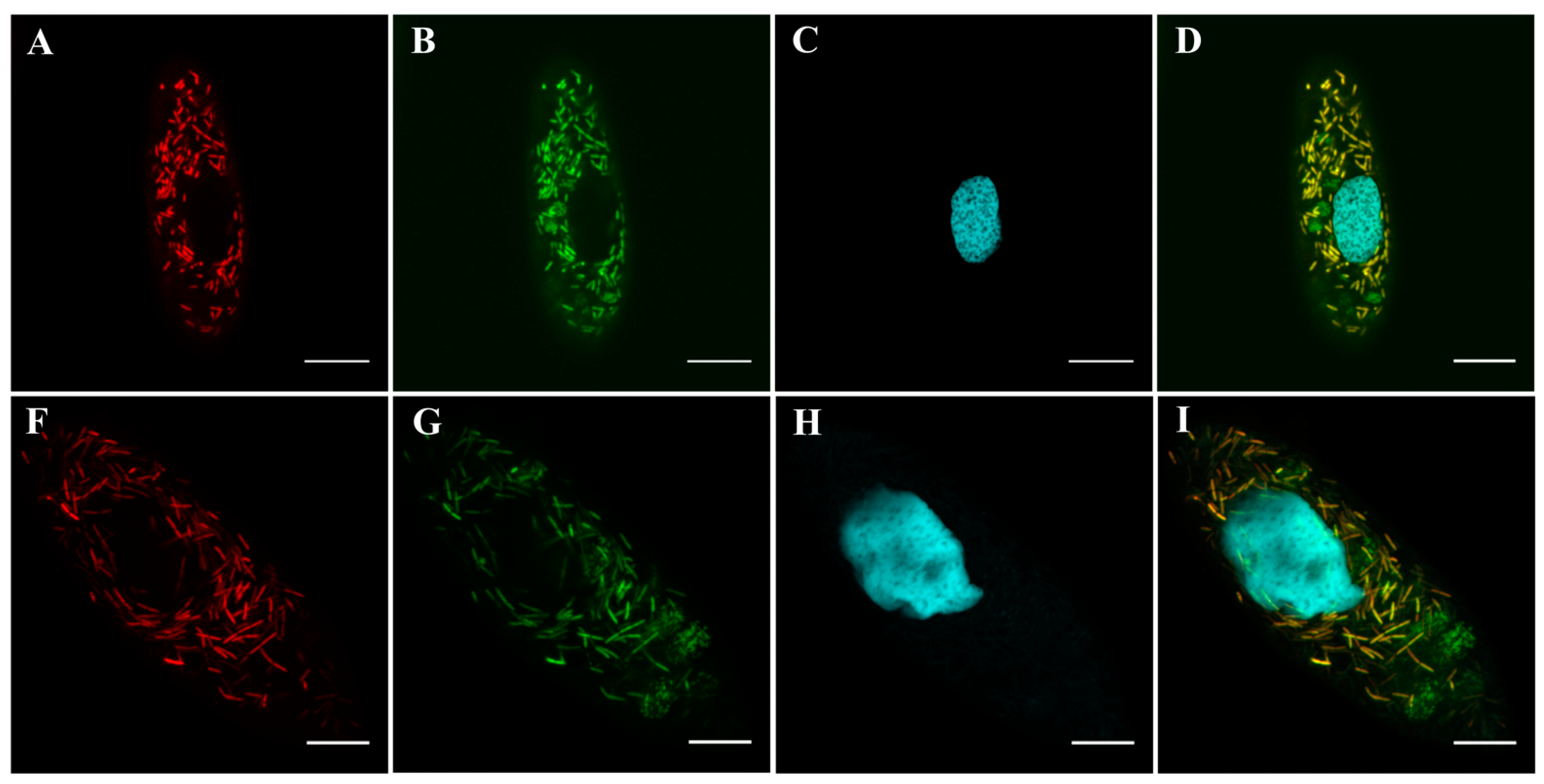
2.3. Fluorescence In Situ Hybridization (FISH) and Probe Design
2.4. Phylogenetic Analysis
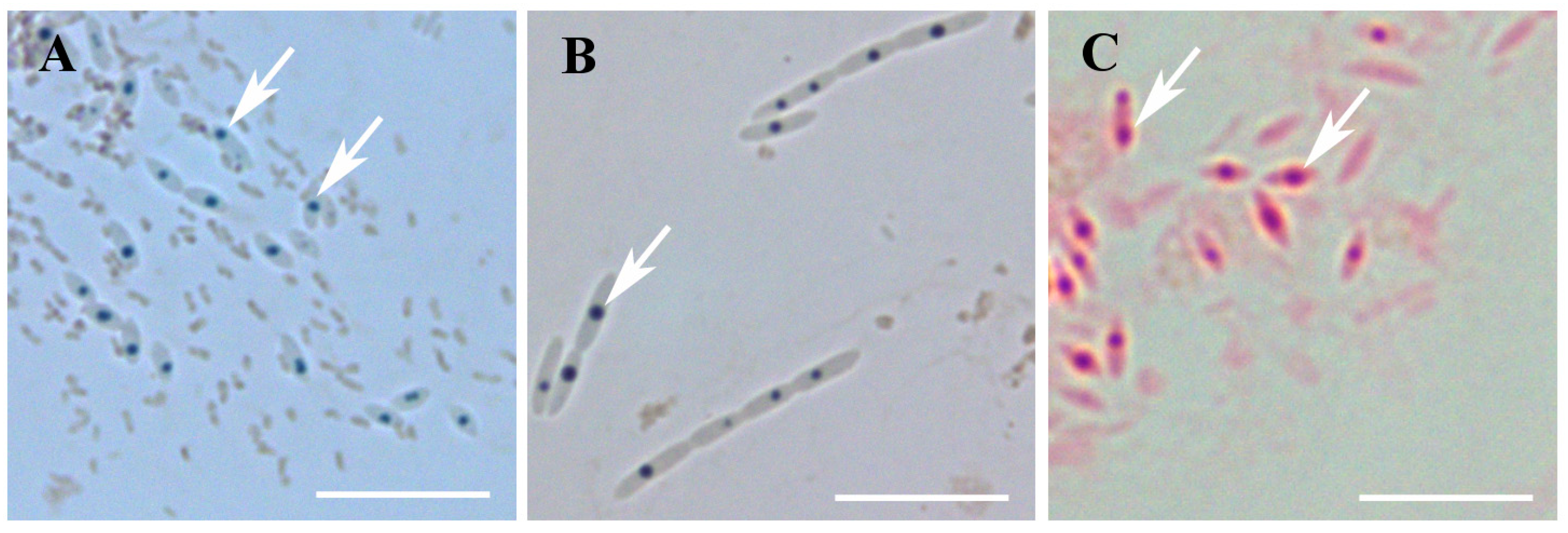
2.5. Cytochemical Staining of Endosymbiotic Bacteria
2.6. Transmission Electron Microscopy (TEM)
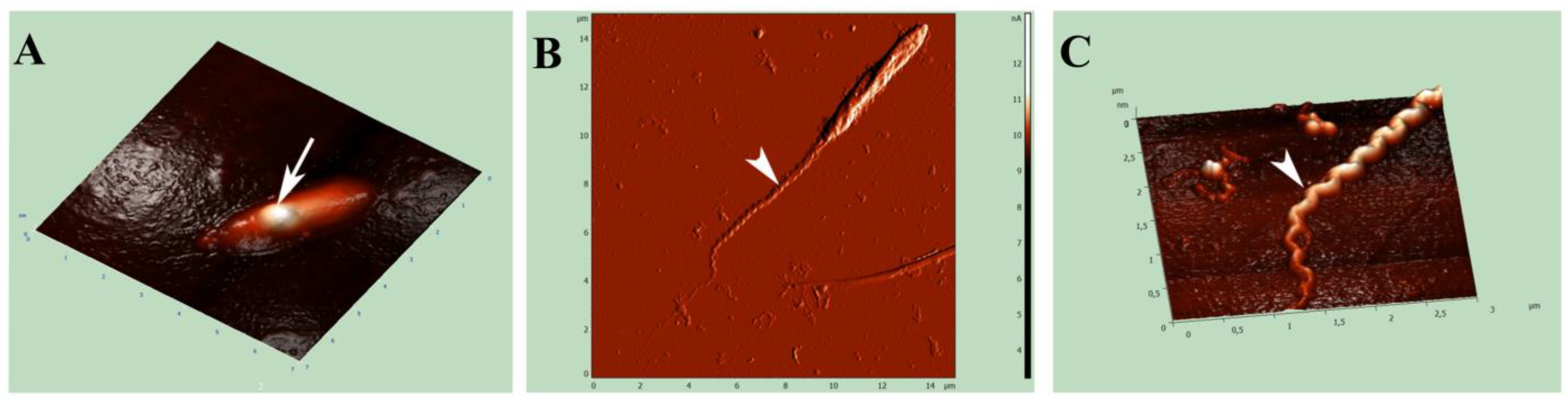
2.7. Atomic Force Microscopy (AFM)
2.8. Killer-Trait Assessment and Experimental Infection
2.9. Data Availability
3. Results
3.1. Host Identification
3.2. Living Host Cells Observations
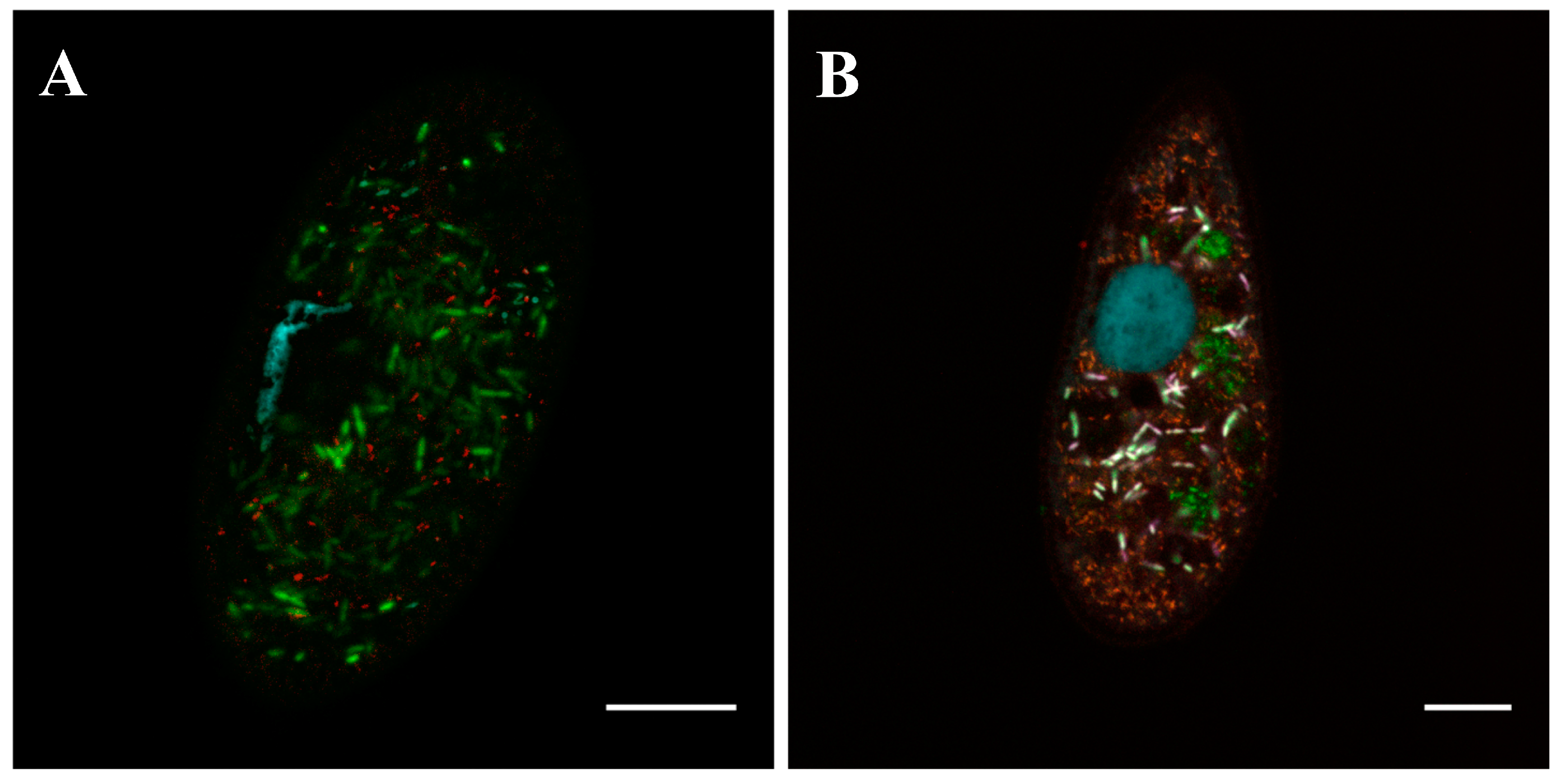
3.3. Spindle-Like Endosymbiont with a Refractive Granule Inhabiting the Cytoplasm of Paramecium caudatum and Paramecium nephridiatum Strains
3.3.1. Morphology and Biology
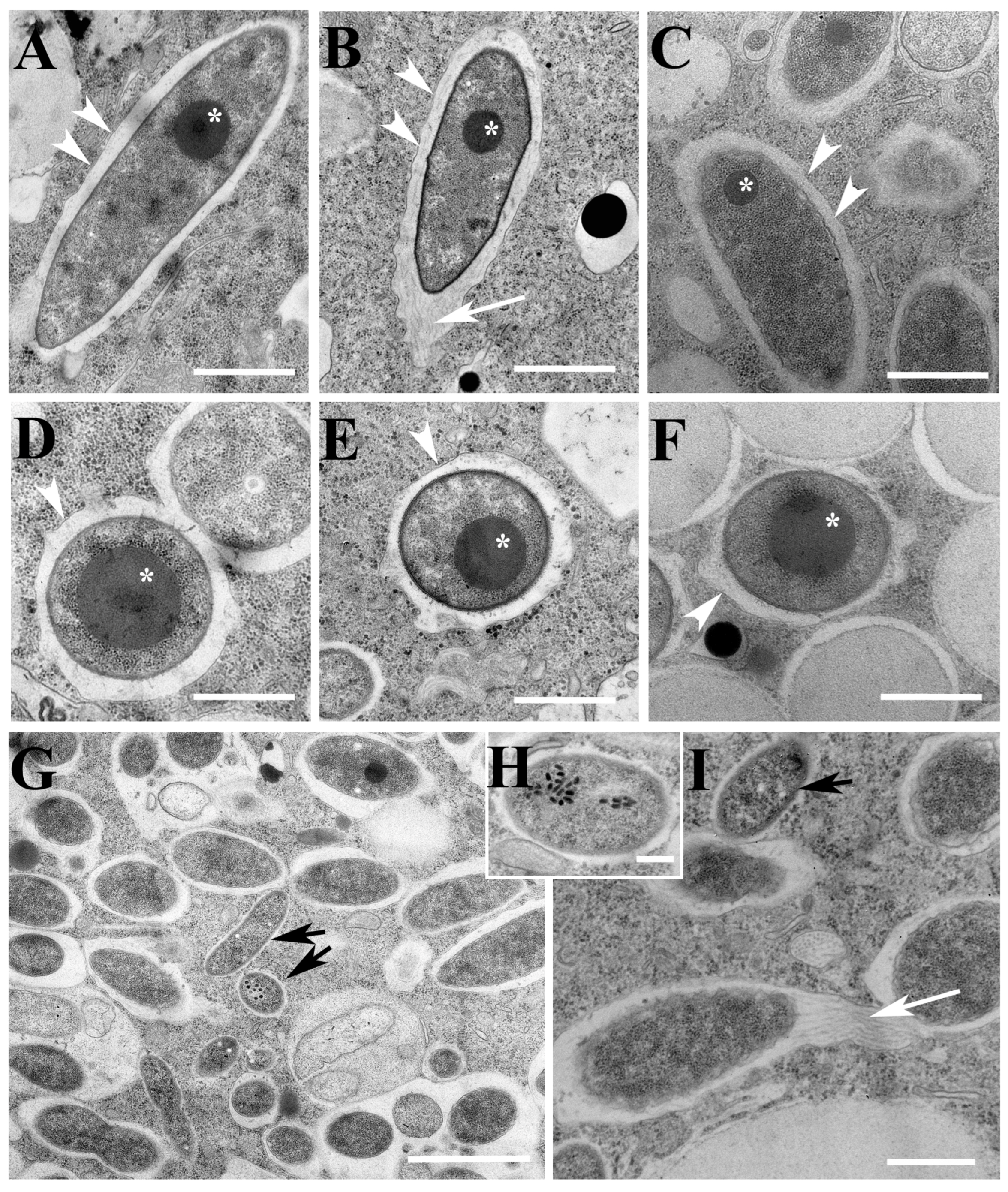
3.3.2. Fine Structure
3.3.3. Chemical Nature of the Inclusion Bodies
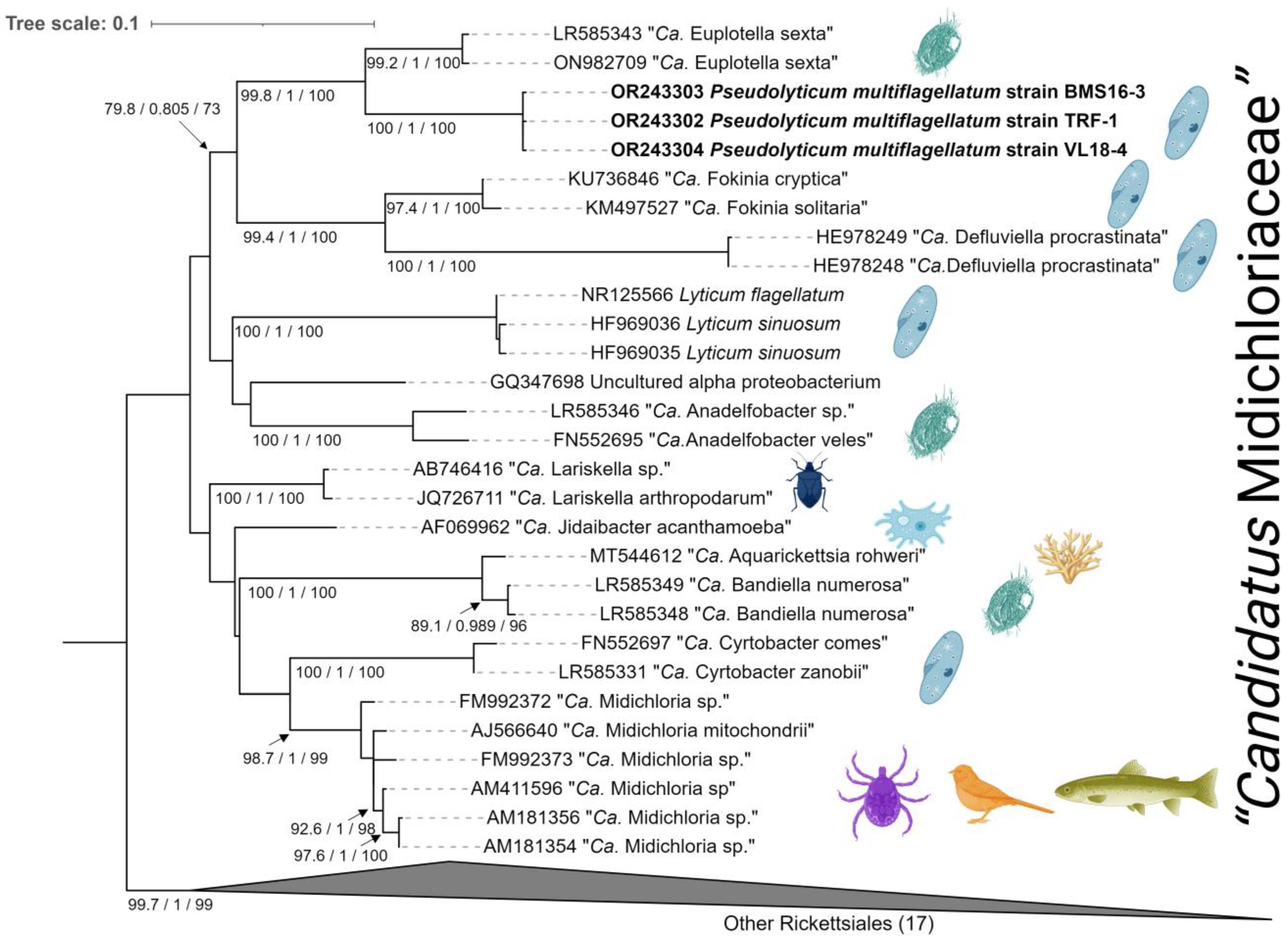
3.3.4. Molecular Identification and Phylogeny
3.3.5. Killer-Trait Assessment
3.4. Rod-Shaped Small Motile Endosymbionts Inhabiting the Cytoplasm of Paramecium nephridiatum Strains
3.4.1. Biology and Morphology
3.4.2. Fine Structure
3.4.3. Molecular Identification and Phylogeny
4. Discussion
4.1. Phylogeny and Taxonomy of the Spindle-like Endosymbiont
4.2. Polymorphism of Pseudolyticum multiflagellatum
4.3. The Presence of Flagella in Pseudolyticum multiflagellatum
4.4. The Nature of the Inclusion Bodies and Its Functions
4.5. Polymorphism in “Ca. Megaira venefica”
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Husnik, F.; Tashyreva, D.; Boscaro, V.; George, E.E.; Lukeš, J.; Keeling, P.J. Bacterial and Archaeal Symbioses with Protists. Curr. Biol. 2021, 31, R862–R877. [Google Scholar] [CrossRef] [PubMed]
- Dziallas, C.; Allgaier, M.; Monaghan, M.G. Act Together—Implications of Symbioses in Aquatic Ciliates. Front. Microbiol. 2012, 3, 288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fokin, S. Bacterial Endocytobionts of Ciliophora and Their Interactions with the Host Cell. Int. Rev. Cytol. 2004, 236, 181–249. [Google Scholar]
- Görtz, H.-D. Symbiotic Associations Between Ciliates and Prokaryotes. In The Prokaryotes: Volume 1: Symbiotic Associations, Biotechnology, Applied Microbiology; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 364–402. ISBN 978-0-387-30741-1. [Google Scholar]
- Fokin, S. Frequency and Biodiversity of Symbionts in Representatives of the Main Classes of Ciliophora. Eur. J. Protistol. 2012, 48, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A.; Bellone, A.; Fokin, S.I.; Boscaro, V.; Vannini, C. Detecting Associations Between Ciliated Protists and Prokaryotes with Culture-Independent Single-Cell Microbiomics: A Proof-of-Concept Study. Microb. Ecol. 2019, 78, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Beale, G.H.; Jurand, A.; Preer, J.R. The Classes of Endosymbiont of Paramecium aurelia. J. Cell Sci. 1969, 5, 65–91. [Google Scholar] [CrossRef]
- Preer, J.R.; Preer, L.B.; Jurand, A. Kappa and Other Endosymbionts in Paramecium aurelia. Bacteriol. Rev. 1974, 38, 113–163. [Google Scholar] [CrossRef]
