Co-Evolution of Iolana Blues with Their Host Plants and the Higher Phylogeny of Subtribe Scolitantidina (Lepidoptera, Lycaenidae)
Abstract
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Taxonomic Considerations
- Our results confirm many of the previously proposed synonymies (i.e., Glaucopsyche (=Shijimiaeoides); Turanana (=Otnjukovia; =Micropsyche); and Pseudophilotes (=Inderskia)) and taxonomic arrangements (Apelles, Bajluana, and Sinia as subgenera of Glaucopsyche; Palaeophilotes as the senior generic name for the Pseudophilotes clade; and Philotiella as the subgenus of Euphilotes) [9,15,17,45].
- Ugelvig et al. [9] separated Glaucopsyche (Phaedrotes) piasus from other Glaucopsyche species due to the paraphyly of the clade that also included Iolana spp. and S. divina. We also found G. piasus to belong to a distinct lineage closer to Euphilotes and far from the Glaucopsyche group (type species: G. lygdamus Doubleday, 1841). Nuclear genomic data from Zhang et al. [17] place this taxon as sister to all the rest of Glaucopsyche (including Sinia), but their mitochondrial phylogeny is supporting a different position, similar to the one we obtained. The same ambiguous situation was obtained in the study by Lukhtanov and Gagarina [15]. Thus, some kind of mito-nuclear discordance seems to be involved in this case. Even though Phaedrotes has been previously used as a stand-alone genus (cf. [46]), given its unstable phylogenetic position, and until better and more comprehensive methods are employed to address this question appropriately, we refrain from recognizing Phaedrotes as a monotypic genus.
- In our analyses, the monotypic Chinese endemic Subsolanoides nagata appears to be a distinct genus and species most closely related to Scolitantides orion. In the phylogeny presented by Kawahara et al. [16], Subsolanoides appears as sister to a clade consisting of Praephilotes, Scolitantides, Philotes, Iolana, and Glaucopsyche. Since the latter study used a larger amount of genomic information, we cannot exclude that the pattern observed in our study can change once additional genes become available.
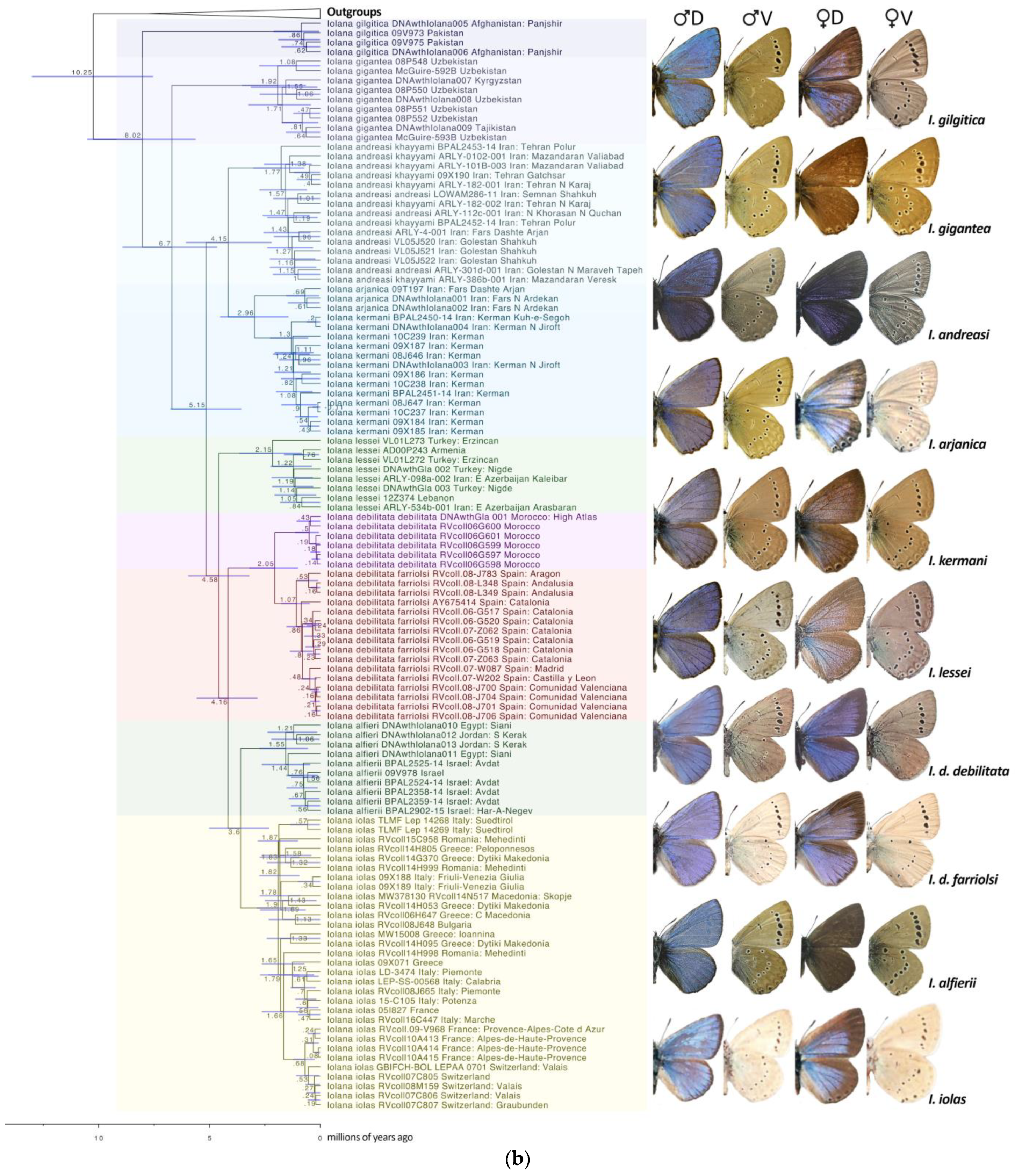
- Lukhtanov and Gagarina [15] suggested that the African and Iberian populations attributed to Iolana debilitata Schultz, 1905 (TL: Algeria) may perhaps represent different species. García-Barros et al. [47] listed three subspecies present in the Iberian Peninsula: ssp. debilitata in Andalusia; ssp. thomasi Hemming, 1931 in Central Spain; and ssp. farriolsi Sagarra, 1931 in the northeast of Iberia. These