Translation Elongation Factor 1-Alpha Sequencing Provides Reliable Tool for Identification of Fusarium graminearum Species Complex Members
Abstract
1. Introduction
2. Materials and Methods
2.1. FGSC Isolate Sequences, Format Converting, and Preparing Samples for Alignment
- (i)
- The first of these three aligned “.fasta” files, named Set I, included F. asiaticum alignment with 107 isolates (not containing Fgss).
- (ii)
- The second one, named Set II, included Fgss alignment with 115 isolates (not containing F. asiaticum).
- (iii)
- The third one, named Set III, included all 248 isolates.
2.2. Distance-Based Phylogenetic Methods
2.3. Character-Based Phylogenetic Methods
2.4. Principal Component Analysis (PCA) by RStudio
3. Results and Discussion
3.1. Distance-Based Phylogenetic Analysis
3.2. Character-Based Phylogenetic Analysis
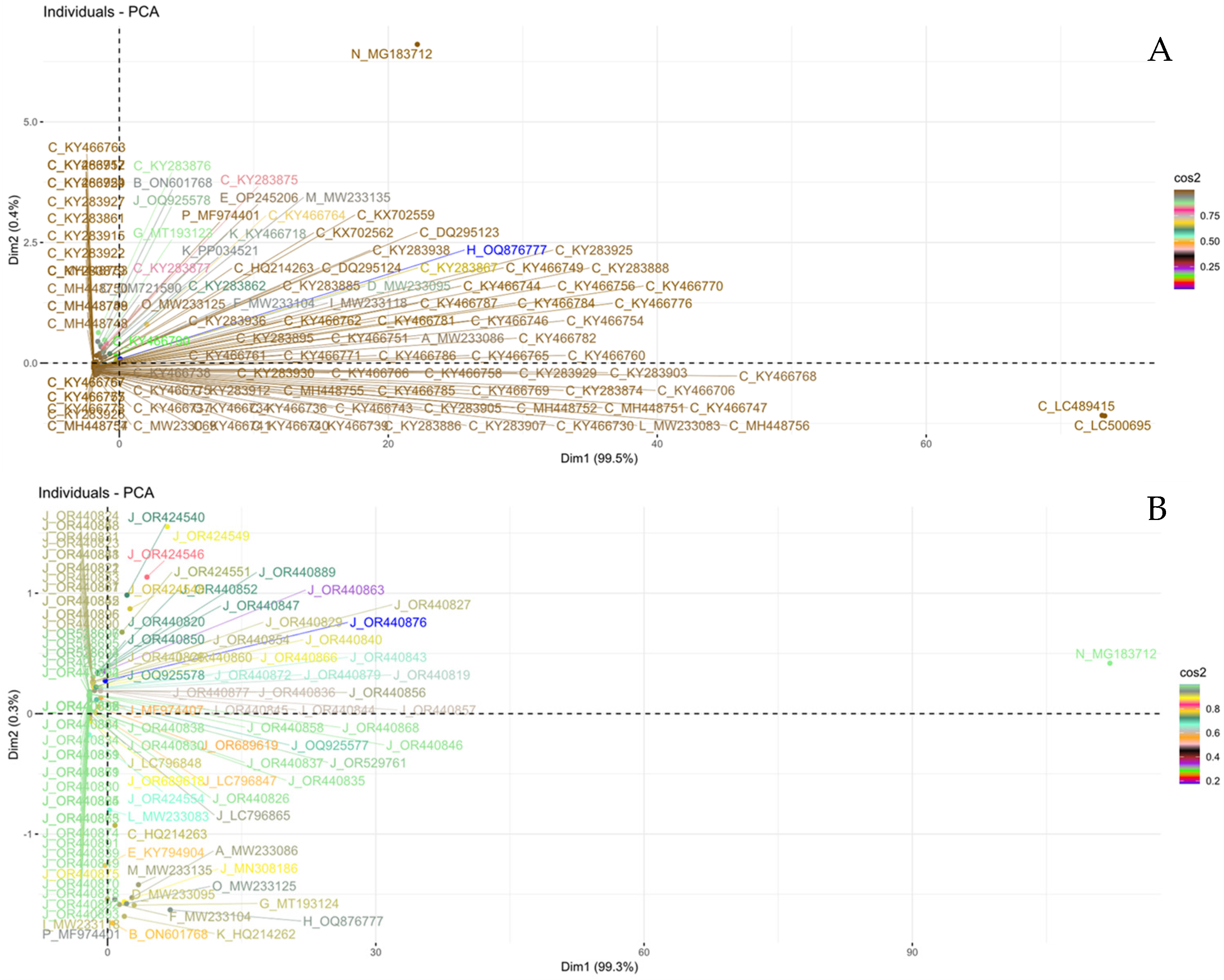
3.3. PCA-Based Similarity Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- McMullen, M.; Jones, R.; Gallenberg, D. Scab of Wheat and Barley: A Re-emerging Disease of Devastating Impact. Plant Dis. 1997, 81, 1340–1348. [Google Scholar] [CrossRef] [PubMed]
- Parry, D.W.; Jenkinson, P.; McLE, L. Fusarium ear blight (scab) in small grain cereals—A review. Plant Pathol. 1995, 44, 207–238. [Google Scholar] [CrossRef]
- Yli-Manila, T.; Gagkaeva, T.Y. Fusarium toxins in cereals in northern Europe and Asia. In Fungi; CRC Press: Boca Raton, FL, USA, 2018; pp. 293–317. [Google Scholar]
- Desjardins, A.E. Fusarium Mycotoxins: Chemistry, Genetics, and Biology. 2006. Available online: https://www.cabidigitallibrary.org/doi/full/10.5555/20063036927 (accessed on 14 July 2024).
- Desjardins, A.E.; Proctor, R.H. Molecular biology of Fusarium mycotoxins. Int. J. Food Microbiol. 2007, 119, 47–50. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- O’donnell, K.; Humber, R.A.; Geiser, D.M.; Kang, S.; Park, B.; Robert, V.A.R.G.; Crous, P.W.; Johnston, P.R.; Aoki, T.; Rooney, A.P.; et al. Phylogenetic diversity of insecticolous fusaria inferred from multilocus DNA sequence data and their molecular identification via FUSARIUM-ID and Fusarium MLST. Mycologia 2012, 104, 427–445. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, P.; Chandler, E.; Draeger, R.C.; Gosman, N.E.; Simpson, D.R.; Thomsett, M.; Wilson, A.H. Molecular tools to study epidemiology and toxicology of Fusarium head blight of cereals. Eur. J. Plant Pathol. 2003, 109, 691–703. [Google Scholar] [CrossRef]
- Schilling, A. Polymerase chain reaction-based assays for species-specific detection of Fusarium culmorum, F. graminearum, and F. avenaceum. Phytopathology 1996, 86, 515–522. [Google Scholar] [CrossRef]