- Fujishima, M. (Ed.) Endosymbionts in Paramecium; Microbiology Monographs; Springer: Berlin/Heidelberg, Germany, 2009; Volume 12, ISBN 978-3-540-92676-4. [Google Scholar]
- Fujishima, M.; Kodama, Y. Endosymbionts in Paramecium. Eur. J. Protistol. 2012, 48, 124–137. [Google Scholar] [CrossRef]
- Lohse, K.; Gutierrez, A.; Kaltz, O. Experimental Evolution of Resistance in Paramecium caudatum against the Bacterial Parasite Holospora undulata. Evolution 2006, 60, 1177–1186. [Google Scholar]
- Grosser, K.; Ramasamy, P.; Amirabad, A.D.; Schulz, M.H.; Gasparoni, G.; Simon, M.; Schrallhammer, M. More than the “Killer Trait”: Infection with the Bacterial Endosymbiont Caedibacter taeniospiralis Causes Transcriptomic Modulation in Paramecium Host. Genome Biol. Evol. 2018, 10, 646–656. [Google Scholar] [CrossRef] [Green Version]
- Castelli, M.; Sabaneyeva, E.L.; Lanzoni, O.; Lebedeva, N.; Floriano, A.-M.; Gaiarsa, S.; Benken, K.; Modeo, L.; Bandi, C.; Potekhin, A.; et al. Deianiraea, an Extracellular Bacterium Associated with the Ciliate Paramecium, Suggests an Alternative Scenario for the Evolution of Rickettsiales. ISME J. 2019, 13, 2280–2294. [Google Scholar] [CrossRef] [PubMed]
- Plotnikov, A.; Balkin, A.; Gogoleva, N.; Lanzoni, O.; Khlopko, Y.; Cherkasov, S.; Potekhin, A. High-Throughput Sequencing of the 16S RRNA Gene as a Survey to Analyze the Microbiomes of Free-Living Ciliates Paramecium. Microb. Ecol. 2019, 78, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Korotaev, A.; Benken, K.; Sabaneyeva, E. “Candidatus Mystax nordicus” Aggregates with Mitochondria of Its Host, the Ciliate Paramecium nephridiatum. Diversity 2020, 12, 251. [Google Scholar] [CrossRef]
- Castelli, M.; Lanzoni, O.; Nardi, T.; Lometto, S.; Modeo, L.; Potekhin, A.; Sassera, D.; Petroni, G. ‘Candidatus Sarmatiella mevalonica’ Endosymbiont of the Ciliate Paramecium Provides Insights on Evolutionary Plasticity among Rickettsiales. Environ. Microbiol. 2021, 23, 1684–1701. [Google Scholar] [CrossRef] [PubMed]
- Castelli, M.; Lanzoni, O.; Giovannini, M.; Lebedeva, N.; Gammuto, L.; Sassera, D.; Melekhin, M.; Potekhin, A.; Fokin, S.; Petroni, G. “Candidatus Gromoviella Agglomerans”, a Novel Intracellular Holosporaceae Parasite of the Ciliate Paramecium Showing Marked Genome Reduction. Env. Environ. Microbiol. Rep. 2022, 14, 34–49. [Google Scholar] [CrossRef]
- Castelli, M.; Serra, V.; Senra, M.; Basuri, C.; Soares, C.; Fokin, S.; Modeo, L.; Petroni, G. The Hidden World of Rickettsiales Symbionts: “Candidatus Spectririckettsia obscura”, a Novel Bacterium Found in Brazilian and Indian Paramecium caudatum. Microb. Ecol. 2019, 77, 748–758. [Google Scholar] [CrossRef] [PubMed]
- Schrallhammer, M.; Castelli, M.; Petroni, G. Phylogenetic Relationships among Endosymbiotic R-Body Producer: Bacteria Providing Their Host the Killer Trait. Syst. Appl. Microbiol. 2018, 41, 213–220. [Google Scholar] [CrossRef]
- Szokoli, F.; Sabaneyeva, E.; Castelli, M.; Krenek, S.; Schrallhammer, M.; Soares, C.; da Silva-Neto, I.; Berendonk, T.; Petroni, G. “Candidatus Fokinia solitaria”, a Novel “Stand-alone” Symbiotic Lineage of Midichloriaceae (Rickettsiales). PLoS ONE 2016, 11, e0145743. [Google Scholar]