authors, as well as Dumont [14], describe subtle differences in the male genitalia and the external morphology across the range of these populations. However, our molecular clock analysis, which estimates that the split between the Spanish populations and those in Morocco occurred around 2 mya (Figure 1), does not support the presence of the North African lineage (taxon debilitata) in Spain. On the other hand, a genetic structure roughly correlating with geography is observed within the Iberian populations. Here, we maintain Spanish and African populations under a single species, Iolana debilitata, but with the nominotypical populations restricted to North Africa, and tentatively consider a single subspecies in Spain, I. debilitata farriolsi Sagarra, 1931, possibly with a latitudinal morphological differentiation. A finer population genetics and morphometric study may shed light on the variation within the Iberian Peninsula.
- The distribution of I. iolas in Southern Europe is not continuous, and there are gaps, for example, between Südtirol and Slovenia [48]. The species is absent in Austria. The currently recognized subspecies include ssp. iolas (TL. Hungary) in Southeastern Europe, the Eastern Alps, and Italy; ssp. wullschlegeli (Oberthür, 1914) [49] (TL. Martigny, Switzerland) in France (Western Alps), Italy (Val d’Aoste), and Switzerland (Valais); and ssp. protogenes Fruhstorfer, 1917 [50] (TL. Digne, France) in Southern France [14]. Martigny and Digne are both in the Western Alps, about 200 km apart. In addition to the localities and haplotypes investigated by Groza et al. [42], we obtained sequences from two populations in Northern Italy (Südtirol [COI] and Friuli-Venezia Giulia [ND1]), both of which showed new and unique haplotypes (Figure 1). Based on current evidence, the presence of two subspecies in the Western Alps makes little sense. Dumont [14] mentions that these two taxa are weakly characterized, with the variation within subspecies being larger than that between subspecies. Our I. iolas haplogroup includes individuals from both ssp. wullschlegeli and ssp. protogenes. Therefore, here, we propose a synonymy: Iolana iolas wullschlegeli (Oberthür, 1914) (=protogenes Fruhstorfer, 1917 syn. nov.).
- We also did not find any reliable diagnostic differences in the morphology, genitalia, or DNA sequences between the nominotypical Iolana andreasi Sheljuzhko, 1919 [51] and the subspecies khayyami Bernardi, 1964 [52]. Therefore, here, we propose a synonymy: I. andreasi Sheljuzhko, 1919 (=khayyami Bernardi, 1964, syn. nov.).