- Zhang, X.; Ma, H.; Zhou, Y.; Xing, J.; Chen, J.; Yu, G.; Sun, X.; Wang, L. Identification and Genetic Division of Fusarium graminearum and Fusarium asiaticum by Species-Specific SCAR Markers. J. Phytopathol. 2014, 162, 81–88. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.; Tacke, B.K.; Casper, H.H. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc. Natl. Acad. Sci. USA 2000, 97, 7905–7910. [Google Scholar] [CrossRef]
- O’Donnell, K.; Sarver, B.A.J.; Brandt, M.; Chang, D.C.; Noble-Wang, J.; Park, B.J.; Sutton, D.A.; Benjamin, L.; Lindsley, M.; Padhye, A.; et al. Phylogenetic Diversity and Microsphere Array-Based Genotyping of Human Pathogenic Fusaria, Including Isolates from the Multistate Contact Lens-Associated U.S. Keratitis Outbreaks of 2005 and 2006. J. Clin. Microbiol. 2007, 45, 2235–2248. [Google Scholar] [CrossRef]
- O’donnell, K.; Ward, T.J.; Aberra, D.; Kistler, H.C.; Aoki, T.; Orwig, N.; Kimura, M.; Bjørnstad, S.; Klemsdal, S.S. Multilocus genotyping and molecular phylogenetics resolve a novel head blight pathogen within the Fusarium graminearum species complex from Ethiopia. Fungal Genet. Biol. 2008, 45, 1514–1522. [Google Scholar] [CrossRef] [PubMed]
- Sarver, B.A.J.; Ward, T.J.; Gale, L.R.; Broz, K.; Kistler, H.C.; Aoki, T.; Nicholson, P.; Carter, J.; O’donnell, K. Novel Fusarium head blight pathogens from Nepal and Louisiana revealed by multilocus genealogical concordance. Fungal Genet. Biol. 2011, 48, 1096–1107. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, S.; Hou, R.; Zhao, Z.; Zheng, Q.; Xu, Q.; Zheng, D.; Wang, G.; Liu, H.; Gao, X.; et al. Functional analysis of the kinome of the wheat scab fungus Fusarium graminearum. PLoS Pathog. 2011, 7, e1002460. [Google Scholar] [CrossRef] [PubMed]
- Garmendia, G.; Umpierrez-Failache, M.; Ward, T.J.; Vero, S. Development of a PCR-RFLP method based on the transcription elongation factor 1-gene to differentiate Fusarium graminearum from other species within the Fusarium graminearum species complex. Food Microbiol. 2018, 70, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Tunal, B.; Yrk, E.; Sefer, Ö.; Kansu, B.; Sharifnabi, B. First Report on Identification of Fusarium graminearum Species Complex Members from Turkey and Iran. Turk. J. Agric. Food Sci. Technol. 2019, 7, 1040–1045. [Google Scholar] [CrossRef]
- Qu, B.; Li, H.P.; Zhang, J.B.; Huang, T.; Carter, J.; Liao, Y.C.; Nicholson, P. Comparison of genetic diversity and pathogenicity of fusarium head blight pathogens from China and Europe by SSCP and seedling assays on wheat. Plant Pathol. 2008, 57, 642–651. [Google Scholar] [CrossRef]
- Wang, C.-L.; Cheng, Y.-H. Identification and trichothecene genotypes of Fusarium graminearum species complex from wheat in Taiwan. Bot. Stud. 2017, 58, 4. [Google Scholar] [CrossRef]