- Lanzoni, O.; Sabaneyeva, E.; Modeo, L.; Castelli, M.; Lebedeva, N.V.; Verni, F.; Schrallhammer, M.; Potekhin, A.; Petroni, G. Diversity and Environmental Distribution of the Cosmopolitan Endosymbiont “Candidatus Megaira”. Sci. Rep. 2019, 9, 1179. [Google Scholar] [CrossRef] [Green Version]
- Mironov, T.; Sabaneyeva, E. A Robust Symbiotic Relationship Between the Ciliate Paramecium multimicronucleatum and the Bacterium Ca. Trichorickettsia mobilis. Front. Microbiol. 2020, 11, 603335. [Google Scholar] [CrossRef]
- Giannotti, D.; Boscaro, V.; Husnik, F.; Vannini, C.; Keeling, P.J. The “Other” Rickettsiales: An Overview of the Family “Candidatus Midichloriaceae”. Appl. Environ. Microbiol. 2022, 88, e02432-21. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Alexander, W.; Gilligan, J.; Rikihisa, Y. The Importance of Rickettsiales Infections. In Rickettsiales: Biology, Molecular Biology, Epidemiology, and Vaccine Development; Thomas, S., Ed.; Springer International Publishing: Cham, Germany, 2016; pp. 3–21. ISBN 978-3-319-46859-4. [Google Scholar]
- Boss, A.; Borkhsenius, O.O.; Ossipov, D. Pseudolyticum multiflagellatum ng, n. sp. a New Symbiotic Bacterium in the Cytoplasm of Paramecium caudatum (Ciliata, Protozoa). Cytologia 1987, 29, 94–99. [Google Scholar]
- Preer, J.R.; Preer, L.B. Revival of Names of Protozoan Endosymbionts and Proposal of Holospora caryophila nom. nov. Int. J. Syst. Bacteriol. 1982, 32, 140–141. [Google Scholar] [CrossRef] [Green Version]
- Fokin, S. Bacterial endobionts in the ciliate Paramecium woodruffi. III. Endobionts of the cytoplasm. Cytologia 1989, 31, 964–970. [Google Scholar]
- Fokin, S.; Stoeck, T.S.; Schmidt, H.J. Rediscovery of Paramecium nephridiatum Gelei, 1925 and its Characteristics. J. Eukaryot. Microbiol. 1999, 46, 416–426. [Google Scholar] [CrossRef]
- Fokin, S.I.; Görtz, H.-D. Diversity of Holospora Bacteria in Paramecium and Their Characterization. In Endosymbionts in Paramecium; Fujishima, M., Ed.; Microbiology Monographs; Springer: Berlin/Heidelberg, Germany, 2009; pp. 161–199. ISBN 978-3-540-92677-1. [Google Scholar]
- Schweikert, M.; Fujishima, M.; Görtz, H.-D. Symbiotic Associations Between Ciliates and Prokaryotes. In The Prokaryotes: Prokaryotic Biology and Symbiotic Associations; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 427–463. ISBN 978-3-642-30194-0. [Google Scholar]
- Sonneborn, T.M. Methods in Paramecium Research. In Methods in Cell Biology; Elsevier: Amsterdam, The Netherlands, 1970; Volume 4, pp. 241–339. [Google Scholar]
- Fokin, S. Paramecium Genus: Biodiversity, Some Morphological Features and the Key to the Main Morphospecies Discrimination. Protistology 2010, 6, 227–235. [Google Scholar]
- Medlin, L.; Elwood, H.; Stickel, S.S.; Sogin, M.L. The Characterization of Enzymatically Amplified Eukaryotic 16S-like RRNA-Coding Regions. Gene 1988, 71, 491–499. [Google Scholar] [CrossRef] [Green Version]