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tolman, T.; Lewington, R. Collins Field Guide: Butterflies of Britain and Europe; Harper Collins Publications: New York, NY, USA, 1997; 320p. [Google Scholar]
- Fiedler, K. European and North West African Lycaenidae (Lepidoptera) and their associations with ants. J. Res. Lepid. 1991, 28, 239–257. [Google Scholar] [CrossRef]
- Gonseth, Y. Liste rouge des lépidoptères diurnes menacés de Suisse. In Listes Rouges des Espèces Animales Menacées de SUISSE; Duelli, P., Ed.; Office Fédéral de l’environnement, des Forêts et du Paysage (OFEFP): Berne, Switzerland, 1994; pp. 48–51. [Google Scholar]
- Carron, G.; Wermeille, E.; Schiesse, H.; Patocchi, N. Programme National de Conservation des Espèces Prioritaires de Papillons diurnes (Rhopalocera et Hesperiidae). Canton du Valais. 2001. 52p. Available online: http://home.page.ch/pub/[email protected]/ (accessed on 10 November 2023).
- Rákosy, L.; Corduneanu, C.; Crișan, A.; Dincă, V.; Kovács, S.; Stănescu, M.; Székely, L. Romanian Red List of Lepidoptera; Presa Universitară Clujeană: Cluj-Napoca, Romania, 2021; 187p. [Google Scholar]
- Vila, R.; Stefanescu, C.; Sesma, J.M. Guia de les Papallones Diürnes de Catalunya; Lynx Edicions: Barcelona, Spain, 2018; 509p. [Google Scholar]
- Tarrier, M.; Delacre, J. Les Papillons de Jour du Maroc; Mèze, Biotope, Ed.; Muséum National d’Histoire Naturelle: Paris, France, 2008; 480p. [Google Scholar]
- Bethune-Baker, G.T. Synonymic notes on the Ruralidae. Entomol. Rec. J. Var. 1914, 26, 159–164. [Google Scholar]
- Ugelvig, L.V.; Vila, R.; Pierce, N.E.; Nash, D.R. A phylogenetic revision of the Glaucopsyche section (Lepidoptera: Lycaenidae), with special focus on the Phengaris–Maculinea clade. Mol. Phylogenetics Evol. 2011, 61, 237–243. [Google Scholar] [CrossRef]
- Hemming, A.F. Revision of the genus Iolana, Bethune-Baker (Lepidoptera, Lycaenidae). Trans. Entomol. Soc. Lond. 1931, 79, 323–333. [Google Scholar] [CrossRef]
- Bernardi, G. Note sur la variation géographique de l’armure génilale mâle des Iolana (Lep. Lycaenidae). Bull. De La Société Entomol. De Fr. 1972, 77, 160–167. [Google Scholar] [CrossRef]
