Molecular and Morphological Phylogenies of Spirorbinae (Serpulidae, Polychaeta, Annelida) and the Evolution of Brooding Modes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Taxon Sampling
2.2. Collection and Preservation
2.3. DNA Extraction, Amplification, and Sequencing
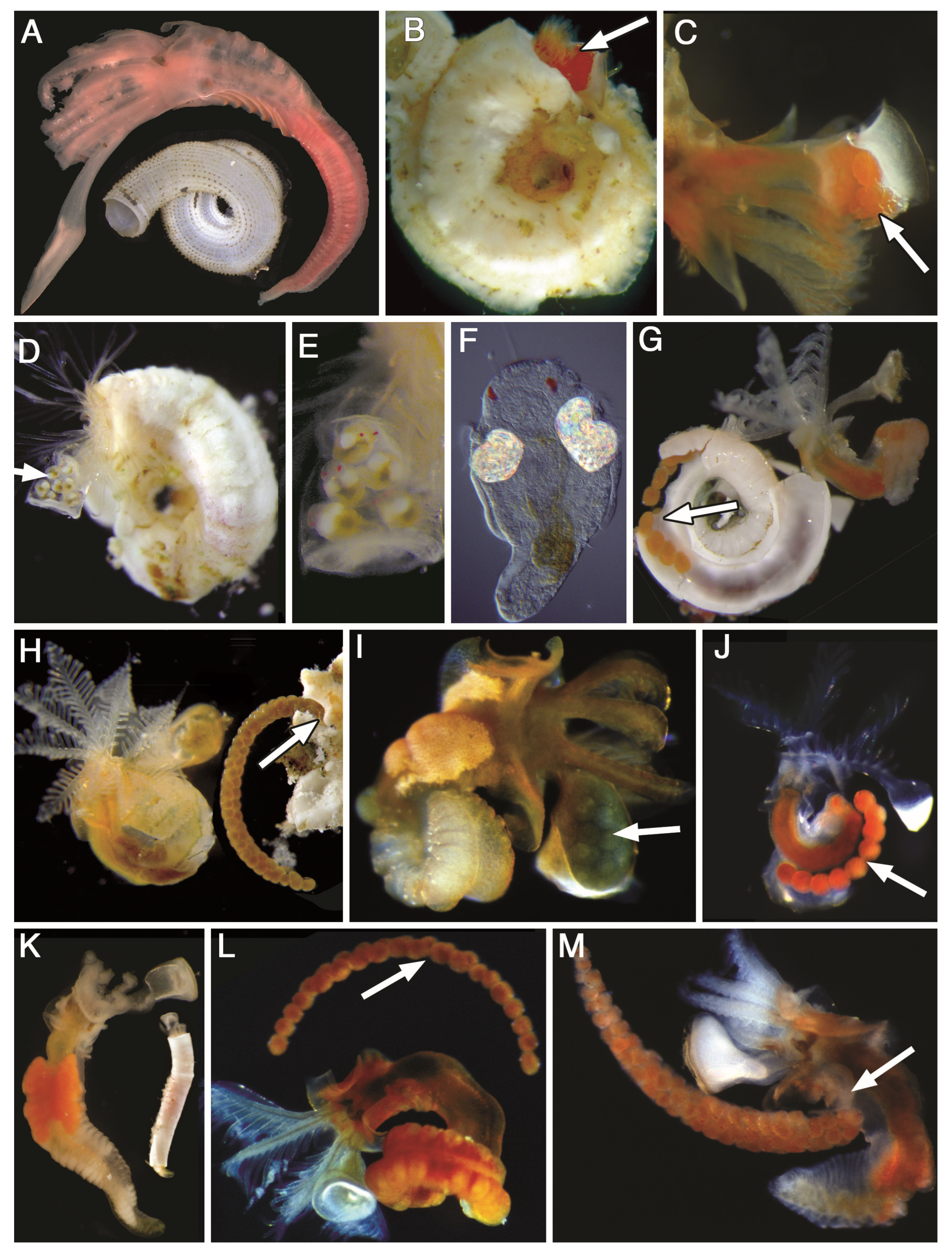
2.4. Morphological Data
2.5. Phylogenetic Analyses
2.5.1. Molecular and Molecular + Morphology Analyses
2.5.2. Transformations
3. Results
3.1. Morphology Data Analysis
3.2. Molecular-Only and Combined DNA + Morphology Data Analyses
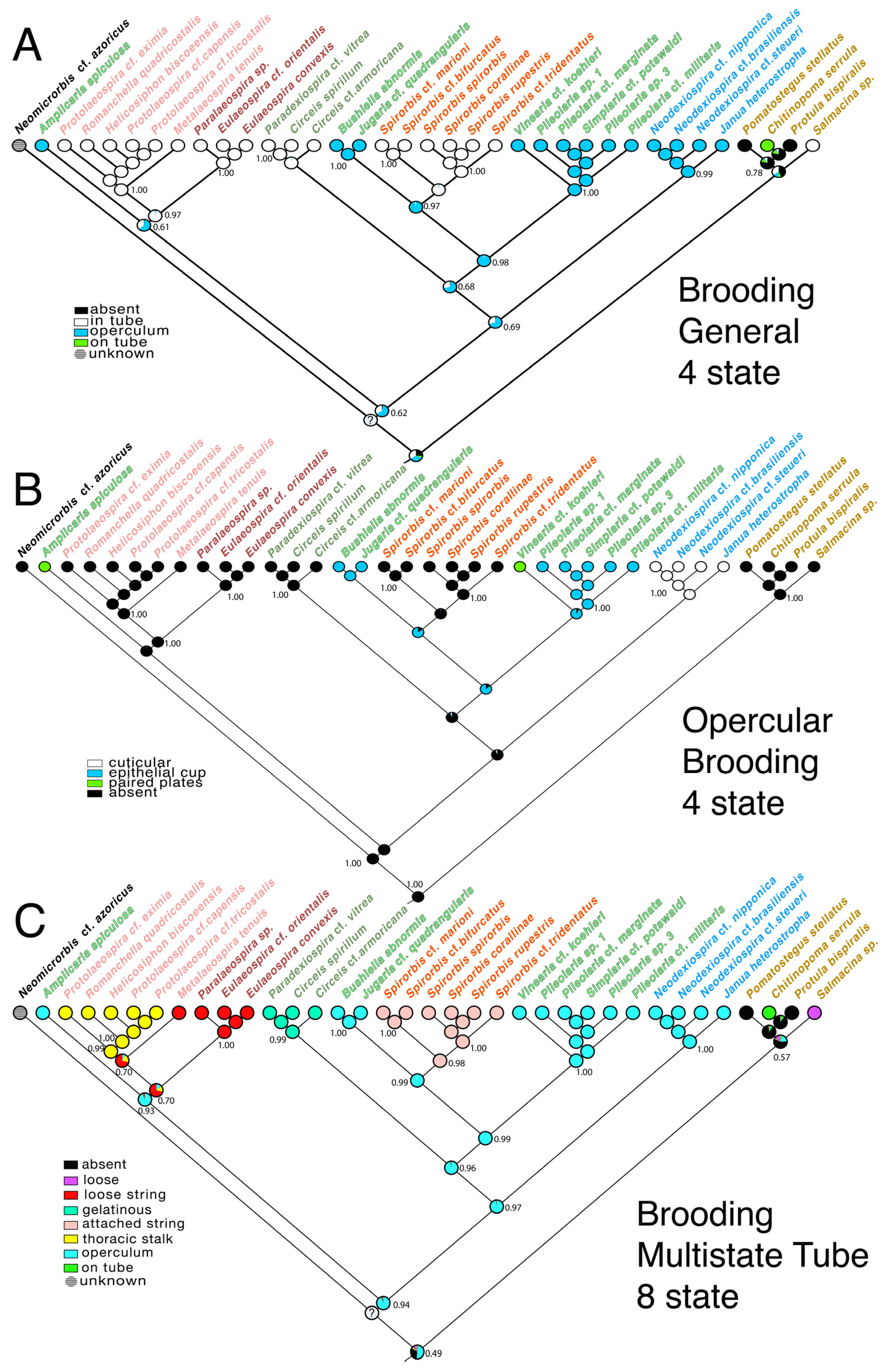
3.3. Transformations
4. Discussion
4.1. Phylogeny of the Spirorbinae
4.2. Evolution of Brooding Modes
4.3. Future Studies
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
- (1)
- Tube coiling: 0. absent; 1. flat spiral; 2. flat spiral posteriorly, uncoiled anteriorly;
- (2)
- Coiling direction: 0. dextral; 1. sinistral; 2. both;
- (3)
- Calcareous tube type: 0. opaque; 1. porcellanous: 2. vitreous;
- (4)
- Longitudinal ridges: 0. absent; 1. present;
- (5)
- Growth rings: 0. absent; 1. present;
- (6)
- Peripheral flange: 0. absent; 1. present;
- (7)
- Crystalline patch: 0. absent; 1. present;
- (8)
- Form of crystalline patch: 2. paired; 3. single; 4. diffuse;
- (9)
- Sexuality pattern: 0. simultaneous hermaphrodite; 1. gonochoric or sequential hermaphrodites;
- (10)
- Sperm head: 0. spherical; 1. elongate;
- (11)
- Larval feeding: 0. lecithotrophic; 1. planktotrophic;
- (12)
- Embryo/larvae incubation: 0. absent; 1. present;
- (13)
- Location of embryo/larvae incubation: 0. tube; 1. operculum. 2. on tube;
- (14)