- Petroni, G.; Dini, F.; Verni, F.; Rosati, G. A Molecular Approach to the Tangled Intrageneric Relationships Underlying Phylogeny in Euplotes (Ciliophora, Spirotrichea). Mol. Phylogenetics Evol. 2002, 22, 118–130. [Google Scholar] [CrossRef]
- Rosati, G.; Modeo, L.; Melai, M.; Petroni, G.; Verni, F. A Multidisciplinary Approach to Describe Protists: A Morphological, Ultrastructural, and Molecular study on Peritromus kahli Villeneuve-Brachon, 1940 (Ciliophora, Heterotrichea). J. Eukaryot. Microbiol. 2004, 51, 49–59. [Google Scholar] [CrossRef]
- Strueder-Kypke, M.L.; Lynn, D. Comparative Analysis of the Mitochondrial Cytochrome c Oxidase Subunit I (COI) Gene in Ciliates (Alveolata, Ciliophora) and Evaluation of Its Suitability as a Biodiversity Marker. Syst. Biodivers. 2010, 8, 131–148. [Google Scholar] [CrossRef]
- Vannini, C.; Rosati, G.; Verni, F.; Petroni, G. Identification of the Bacterial Endosymbionts of the Marine Ciliate Euplotes magnicirratus (Ciliophora, Hypotrichia) and Proposal of “Candidatus Devosia Euplotis”. Int. J. Syst. Evol. Microbiol. 2004, 54, 1151–1156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Don, R.H.; Cox, P.T.; Wainwright, B.J.; Baker, K.; Mattick, J.S. ‘Touchdown’ PCR to Circumvent Spurious Priming during Gene Amplification. Nucleic Acids Res. 1991, 19, 4008. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, W.; Strunk, O.; Westram, R.; Richter, L.; Meier, H.; Yadhukumar; Buchner, A.; Lai, T.; Steppi, S.; Jobb, G.; et al. ARB: A Software Environment for Sequence Data. Nucleic Acids Res. 2004, 32, 1363–1371. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and Tools for High Throughput RRNA Analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greuter, D.; Loy, A.; Horn, M.; Rattei, T. ProbeBase—An Online Resource for RRNA-Targeted Oligonucleotide Probes and Primers: New Features 2016. Nucleic Acids Res. 2016, 44, D586–D589. [Google Scholar] [CrossRef] [PubMed]
- Lindrea, K.C.; Seviour, E.M.; Seviour, R.J.; Blackall, L.L.; Soddell, J.A. Practical Methods for the Examination and Characterization of Activated Sludge. In The Microbiology of Activated Sludge; Seviour, R.J., Blackall, L.L., Eds.; Springer: Dordrecht, The Netherlands, 1998; pp. 257–300. ISBN 978-94-011-3951-9. [Google Scholar] [CrossRef]
- Amann, R.I.; Binder, B.J.; Olson, R.J.; Chisholm, S.W.; Devereux, R.S.; Stahl, D. Combination of 16S RRNA-Targeted Oligonucleotide Probes with Flow Cytometry for Analyzing Mixed Microbial Populations. Appl. Environ. Microbiol. 1990, 56, 1919–1925. [Google Scholar] [CrossRef] [Green Version]
- Manz, W.; Amann, R.; Ludwig, W.; Wagner, M.S.; Schleifer, K.-H. Phylogenetic Oligodeoxynucleotide Probes for the Major Subclasses of Proteobacteria: Problems and Solutions. Syst. Appl. Microbiol. 1992, 15, 593–600. [Google Scholar] [CrossRef]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Bourexis, D.; Brister, J.R.; Canese, K.; Comeau, D.C.; Funk, K.; Kim, S.; Klimke, W.; et al. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2020, 49, D10–D17. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A Better Web Interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [Green Version]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anisimova, M.; Gil, M.; Dufayard, J.-F.; Dessimoz, C.; Gascuel, O. Survey of Branch Support Methods Demonstrates Accuracy, Power, and Robustness of Fast Likelihood-Based Approximation Schemes. Syst. Biol. 2011, 60, 685–699. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- BioRender. Available online: https://www.biorender.com/ (accessed on 30 May 2023).