- Lukhtanov, V.A.; Lukhtanov, A.G. Die Tagfalter Nordwestasiens (Lepidoptera, Diurna); U. Eitschberger: Marktleuthen, Germany, 1994; 440p, ISBN 3-923807-02-3. [Google Scholar]
- Hesselbarth, G.; Van Oorschot, H.; Wagener, S. Die Tagfalter der Türkei; Selbstverlag Sigbert Wagener: Bocholt, Germany, 1995; Volume 1, 754p. [Google Scholar]
- Dumont, D. Révision du genre Iolana Bethune-Baker, 1914 (Lepidoptera: Lycaenidae). Description d’une nouvelle espèce: kermani n. sp. Linneana Belg. 2004, 19, 332–357. [Google Scholar]
- Lukhtanov, V.A.; Gagarina, A.V. Molecular Phylogeny and Taxonomy of the Butterfly Subtribe Scolitantidina with Special Focus on the Genera Pseudophilotes, Glaucopsyche and Iolana (Lepidoptera, Lycaenidae). Insects 2022, 13, 1110. [Google Scholar] [CrossRef]
- Kawahara, A.Y.; Storer, C.; Carvalho, A.P.S.; Plotkin, D.M.; Condamine, F.L.; Braga, M.P.; Ellis, E.A.; Laurent, R.A.S.; Li, X.; Barve, V.; et al. A global phylogeny of butterflies reveals their evolutionary history, ancestral hosts and biogeographic origins. Nat. Ecol. Evol. 2022, 7, 903–913. [Google Scholar] [CrossRef]
- Zhang, J.; Cong, Q.; Shen, J.; Song, L.; Opler, P.A.; Grishin, N.V. Additional taxonomic refinements suggested by genomic analysis of butterflies. The Taxonomic Report of the International Lepidoptera Survey 2023, 11, 1–46. [Google Scholar] [CrossRef]
- Eliot, J.N. The higher classification of the Lycaenidae (Lepidoptera): A tentative arrangement. Bull. Nat. Hist. Mus. Entomol. Ser. 1973, 28, 371–505. [Google Scholar] [CrossRef]
- Hajibabaei, M.; Dewaard, J.R.; Ivanova, N.V.; Ratnasingham, S.; Dooh, R.T.; Kirk, S.L.; Mackie, P.M.; Hebert, P.D.N. Critical factors for assembling a high volume of DNA barcodes. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1959–1967. [Google Scholar] [CrossRef]
- Tamura, K.; Stechrer, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large data sets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Bandelt, H.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. Available online: http://popart.otago.ac.nz (accessed on 10 December 2023). [CrossRef]