- Tube incubation: 0. loose in tube; 1. unattached string; 2. gelatinous matrix; 3. posterior filament; 4. thoracic stalk;
- (15)
- Opercular brood chamber; 0. distal cuticular plate; 1. epithelial cup; 2. paired plates;
- (16)
- Dorsal convex collar flap: 0. absent; 1. present;
- (17)
- Number of radioles: 0. <10; 1. >10;
- (18)
- Larval attachment gland: 0. absent; 1. single; 2. paired;
- (19)
- Position of larval attachment gland: 0. anterior; 1. posterior;
- (20)
- Operculum: 0. absent; 1. present;
- (21)
- Opercular calcification: 0. absent; 1. present;
- (22)
- Opercular peduncle: 0. smooth; 1. with pinnules;
- (23)
- Opercular plate: 0. absent; 1. present;
- (24)
- Distal opercular plate calcified: 0. absent; 1. present;
- (25)
- Orientation of opercular plate relative to tube mouth: 0. perpendicular; 1. oblique;
- (26)
- Opercular plate spines: 0. absent; 1. present;
- (27)
- Secondary opercular plate below embryos: 0. absent: 1. present;
- (28)
- Primary operculum becomes brood chamber: 0. absent; 1. present;
- (29)
- Opercular plates retained after molting: 0. absent; 1. present;
- (30)
- Brood chamber-talon fusion: 0. absent: 1. present;
- (31)
- Primary opercular plate rim: 0. absent; 1. present;
- (32)
- Secondary opercular plate rim: 0. absent; 1. present;
- (33)
- Fiber connecting talon and tori: 0. absent; 1. present;
- (34)
- Primary talon: 0. absent; 1. present;
- (35)
- Secondary talon: 0. absent; 1. present;
- (36)
- Primary talon type: 0. spatulate; 1. vestigial; 2. tooth;
- (37)
- Terminal talon bifurcation: 0. absent; 1. present;
- (38)
- Talon projection: 0. absent; 1. present;
- (39)
- Primary talon location: 0. eccentric; 1. peripheral;
- (40)
- Primary talon external: 0. absent; 1. present;