- Inkscape. Available online: https://inkscape.org (accessed on 30 May 2023).
- Dévényi, T.; Gergely, J. Amino Acids, Peptides and Proteins: Biochemical and Immunochemical Techniques in Protein Chemistry; Amsterdam; Elsevier Scientific Pub. Co.: New York, NY, USA, 1974; ISBN 978-0-444-41127-3. [Google Scholar]
- Elghetany, M.T.; Saleem, A. Methods for Staining Amyloid in Tissues: A Review. Stain. Technol. 1988, 63, 201–212. [Google Scholar] [CrossRef]
- Burdon, K.L. Fatty Material in Bacteria and Fungi Revealed by Staining Dried, Fixed Slide Preparations. J. Bacteriol. 1946, 52, 665–678. [Google Scholar] [CrossRef]
- Strüder-Kypke, M.C.; Wright, A.D.; Fokin, S.I.; Lynn, D.H. Phylogenetic Relationships of the Subclass Peniculia (Oligohymenophorea, Ciliophora) Inferred from Small Subunit RRNA Gene Sequences. J. Eukaryot Microbiol. 2000, 47, 419–429. [Google Scholar] [CrossRef]
- Hoshina, R.; Hayashi, S.; Imamura, N. Intraspecific Genetic Divergence of Paramecium bursaria and Re-Construction of the Paramecian Phylogenetic Tree. Acta Protozool. 2006, 45, 377–386. [Google Scholar]
- Fokin, S.I.; Lebedeva, N.A.; Potekhin, A.; Gammuto, L.; Petroni, G.; Serra, V. Holospora-like Bacteria “Candidatus Gortzia yakutica” and Preeria caryophila: Ultrastructure, Promiscuity, and Biogeography of the Symbionts. Eur. J. Protistol. 2023, 90, 125998. [Google Scholar] [CrossRef]
- Barth, D.; Berendonk, T.U. The Mitochondrial Genome Sequence of the Ciliate Paramecium caudatum Reveals a Shift in Nucleotide Composition and Codon Usage within the Genus Paramecium. BMC Genom. 2011, 12, 272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boscaro, V.; Husnik, F.; Vannini, C.; Keeling, P.J. Symbionts of the Ciliate Euplotes: Diversity, Patterns and Potential as Models for Bacteria–Eukaryote Endosymbioses. Proc. Biol. Sci. 2019, 286, 20190693. [Google Scholar] [CrossRef]
- Szokoli, F.; Castelli, M.; Sabaneyeva, E.; Schrallhammer, M.; Krenek, S.; Doak, T.G.; Berendonk, T.U.; Petroni, G. Disentangling the Taxonomy of Rickettsiales and Description of Two Novel Symbionts (“Candidatus Bealeia paramacronuclearis” and “Candidatus Fokinia cryptica”) Sharing the Cytoplasm of the Ciliate Protist Paramecium biaurelia. Appl. Environ. Microbiol. 2016, 82, 7236–7247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boscaro, V.; Petroni, G.; Ristori, A.; Verni, F.; Vannini, C. “Candidatus Defluviella procrastinata” and “Candidatus Cyrtobacter zanobii”, Two Novel Ciliate Endosymbionts Belonging to the “Midichloria Clade”. Microb. Ecol. 2013, 65, 302–310. [Google Scholar] [CrossRef]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.-H.; Whitman, W.B.; Euzéby, J.; Amann, R.; Rosselló-Móra, R. Uniting the Classification of Cultured and Uncultured Bacteria and Archaea Using 16S RRNA Gene Sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef]