- Wiemers, M.; Chazot, N.; Wheat, C.W.; Schweiger, O.; Wahlberg, N. A complete time-calibrated multi-gene phylogeny of the European butterflies. ZooKeys 2020, 938, 97–124. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1.3.1. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh, 2010. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 10 December 2023).
- Van Dam, M.H.; Matzke, N.J. Evaluating the influence of connectivity and distance on biogeographical patterns in the south-western deserts of North America. J. Biogeogr. 2016, 43, 1514–1532. [Google Scholar] [CrossRef]
- Sanderson, M.J.; Wojciechowski, M.F. Diversification rates in a temperate legume clade: Are there so many species of Astragalus (Fabaceae)? Am. J. Bot. 1996, 83, 1488–1502. [Google Scholar] [CrossRef]
- Wojciechowski, M.F.; Lavin, M.; Sanderson, M.J. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am. J. Bot. 2004, 91, 1846–1862. [Google Scholar] [CrossRef]
- Duan, L.; Wen, J.; Yang, X.; Liu, P.L.; Arslan, E.; Ertugrul, K.; Chang, Z.Y. Phylogeny of Hedysarum and tribe Hedysareae (Leguminosae: Papilionoideae) inferred from sequence data of ITS, matK, trnL-F and psbA-trnH. Taxon 2015, 64, 49–64. [Google Scholar] [CrossRef]
- Moghaddam, M.; Kazempour Osaloo, S.; Hosseiny, H.; Azimi, F. Phylogeny and divergence times of the Coluteoid clade with special reference to Colutea (Fabaceae) inferred from nrDNA ITS and two cpDNAs, matK and rpl32-trnL(UAG) sequences data. Plant Biosyst. 2017, 151, 1082–1093. [Google Scholar] [CrossRef]
- Azani, N.; Bruneau, A.; Wojciechowski, M.; Zarre, S. Miocene climate change as a driving force for multiple origins of annual species in Astragalus (Fabaceae, Papilionoideae). Mol. Phylogenetics Evol. 2019, 137, 210–221. [Google Scholar] [CrossRef]
- Nafisi, H.; Kazempour-Osaloo, S.; Mozaffarian, V.; Schneeweiss, G.M. Molecular phylogeny and divergence times of the genus Hedysarum (Fabaceae) with special reference to section Multicaulia in Southwest Asia. Plant Syst. Evol. 2019, 305, 1001–1017. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, C.; Sun, Y.; Wu, P.; Chen, X.; Dong, W.; Yang, X.; Zhou, S. Methods for Quick DNA Barcode Reference Library Construction. Ecol. Evol. 2021, 11, 11627–11638. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Higgins, L.G. The Classification European Butterflies; Collins: London, UK, 1975; 320p. [Google Scholar]