- (41)
- Collar margin fused: 0. absent; 1. present;
- (42)
- Number of segments; concave side: 0. three; 1. four; 2. five; 3. >five;
- (43)
- Number of thoracic tori, concave side: 0. two; 1. three; 2. four; 3. >five;
- (44)
- Number of segments, convex side: 0. Three; 1. four; 2. five; 3. >five;
- (45)
- Number of thoracic tori, convex side: 0. two; 1. three; 2. >five;
- (46)
- Thoracic tori symmetry: 0. symmetric; 1. asymmetric;
- (47)
- Collar chaetae type: 0. fin-and-blade; 1. capillary and limbate;
- (48)
- Collar chaetae distribution: 0. symmetric; 1. more on convex; 2. more on concave;
- (49)
- Same collar chaetae form convex and concave side: 0. present; 1. absent;
- (50)
- Capillary collar chaetae: 0. absent; 1. present;
- (51)
- Capillary collar chaetae distribution: 1. both sides; 2. convex only;
- (52)
- Gap between fin and blade: 0. absent; 1. present;
- (53)
- Collar chaetae blade teeth: 0. fine; 1. coarse;
- (54)
- Form of collar chaetae fin teeth: 0. fine; 1. coarse;
- (55)
- Collar chaetae cross-striations: 0. absent; 1. present;
- (56)
- Collar chaetae cross striation distribution: 0. present both sides; 1. absent concave side;
- (57)
- Capillary chaetae in second thoracic fascicle: 0. absent; 1. present;
- (58)
- Capillary chaetae in third thoracic fascicle: 0. absent; 1. present;
- (59)
- Sickle (Apomatus) chaetae in third thoracic fascicles: 0. absent; 1. present;
- (60)
- Shape of sickle (Apomatus) chaetae: 0. parallel-sided; 1. pennant-shaped;
- (61)
- Thoracic uncini distribution: 0. more on concave; 1. more on convex; 2. same number on both;
- (62)
- Multiple rows of thoracic uncinal teeth (rasp-shaped): 0. absent; 1. present;