- Karasz, D.C.; Weaver, A.I.; Buckley, D.H.; Wilhelm, R.C. Conditional Filamentation as an Adaptive Trait of Bacteria and Its Ecological Significance in Soils. Environ. Microbiol. 2022, 24, 1–17. [Google Scholar] [CrossRef] [PubMed]
- van Teeseling, M.C.F.; de Pedro, M.A.; Cava, F. Determinants of Bacterial Morphology: From Fundamentals to Possibilities for Antimicrobial Targeting. Front. Microbiol. 2017, 8, 1264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corno, G.; Jürgens, K. Direct and Indirect Effects of Protist Predation on Population Size Structure of a Bacterial Strain with High Phenotypic Plasticity. Appl. Environ. Microbiol. 2006, 72, 78–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, T.D.; Ali, M.A.; Lee, D.; Félix, M.-A.; Luallen, R.J. Bacterial Filamentation as a Mechanism for Cell-to-Cell Spread within an Animal Host. Nat. Commun. 2022, 13, 693. [Google Scholar] [CrossRef]
- Boscaro, V.; Schrallhammer, M.B.; Benken, K.A.; Krenek, S.; Szokoli, F.; Berendonk, T.U.; Schweikert, M.; Verni, F.; Sabaneyeva, E.V.; Petroni, G. Rediscovering the Genus Lyticum, Multiflagellated Symbionts of the Order Rickettsiales. Sci. Rep. 2013, 3, 3305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neubauer, P.; Fahnert, B.; Lilie, H.; Villaverde, A. Protein Inclusion Bodies in Recombinant Bacteria. In Inclusions in Prokaryotes; Shively, J.M., Ed.; Microbiology Monographs; Springer: Berlin/Heidelberg, Germany, 2006; pp. 237–292. ISBN 978-3-540-33774-4. [Google Scholar]
- Park, H.-W.; Federici, B.A.; Sakano, Y. Inclusion Proteins from Other Insecticidal Bacteria. In Inclusions in Prokaryotes; Shively, J.M., Ed.; Microbiology Monographs; Springer: Berlin/Heidelberg, Germany, 2006; pp. 321–330. ISBN 978-3-540-33774-4. [Google Scholar]
- Pötter, M.; Steinbüchel, A. Biogenesis and Structure of Polyhydroxyalkanoate Granules. In Inclusions in Prokaryotes; Shively, J.M., Ed.; Microbiology Monographs; Springer: Berlin/Heidelberg, Germany, 2006; pp. 109–136. ISBN 978-3-540-33774-4. [Google Scholar]
- Wältermann, M.; Steinbüchel, A. Wax Ester and Triacylglycerol Inclusions. In Inclusions in Prokaryotes; Shively, J.M., Ed.; Microbiology Monographs; Springer: Berlin/Heidelberg, Germany, 2006; pp. 137–166. ISBN 978-3-540-33774-4. [Google Scholar]
- Schrallhammer, M.; Schweikert, M. The Killer Effect of Paramecium and Its Causative Agents. In Endosymbionts in Paramecium; Fujishima, M., Ed.; Microbiology Monographs; Springer: Berlin/Heidelberg, Germany, 2009; pp. 227–246. ISBN 978-3-540-92677-1. [Google Scholar]
- Pond, F.R.; Gibson, I.; Lalucat, J.; Quackenbush, R.L. R-Body-Producing Bacteria. Microbiol. Rev. 1989, 53, 25–67. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Amat, A. R-Bodies. In Inclusions in Prokaryotes; Shively, J.M., Ed.; Microbiology Monographs; Springer: Berlin/Heidelberg, Germany, 2006; pp. 331–341. ISBN 978-3-540-33774-4. [Google Scholar]