- Muto-Fujita, A.; Takemoto, K.; Kanaya, S.; Nakazato, T.; Tokimatsu, T.; Matsumoto, N.; Kono, M.; Chubachi, Y.; Ozaki, K.; Kotera, M. Data integration aids understanding of butterfly–host plant networks. Sci. Rep. 2017, 7, 43368. [Google Scholar] [CrossRef]
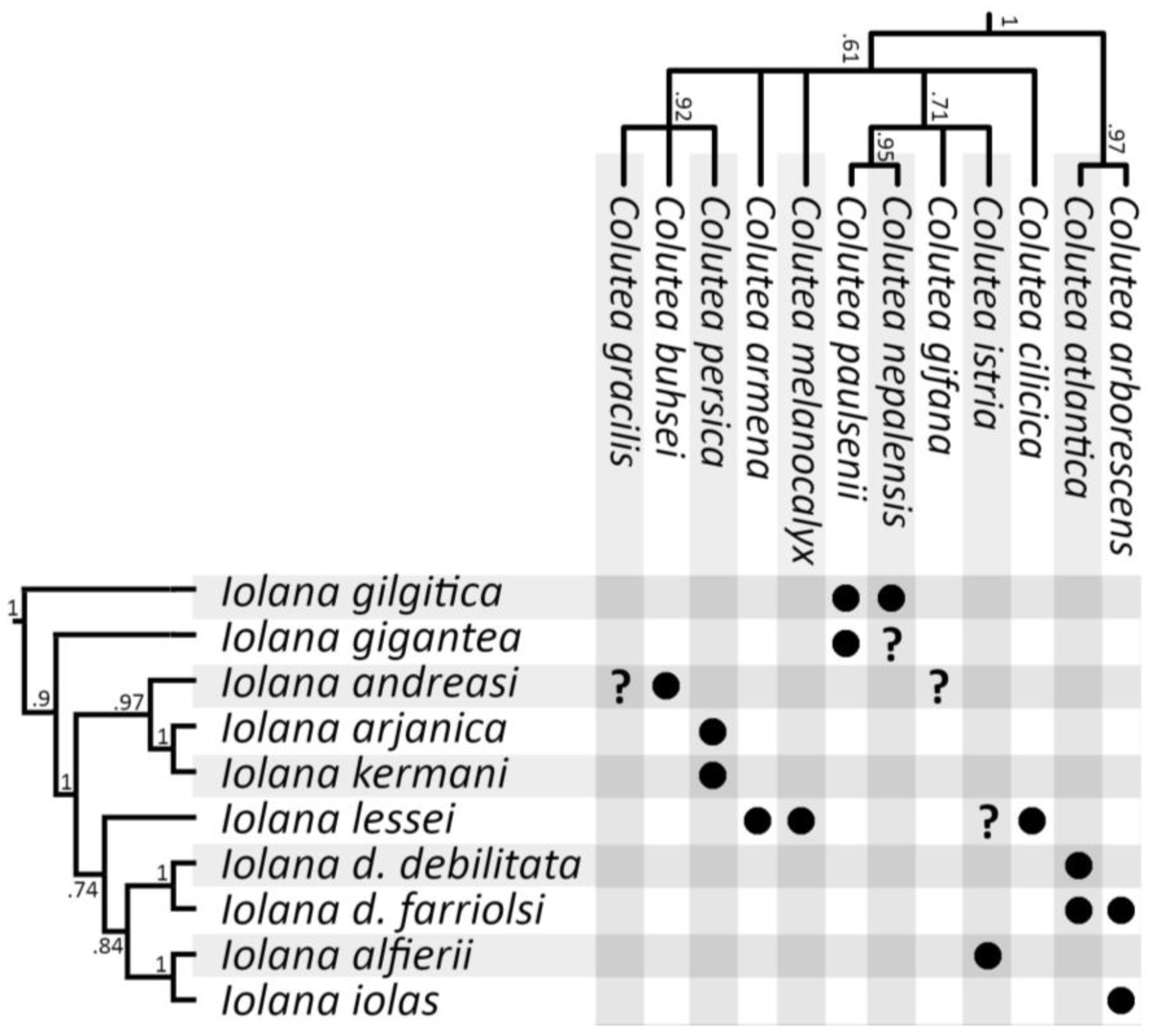
- Mirzaei, L.; Mehregan, I.; Nejadsatari, T.; Assadi, M. Phylogeny analysis of Colutea L. (Fabaceae) from Iran based on ITS sequence data. Biodiversitas 2015, 16, 168–172. [Google Scholar] [CrossRef]
- Quek, S.-P.; Davies, S.J.; Itino, T.; Pierce, N.E. Codiversification in an ant-plant mutualism: Stem texture and the evolution of host use in Crematogaster (Formicidae: Myrmicinae) inhabitants of Macaranga (Euphorbiaceae). Evolution 2004, 58, 554–570. [Google Scholar] [CrossRef]
- Groza, B.; Vodă, R.; Székely, L.; Vila, R.; Dincă, V. Genetics and extreme confinement of three overlooked butterfly species in Romania call for immediate conservation actions. J. Insect Conserv. 2021, 25, 137–146. [Google Scholar] [CrossRef]
- Thomas, J.A.; Schönrogge, K. Conservation of co-evolved interactions: Understanding the Maculinea–Myrmica complex. Insect Conserv. Divers. 2019, 12, 459–466. [Google Scholar] [CrossRef]
- Benyamini, D. Butterflies of the Levant and Neighbouring Areas (Southern Turkey, Syria, Lebanon, Israel, Jordan, Egypt, North-West Saudi Arabia & Cyprus); Volume IV: Lycaenidae; 4D Microrobotics: Beit-Aryeh, Israel, 2023; 192p. [Google Scholar]
- Zhang, J.; Cong, Q.; Shen, J.; Opler, P.A.; Grishin, N.V. Changes to North American butterfly names. Taxon. Rep. Int. Lepid. Surv. 2019, 8, 1–12. [Google Scholar]
- Coolidge, K.L. The life history of Phaedrotes piasus Boisd. (Lepidoptera: Lycaenidae). Entomol. News 1923, 34, 295–300. [Google Scholar]
- García-Barros, E.; Munguira, M.L.; Stefanescu, C.; Vives Moreno, A. Fauna Ibérica Volumen 37: Lepidoptera: Papilionoidea; Consejo Superior de Investigaciones Científicas: Madrid, Spain, 2013. [Google Scholar]