- (63)
- Transverse uncini rows: 0. straight;1. diagonal;
- (64)
- Thoracic uncini peg: 0. blunt; 1. pointed;
- (65)
- Thoracic uncinal peg lateral teeth: 0. absent; 1. present;
- (66)
- Chaetiger with smallest number of thoracic uncini: 0. last thoracic chaetiger; 1. 2nd convex; 2. 3rd convex: 3. first thoracic chaetiger;
- (67)
- Number of abdominal chaetigers: 0. 0–10; 1. 11–20; 2. 21–30; 3. 30+;
- (68)
- Abdominal uncini on convex side: 0. absent; 1. present;
- (69)
- Location of largest abdominal tori: 0. anterior; 1. posterior; 2. even distribution; 3. middle;
- (70)
- Abdominal uncini symmetry: 0. symmetric; 1. asymmetric;
- (71)
- Abdominal uncinal tooth transverse rows: 0. straight; 1. diagonal;
- (72)
- Multiple abdominal uncini tooth rows: 0. absent; 1. present;
- (73)
- Number of multiple abdominal uncini rows: 0. <ten; 1. >ten;
- (74)
- Abdominal uncinal peg: 0. flat; 1. gouge-shaped: 2. pointed;
- (75)
- Flat geniculate abdominal chaetae type: 0. pennant-shaped; 1. parallel-sided; 2. brush-like;
- (76)
- Paired abdominal chaetae: 0. absent; 1. present;
- (77)
- Distribution of abdominal chaetae: 0. entire abdomen; 1. posterior; 2. anterior;
- (78)
- Capillary abdominal chaetae: 0. absent; 1. present;
- (79)
- Capillary abdominal chaetae left/right distribution: 0. concave; 1. both sides;
- (80)
- Abdominal chaetal teeth: 0. fine; 1. coarse;
- (81)
- First abdominal chaetal tooth size: 0. same as other teeth; 1. first two small; 2. larger than other teeth; 3. first tooth small;
- (82)
- Abdominal chaetae heel projection: 0. absent; 1. present;
- (83)
- Size of abdominal chaetae vs. collar chaetae: 0. same; 1. larger; 2. Smaller.