- Wang, B.; Lin, Y.-C.; Vasquez-Rifo, A.; Jo, J.; Price-Whelan, A.; McDonald, S.T.; Brown, L.M.; Sieben, C.; Dietrich, L.E.P. Pseudomonas aeruginosa PA14 Produces R-Bodies, Extendable Protein Polymers with Roles in Host Colonization and Virulence. Nat. Commun. 2021, 12, 4613. [Google Scholar] [CrossRef] [PubMed]
- Fiebig, A.; Pradella, S.; Petersen, J.; Päuker, O.; Michael, V.; Lünsdorf, H.; Göker, M.; Klenk, H.-P.; Wagner-Döbler, I. Genome of the R-Body Producing Marine Alphaproteobacterium Labrenzia alexandrii Type Strain (DFL-11T). Stand. Genom. Sci. 2013, 7, 413–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuoka, J.; Ishizuna, F.; Kurumisawa, K.; Morohashi, K.; Ogawa, T.; Hidaka, M.; Saito, K.; Ezawa, T.; Aono, T. Stringent Expression Control of Pathogenic R-Body Production in Legume Symbiont Azorhizobium caulinodans. mBio 2017, 8, e00715-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raymann, K.; Bobay, L.-M.; Doak, T.G.; Lynch, M.; Gribaldo, S. A Genomic Survey of Reb Homologs Suggests Widespread Occurrence of R-Bodies in Proteobacteria. G3 Genes Genomes Genet. 2013, 3, 505–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Federici, B.A.; Lüthy, P.; Ibarra, J.E. Parasporal Body of Bacillus thuringiensis israelensis. In Bacterial Control of Mosquitoes & Black Flies: Biochemistry, Genetics & Applications of Bacillus Thuringiensis israelensis and Bacillus sphaericus; de Barjac, H., Sutherland, D.J., Eds.; Springer: Dordrecht, The Netherlands, 1990; pp. 16–44. ISBN 978-94-011-5967-8. [Google Scholar]
- Schönherr, R.; Rudolph, J.M.; Redecke, L. Protein Crystallization in Living Cells. Biol. Chem. 2018, 399, 751–772. [Google Scholar] [CrossRef]
- Glare, T.R.; Jurat-Fuentes, J.-L.; O’Callaghan, M. Chapter 4—Basic and Applied Research: Entomopathogenic Bacteria. In Microbial Control of Insect and Mite Pests; Lacey, L.A., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 47–67. ISBN 978-0-12-803527-6. [Google Scholar]
- Cao, B.; Nie, Y.; Guan, Z.; Chen, C.; Wang, N.; Wang, Z.; Shu, C.; Zhang, J.; Zhang, D. The Crystal Structure of Cry78Aa from Bacillus Thuringiensis Provides Insights into Its Insecticidal Activity. Commun. Biol. 2022, 5, 801. [Google Scholar] [CrossRef] [PubMed]
- Bowen, D.J.; Ensign, J.C. Isolation and Characterization of Intracellular Protein Inclusions Produced by the Entomopathogenic Bacterium Photorhabdus luminescens. Appl. Env. Environ. Microbiol. 2001, 67, 4834–4841. [Google Scholar] [CrossRef] [Green Version]
- You, J.; Liang, S.; Cao, L.; Liu, X.; Han, R. Nutritive Significance of Crystalline Inclusion Proteins of Photorhabdus luminescens in Steinernema Nematodes. FEMS Microbiol. Ecol. 2006, 55, 178–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fokin, S.I.; Boss, A.O.-L.; Ossipov, D.V. A virus-containing cytoplasmic symbiont of the ciliate Paramecium woodruffi. Tsitologiya 1987, 29, 1303–1306. [Google Scholar]
- Fokin, S.; Sabaneyeva, E. Bacterial Endocytobionts of the Ciliate Paramecium calkinsi. Eur. J. Protistol. 1993, 29, 390–395. [Google Scholar] [CrossRef] [PubMed]