- Yörük, E.; Albayrak, G. Genetic characterization of Fusarium graminearum and F. culmorum isolates from Turkey by using random-amplified polymorphic DNA. Genetics and Molecular Research 2013, 12, 1360–1372. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, 78–82. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Hafez, M.; Abdelmagid, A.; Adam, L.R.; Daayf, F. Specific Detection and Identification of Fusarium graminearum Sensu Stricto Using a PCR-RFLP Tool and Specific Primers Targeting the Translational Elongation Factor 1α Gene. Plant Dis. 2020, 104, 1076–1086. [Google Scholar] [CrossRef] [PubMed]
- Boutigny, A.-L.; Gautier, A.; Basler, R.; Dauthieux, F.; Leite, S.; Valade, R.; Aguayo, J.; Ioos, R.; Laval, V. Metabarcoding targeting the EF1 alpha region to assess Fusarium diversity on cereals. PLoS ONE 2019, 14, e0207988. [Google Scholar] [CrossRef] [PubMed]
- Geiser, D.M.; Aoki, T.; Bacon, C.W.; Baker, S.E.; Bhattacharyya, M.K.; Brandt, M.E.; Brown, D.W.; Burgess, L.W.; Chulze, S.; Coleman, J.J.; et al. One Fungus, One Name: Defining the Genus Fusarium in a Scientifically Robust Way That Preserves Longstanding Use. Phytopathology 2013, 103, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Geiser, D.M.; Al-Hatmi, A.M.S.; Ao, T.; Arie, T.; Balmas, V.; Barnes, I.; Viljoen, A. Phylogenomic Analysis of a 55. 1-kb 19-Gene Dataset Resolves a Monophyletic Fusarium that Includes the Fusarium solani Species Complex. Phytopathology 2021, 111, 1064–1079. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, C.L.; Somma, S.; Proctor, R.H.; Stea, G.; Mulè, G.; Logrieco, A.F.; Pinto, V.F.; Moretti, A. Genetic Diversity in Fusarium graminearum from a Major Wheat-Producing Region of Argentina. Toxins 2011, 3, 1294–1309. [Google Scholar] [CrossRef] [PubMed]
- Bentley, A.R.; Cromey, M.G.; Farrokhi-Nejad, R.; Leslie, J.F.; Summerell, B.A.; Burgess, L.W. Fusarium crown and root rot pathogens associated with wheat and grass stem bases on the South Island of New Zealand. Australas. Plant Pathol. 2006, 35, 495. [Google Scholar] [CrossRef]
- Minati, M.H.; Mohammed-Ameen, M.K. Novel report on six Fusarium species associated with head blight and crown rot of wheat in Basra province, Iraq. Bull. Natl. Res. Cent. 2019, 43, 139. [Google Scholar] [CrossRef]
- Stępniewska, H.; Jankowiak, R.; Bilański, P.; Hausner, G. Structure and Abundance of Fusarium Communities Inhabiting the Litter of Beech Forests in Central Europe. Forests 2021, 12, 811. [Google Scholar] [CrossRef]
- Fernández-Ortuño, D.; Loza-Reyes, E.; Atkins, S.L.; Fraaije, B.A. The CYP51C gene, a reliable marker to resolve interspecific phylogenetic relationships within the Fusarium species complex and a novel target for species-specific PCR. Int. J. Food Microbiol. 2010, 144, 301–309. [Google Scholar] [CrossRef]