References
- Bailey, J.H. Methods of brood protection as a basis for reclassification of the Spirorbinae (Serpulidae). Zool. J. Linn. Soc. 1969, 48, 387–407. [Google Scholar] [CrossRef]
- Kupriyanova, E.K.; Nishi, E.; ten Hove, H.A.; Rzhavsky, A.V. Life history patterns in serpulimorph polychaetes: Ecological and evolutionary perspectives. Ocean. Mar. Biol. Ann. Rev. 2001, 39, 1–101. [Google Scholar]
- Kupriyanova, E.K.; Rzhavsky, A.V.; ten Hove, H.A. Family Serpulidae Rafinesque, 1815. In Handbook of Zoology. A Natural History of the Phyla of the Animal Kingdom; Schmidt–Rhaesa, A., Ed.; De Gruyter Publishers: Berlin, Germany, 2020; pp. 213–275. [Google Scholar]
- Pillai, T.G. Studies on a collection of spirorbids from Ceylon, together with a critical review and revision of spirorbid systematics and an account of their phylogeny and zoogeography. Ceylon J. Sci. Biol. Sci. 1970, 8, 100–172. [Google Scholar]
- Macdonald, T.A. Phylogenetic relations among spirorbid subgenera and the evolution of opercular brooding. Hydrobiologia 2003, 496, 125–143. [Google Scholar] [CrossRef]
- Kupriyanova, E.K.; McDonald, T.A.; Rouse, G.W. Phylogenetic relationships within Serpulidae (Annelida: Polychaeta) inferred from molecular and morphological data. Zool. Scr. 2006, 35, 421–439. [Google Scholar] [CrossRef]
- Kupriyanova, E.K.; ten Hove, H.A.; Sket, B.; Zakšek, V.; Trontelj, P.; Rouse, G.W. Evolution of a unique freshwater cave-dwelling serpulid polychaete Marifugia cavatica Absolon and Hrabĕ, 1930. Syst. Biodivers. 2009, 7, 389–401. [Google Scholar] [CrossRef]
- Kupriyanova, E.K.; Rouse, G.W. Yet another example of paraphyly in Annelida: Molecular evidence that Sabellidae contains Serpulidae. Mol. Phylogenet. Evol. 2008, 46, 1174–1181. [Google Scholar] [CrossRef]
- Kupriyanova, E.K.; ten Hove, H.A.; Rouse, G.W. Phylogenetic relationships of Serpulidae (Annelida, Polychaeta) inferred from morphology and molecular data: Re-classification of Serpulidae. Diversity 2023, 15, 398. [Google Scholar] [CrossRef]
- Knight-Jones, P. New Spirorbidae (Polychaeta: Sedentaria) from the east Pacific, Atlantic, Indian and Southern Oceans. Zool. J. Linn. Soc. 1978, 64, 201–240. [Google Scholar] [CrossRef]
- Knight-Jones, P.; Fordy, M.R. Setal structure, function and interrelationships in Spirorbidae (Polychaeta, Sedentaria). Zool. Scr. 1979, 8, 119–138. [Google Scholar] [CrossRef]
- Fitzhugh, K. A systematic revision of the Sabellidae-Caobangiidae-Sabellongidae complex (Annelida: Polychaeta). Bull. Am. Mus. Nat. Hist. 1989, 192, 1–104. [Google Scholar]
- Rzhavsky, A.V.; Kupriyanova, E.K.; Sikorski, A.V.; Dahle, S. Calcareous tubeworms (Serpulidae, Polychaeta, Serpulidae) of the Arctic Ocean; KMK Scientific Press: Moscow, Russia, 2014; pp. 1–187. [Google Scholar]
- Rzhavsky, A.V.; Kupriyanova, E.K. Evolution of spirorbin brooding: A phylogenetic analysis and a test of an oxygen limitation hypothesis. Invertebr. Zool. 2019, 16, 409–430. [Google Scholar] [CrossRef]
- Capa, M.; Kupriyanova, E.K.; Nogueira, J.M.; Bick, A.; Tovar-Hernández, M.A. Fanworms: Yesterday, today and tomorrow. Diversity 2021, 13, 130. [Google Scholar] [CrossRef]
- Knight-Jones, P.; Thorp, C.H. The opercular brood chambers of Spirorbidae. J. Linn. Soc. (Zool.) 1984, 80, 121–133. [Google Scholar] [CrossRef]
- Potswald, H.E. The biology of fertilization and brood protection in Spirorbis (Laeospira) moerchi. Biol. Bull. 1968, 135, 208–222. [Google Scholar] [CrossRef]
- Potswald, H.E. Further observations on the structure and function of the operculum in Spirorbis borealis (Serpulidae: Spirorbinae). Biol. Bull. 1977, 152, 209–220. [Google Scholar] [CrossRef]
- Knight-Jones, P.; Knight-Jones, E.W.; Dales, R.P. Spirorbidae (Polychaeta Sedentaria) from Alaska to Panama. J. Zool. Lond. 1979, 189, 419–458. [Google Scholar] [CrossRef]