- Fokin, S.I.; Giamberini, L.; Molloy, D.P.; de Vaate, A.B. Bacterial Endocytobionts within Endosymbiotic Ciliates in Dreissena polymorpha (Lamellibranchia: Mollusca). Acta Protozool. 2003, 42, 31–40. [Google Scholar]
- Berezina, I.G. An Electron Microscope Study of Endosymbionts in Blepharisma japonicum. Acta Protozool. 1974, 13, 365–369. [Google Scholar]
- Fokin, S.I.; Schweikert, M. Bacterial Endocytobionts in the Macronucleus of Frontonia leucas (Ciliophora, Peniculida). Eur. J. Protistol. 2003, 39, 311–318. [Google Scholar] [CrossRef]
- George, E.E.; Barcytė, D.; Lax, G.; Livingston, S.; Tashyreva, D.; Husnik, F.; Lukeš, J.; Eliáš, M.; Keeling, P.J. A Single Cryptomonad Cell Harbors a Complex Community of Organelles, Bacteria, a Phage, and Selfish Elements. Curr. Biol. 2023, 33, 1982–1996.e4. [Google Scholar] [CrossRef]

| Strain Index | Host | Endosymbiont (s) | Sampling Sites | Cultivation Salinity Level (‰) |
|---|---|---|---|---|
| VL18-4 | P. caudatum | Ps. multiflagellatum | Russia, Vladimir: pond (56.106928, 40.428814) | 0 |
| BMS16-3 | P. nephridiatum | Ps. multiflagellatum, Ca. M. venefica | Russia, Karelsky region, Sredniy island: littoral pool (66.282500, 33.709444) | 0 |
| TRF-1 | P. nephridiatum | Ps. multiflagellatum, Ca. M. venefica | Russia, St. Petersburg region, Peterhof: Troitsky stream (59.894074, 29.879725) | 0–5.0 |
| VL18-4 * | P. caudatum | No, aposymbiotic, spontaneous loss | Russia, Vladimir: pond (56.106928, 40.428814) | 0 |
| SD11-1 | P. nephridiatum | No, naïve | Russia, St. Petersburg region, Sestroretsk: pond (60.096911, 29.958959) | 0 |
| Eta11-10 | P. nephridiatum | No, naïve | Estonia, Tallinn, Kopli bay (59.4344225, 24.6766373) | 5.0 |
| 1M15-3 | P. nephridiatum | Ca. M. venefica | Russia, St. Petersburg region, Peterhof: ditch (59.878965, 29.851437) | 0–5.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kursacheva, E.; Korotaev, A.; Benken, K.; Lebedeva, N.; Sabaneyeva, E. Phenotypic Polymorphism in Two Endosymbiotic Bacteria of the Ciliate Paramecium: Pseudolyticum multiflagellatum and “Ca. Megaira venefica”. Diversity 2023, 15, 924. https://doi.org/10.3390/d15080924
Kursacheva E, Korotaev A, Benken K, Lebedeva N, Sabaneyeva E. Phenotypic Polymorphism in Two Endosymbiotic Bacteria of the Ciliate Paramecium: Pseudolyticum multiflagellatum and “Ca. Megaira venefica”. Diversity. 2023; 15(8):924. https://doi.org/10.3390/d15080924
Chicago/Turabian StyleKursacheva, Ekaterina, Alexander Korotaev, Konstantin Benken, Natalia Lebedeva, and Elena Sabaneyeva. 2023. "Phenotypic Polymorphism in Two Endosymbiotic Bacteria of the Ciliate Paramecium: Pseudolyticum multiflagellatum and “Ca. Megaira venefica”" Diversity 15, no. 8: 924. https://doi.org/10.3390/d15080924
APA StyleKursacheva, E., Korotaev, A., Benken, K., Lebedeva, N., & Sabaneyeva, E. (2023). Phenotypic Polymorphism in Two Endosymbiotic Bacteria of the Ciliate Paramecium: Pseudolyticum multiflagellatum and “Ca. Megaira venefica”. Diversity, 15(8), 924. https://doi.org/10.3390/d15080924