- Verovnik, R.; Lasan, M. On the presence of Iolana iolas (Ochsenheimer, 1816) in Slovenia. Nat. Slov. 2003, 5, 43–44. [Google Scholar] [CrossRef]
- Oberthür, C. Etudes de Lépidoptérologie Comparée; Smithsonian Institution: Washington, DC, USA, 1914; Volume 10, p. 392. [Google Scholar]
- Fruhstorfer, H. Neue palaearktische Lycaeniden. Dtsch. Entomol. Z. Iris 1917, 31, 24–43. [Google Scholar]
- Sheljuzhko, L. Neue palaearktische Lepidopteren-Formen. Neue Beiträge Zur Syst. Insektenkunde 1919, 1, 129–132. [Google Scholar]
- Bernardi, G. Lépidoptères Lycaenidae (sauf Agrodiaetus) récoltés en Iran par H. de Lesse en 1955 et 1958. Alexanor 1964, 3, 209–216, 273–278. [Google Scholar]
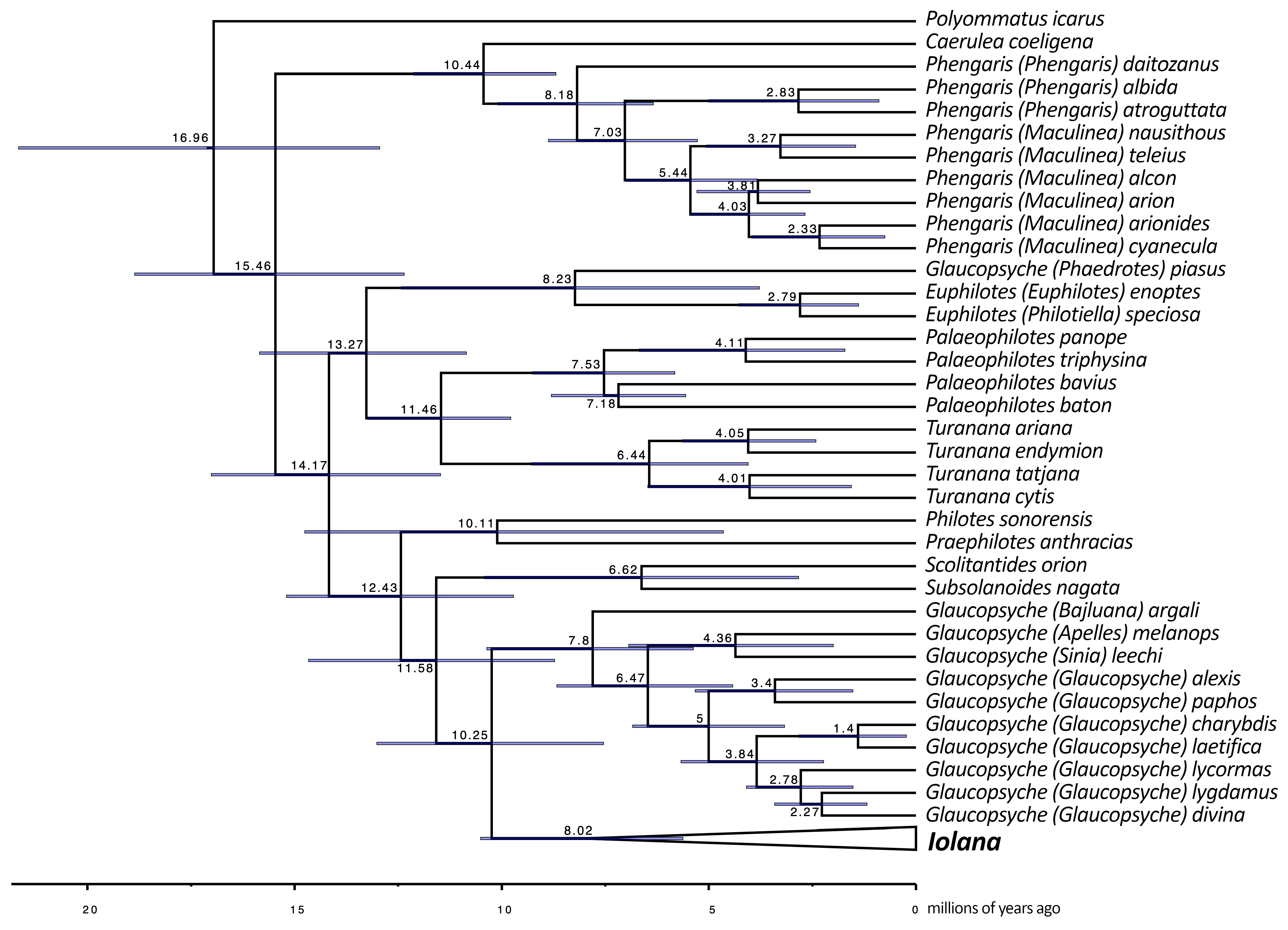


| Calibration Point | Wiemers et al. 2020 | Kawahara et al. 2022 | Results from This Study | |
|---|---|---|---|---|
| 1 | Turanana + Pseudophilotes/Palaeophilotes | 12.26 | - | 11.46 |
| 2 | Caerulea + Phengaris | - | 9.35 | 10.44 |
| 3 | P. bavius + P. baton | 8.12 | - | 7.18 |
| 4 | T. endymion + T. ariana | - | 4.67 | 4.05 |
| 5 | P. alcon + P. arion | 5.12 | - | 3.81 |
| 6 | I. iolas + I. debilitata | 3.39 | - | 4.16 |
| 7 | Euphilotes + Philotiella | - | 2.47 | 2.79 |
| 8 | G. divina + G. lygdamus | - | 2.36 | 2.27 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nazari, V.; Montagud Alario, S.; Spilani, L.; Dincă, V.; Naderi, A.; ten Hagen, W.; Vila, R. Co-Evolution of Iolana Blues with Their Host Plants and the Higher Phylogeny of Subtribe Scolitantidina (Lepidoptera, Lycaenidae). Diversity 2024, 16, 89. https://doi.org/10.3390/d16020089
Nazari V, Montagud Alario S, Spilani L, Dincă V, Naderi A, ten Hagen W, Vila R. Co-Evolution of Iolana Blues with Their Host Plants and the Higher Phylogeny of Subtribe Scolitantidina (Lepidoptera, Lycaenidae). Diversity. 2024; 16(2):89. https://doi.org/10.3390/d16020089
Chicago/Turabian StyleNazari, Vazrick, Sergio Montagud Alario, Loukia Spilani, Vlad Dincă, Alireza Naderi, Wolfgang ten Hagen, and Roger Vila. 2024. "Co-Evolution of Iolana Blues with Their Host Plants and the Higher Phylogeny of Subtribe Scolitantidina (Lepidoptera, Lycaenidae)" Diversity 16, no. 2: 89. https://doi.org/10.3390/d16020089