- Harris, T. Some observations upon methods of incubation in the genus Spirorbis and their possible zoogeographic significance. In Essays in Hydrobiology Presented to Leslie Harvey; Clark, R.B., Wooton, R.J., Eds.; University of Exeter: Exeter, UK, 1972; pp. 107–118. [Google Scholar]
- Hess, H.C. The evolution of parental care in brooding spirorbid polychaetes: The effect of scaling constraints. Am. Nat. 1993, 14, 577–596. [Google Scholar] [CrossRef]
- Knight-Jones, P.; Knight-Jones, E.W. Spirorbidae (Polychaeta) from Signy Island, South Orkneys, including 3 new species. Ophelia 1994, 40, 75–94. [Google Scholar] [CrossRef]
- Caullery, M.; Mesnil, F. Études sur la morphologie comparée et la phylogenie des espèces chez les Spirorbes. Bull. Scient. Fr. Belg. 1897, 30, 185–233. [Google Scholar]
- Gee, J.M. The British Spirorbinae with a description of Spirorbis cuneatus sp. n., and a review of the genus Spirorbis. Proc. Zool. Soc. London 1964, 143, 405–441. [Google Scholar] [CrossRef]
- Thorp, C.H. The structure of the operculum in Pileolaria (Pileolaria) granulata (L.) (Polychaeta, Serpulidae) and related species. J. Exp. Mar. Biol. Ecol. 1975, 20, 215–235. [Google Scholar] [CrossRef]
- Nishi, E. On the origin of brooding characteristics in spirorbids with the phylogeny of sabellids and serpulids (Annelida, Polychaeta, Sedentaria). Proc. Jpn. Soc. Syst. Zool. 1993, 49, 6–12. [Google Scholar]
- Knight-Jones, P.; Knight-Jones, E.W. Systematics, ecology and distribution of southern hemisphere spirorbids (Polychaeta; Spirorbidae). In Proceedings of the First International Polychaete Conference, Sydney, Australia, July 1983; Hutchings, P.A., Ed.; Linnean Society of New South Wales: Sydney, Australia, 1984; pp. 197–210. [Google Scholar]
- Knight-Jones, P.; Knight-Jones, E.W.; Kawahara, T.A. Review of the genus Janua, including Dexiospira (Polychaeta: Spirorbinae). Zool. J. Linn. Soc. 1975, 56, 91–129. [Google Scholar] [CrossRef]
- Knight-Jones, P.; Knight-Jones, E.W.; Buzhinskaya, G. Distribution and interrelationships of northern spirorbid genera. Bull. Mar. Sci. 1991, 48, 189–197. [Google Scholar]
- Nygren, A.; Sundberg, P. Phylogeny and evolution of reproductive modes in Autolytinae (Syllidae, Annelida). Mol. Phylogenet. Evol. 2003, 29, 235–249. [Google Scholar] [CrossRef]
- Medlin, L.; Elwood, H.J.; Stickel, S.; Sogin, M.L. The characterization of enzymatically amplified eukaryotic 16S-like rRNA coding regions. Gene 1988, 71, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Boore, J.L.; Brown, W.M. Mitochondrial genomes of Galathealinum, Helobdella, and Platynereis: Sequence and gene arrangement comparisons indicate that Pogonophora is not a Phylum and Annelida and Arthropoda are not sister taxa. Mol. Biol. Evol. 2000, 17, 87–106. [Google Scholar] [CrossRef] [PubMed]
- Struck, T.H.; Purschke, G.; Halanych, K.M. Phylogeny of Eunicida (Annelida) and exploring data congruence using partition addition bootstrap alteration (PABA) approach. Syst. Biol. 2006, 55, 1–20. [Google Scholar] [CrossRef]
- Pleijel, F. On character coding for phylogeny reconstruction. Cladistics 1995, 11, 309–315. [Google Scholar] [CrossRef]
- Knight-Jones, P. Spirorbinae (Serpulidae: Polychaeta) from southeastern Australia. Pt. 1. A new genus, four new subgenera and seven new species. Bull. Br. Mus. Nat. Hist. Ser. Zool. 1973, 24, 231–259. [Google Scholar]
- Swofford, D.L. PAUP*. Phylogenetic Analysis Using Parsimony (and Other Methods); Version 4; Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar]
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Version 3.70. Available online: http://mesquiteproject.org (accessed on 29 December 2023).
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Edler, D.; Klein, J.; Antonelli, A.; Silvestro, D. RaxmlGUI 2.0: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol. Evol. 2021, 12, 373–377. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, A.M.; Diego, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Lewis, P.O. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol. 2001, 50, 913–925. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Kupriyanova, E.K. Life history evolution in Serpulimorph polychaetes: A phylogenetic analysis. Hydrobiologia 2003, 496, 105–114. [Google Scholar] [CrossRef]
- Lehrke, J.; ten Hove, H.A.; Macdonald, T.A.; Bartolomaeus, T.; Bleidorn, C. Phylogenetic relationships of Serpulidae (Annelida: Polychaeta) based on 18S rDNA sequence data, and implications for opercular evolution. Org. Divers. Evol. 2007, 7, 195–206. [Google Scholar] [CrossRef]
- ten Hove, H.A.; Kupriyanova, E.K. Taxonomy of Serpulidae (Annelida, Polychaeta): The state of affairs. Zootaxa 2009, 2036, 1–126. [Google Scholar]
- Zibrowius, H. Un espèce actuelle du genre Neomicrorbis Rovereto (Polychaeta, Serpulidae) découverte dans l‘étage bathyal aux Açores. Bull. Mus. Natl. Hist. Nat. 1972, 33, 423–430. [Google Scholar]
- Ippolitov, A.P.; Rzhavsky, A.V. Tube morphology, ultrastructures and mineralogy in Recent Spirorbinae (Annelida; Polychaeta; Serpulidae). 2. Tribe Spirorbini. Invert. Zool. 2015, 12, 61–92. [Google Scholar] [CrossRef]
- Ippolitov, A.P.; Rzhavsky, A.V. Tube morphology, ultrastructures and mineralogy in Recent Spirorbinae (Annelida; Polychaeta; Serpulidae). 3. Tribe Circeini. Invert. Zool. 2015, 12, 151–173. [Google Scholar] [CrossRef]
- Pillai, T.G. Some marine and brackish-water serpulid polychaetes from Ceylon, including new genera and species. Ceylon J. Sci. Biol. Sci. 1960, 3, 1–40. [Google Scholar]
- Brown, A.C.; Rouse, G.W.; Hutchings, P.; Colgan, D. Assessing the usefulness of histone H3, U2 snRNA and 28S rDNA in analyses of polychaete relationships. Aus. J. Zool. 1999, 47, 499–516. [Google Scholar]
- Norén, M.; Jondelius, U. Phylogeny of the Prolecithophora (Platyhelminthes) Inferred from 18S rDNA Sequences. Cladistics 1999, 15, 103–112. [Google Scholar] [CrossRef]


| Taxa | Source | Voucher Number | 18S | 28S |
|---|---|---|---|---|
| Filograninae outgroup | ||||
| Chitinopoma serrula | Norway | E3524 | DQ317112 | EU195350 |
| Pomatostegus actinoceras | Mexico | W.42378 | OQ379437 | OQ389670 |
| Protula bispiralis | Japan | E3657 | OQ379439 | OQ389672 |
| Salmacina sp. | Edithburgh, SA, Australia | E3499 | DQ317126 | EU256545 |
| Spirorbinae | ||||
| Neomicrorbis cf. azoricus | Cocos (Keeling) Isl. Australia | W.54342 | PP002544 | PP002543 |
| Pileolariini | ||||
| Amplicaria spiculosa | Whyalla, SA, Australia | E3490 | DQ242560 | DQ242579 |
| Bushiella abnormis | Barkley Sound, BC, Canada | E3488 | DQ242563 | DQ242598 |
| Jugaria cf. quadrangularis | Barkley Sound, BC, Canada | E3479 | DQ242564 | DQ242599 |
| Pileolaria cf. marginata | Barkley Sound, BC, Canada | E3478 | DQ242565 | DQ242594 |
| Pileolaria cf. militaris | Pt. Cartwright, QLD, Australia | E3492 | DQ242567 | DQ242593 |
| Pileolaria sp. 1 (orange eggs) | Whyalla, SA, Australia | E3493 | DQ242562 | DQ242596 |
| Pileolaria sp. 3 (gold eggs) | Rapid Bay, SA, Australia | E3494 | DQ242568 | DQ242597 |
| Simplaria cf. potswaldi | Barkley Sound, Canada | E3504 | DQ242566 | DQ242595 |
| Vinearia cf. koehleri | Pt. Cartwright, QLD, Australia | E3475 | DQ242561 | DQ242592 |
| Januini | ||||
| Janua heterostropha | Sangerdi, Iceland | E3506 | DQ242548 | DQ242585 |
| Neodexiospira cf. brasiliensis | Barkley Sound, BC, Canada | E3498 | DQ242550 | DQ242586 |
| Neodexiospira cf. nipponica | Barkley Sound, BC, Canada | E3486 | DQ242549 | DQ242587 |
| Neodexiospira cf. steueri | Encounter Bay, SA, Australia | E3523 | DQ242551 | DQ242588 |
| Paralaeospirini | ||||
| Paralaeospira sp. | Encounter Bay, SA, Australia | E3485 | DQ242555 | DQ242580 |
| Eulaeospira convexis | North Bondi, NSW, Australia | E3496 | DQ242552 | DQ242582 |
| Eulaeospira cf. orientalis | Encounter Bay, SA, Australia | E3495 | DQ242553 | DQ242581 |
| Romanchellini | ||||
| Helicosiphon biscoensis | Antarctica | BIC A4000 | OQ379432 | OQ392408 |
| Protolaeospira cf. eximia | Barkley Sound, BC, Canada | E3482 | DQ242556 | DQ242584 |
| Protolaeospira cf. tricostalis | Bondi, NSW, Australia | E3487 | DQ242557 | DQ242606 |
| Protolaeospira cf. capensis | Bondi, NSW, Australia | E3484 | DQ242558 | DQ242607 |
| Romanchella quadricostalis | Kangaroo Isl., SA, Australia | E3491 | DQ242559 | DQ242608 |
| Metalaeospira tenuis | Port Lincoln, SA, Australia | E3480 | DQ242554 | DQ242583 |
| Circeini | ||||
| Circeis cf. armoricana | Barkley Sound, BC, Canada | E3476 | DQ242545 | DQ242589 |
| Circeis spirillum | Stykkishlómør, Iceland | E3507 | DQ242546 | DQ242590 |
| Paradexiospira cf. vitrea | Barkley Sound, BC, Canada | E3483 | DQ242547 | DQ242591 |
| Spirorbini | ||||
| Spirorbis cf. bifurcatus | Barkley Sound, BC, Canada | E3489 | DQ242569 | DQ242600 |
| Spirorbis cf. marioni | PJs Pets, Edmonton, Canada | E3481 | DQ242570 | DQ242605 |
| Spirorbis corallinae | Finnøy, Norway | E3497 | DQ242572 | DQ242603 |
| Spirorbis rupestris | Finnøy, Norway | E3500 | DQ242571 | DQ242601 |
| Spirorbis spirorbis | Sangerdi, Iceland | E3357 | AY577887 | DQ242604 |
| Spirorbis cf. tridentatus | Finnøy, Norway | E3477 | DQ242573 | DQ242602 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rouse, G.W.; Macdonald, T.A.; Kupriyanova, E.K. Molecular and Morphological Phylogenies of Spirorbinae (Serpulidae, Polychaeta, Annelida) and the Evolution of Brooding Modes. Diversity 2024, 16, 237. https://doi.org/10.3390/d16040237
Rouse GW, Macdonald TA, Kupriyanova EK. Molecular and Morphological Phylogenies of Spirorbinae (Serpulidae, Polychaeta, Annelida) and the Evolution of Brooding Modes. Diversity. 2024; 16(4):237. https://doi.org/10.3390/d16040237
Chicago/Turabian StyleRouse, Greg W., Tara A. Macdonald, and Elena K. Kupriyanova. 2024. "Molecular and Morphological Phylogenies of Spirorbinae (Serpulidae, Polychaeta, Annelida) and the Evolution of Brooding Modes" Diversity 16, no. 4: 237. https://doi.org/10.3390/d16040237