- Noel, Z.A.; Roze, L.V.; Breunig, M.; Trail, F. Endophytic Fungi as a Promising Biocontrol Agent to Protect Wheat from Fusarium graminearum Head Blight. Plant Dis. 2022, 106, 595–602. [Google Scholar] [CrossRef]
- Sanna, M.; Martino, I.; Guarnaccia, V.; Mezzalama, M. Diversity and Pathogenicity of Fusarium Species Associated with Stalk and Crown Rot in Maize in Northern Italy. Plants 2023, 12, 3857. [Google Scholar] [CrossRef] [PubMed]
- Backeljau, T.; De Bruyn, L.; De Wolf, H.; Jordaens, K.; Van Dongen, S.; Winnepennincks, B. Multiple UPGMA and Neighbor-joining Trees and the Performance of Some Computer Packages. Mol. Biol. Evol. 1996, 13, 309–313. [Google Scholar] [CrossRef]
- Drummond, A.; Rodrigo, A.G. Reconstructing Genealogies of Serial Samples Under the Assumption of a Molecular Clock Using Serial-Sample UPGMA. Mol. Biol. Evol. 2000, 17, 1807–1815. [Google Scholar] [CrossRef][Green Version]
- Michu, E. A short guide to phylogeny reconstruction. Plant Soil Environ. 2007, 53, 442–446. [Google Scholar] [CrossRef]
- Saitou, N. Property and efficiency of the maximum likelihood method for molecular phylogeny. J. Mol. Evol. 1988, 27, 261–273. [Google Scholar] [CrossRef]

| No | Species | Sample Code | No | Species | Sample Code | No | Species | Sample Code | No | Species | Sample Code |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F. acacia-mearnsii | A_OP245201 | 63 | F. asiaticum | C_KY466730 | 125 | Fgss | J_OR440886 | 187 | Fgss | J_OR440824 |
| 2 | F. acacia-mearnsii | A_MW233086 | 64 | F. asiaticum | C_KY466706 | 126 | Fgss | J_OR440885 | 188 | Fgss | J_OR440823 |
| 3 | F. aethiopicum | B_ON601768 | 65 | F. asiaticum | C_KY283938 | 127 | Fgss | J_OR440884 | 189 | Fgss | J_OR440822 |
| 4 | F. aethiopicum | B_MW233126 | 66 | F. asiaticum | C_KY283936 | 128 | Fgss | J_OR440883 | 190 | Fgss | J_OR440821 |
| 5 | F. asiaticum | C_LC489415 | 67 | F. asiaticum | C_KY283930 | 129 | Fgss | J_OR440882 | 191 | Fgss | J_OR440820 |
| 6 | F. asiaticum | C_LC500695 | 68 | F. asiaticum | C_KY283929 | 130 | Fgss | J_OR440881 | 192 | Fgss | J_OR440819 |
| 7 | F. asiaticum | C_MW233069 | 69 | F. asiaticum | C_KY283927 | 131 | Fgss | J_OR440879 | 193 | Fgss | J_OR689619 |
| 8 | F. asiaticum | C_OM721590 | 70 | F. asiaticum | C_KY283926 | 132 | Fgss | J_OR440880 | 194 | Fgss | J_OR689618 |
| 9 | F. asiaticum | C_MH448758 | 71 | F. asiaticum | C_KY283925 | 133 | Fgss | J_OR440878 | 195 | Fgss | J_MN308186 |
| 10 | F. asiaticum | C_MH448757 | 72 | F. asiaticum | C_KY283924 | 134 | Fgss | J_OR440877 | 196 | Fgss | J_MF974407 |
| 11 | F. asiaticum | C_MH448756 | 73 | F. asiaticum | C_KY283922 | 135 | Fgss | J_OR440876 | 197 | Fgss | J_LC796865 |
| 12 | F. asiaticum | C_MH448755 | 74 | F. asiaticum | C_KY283917 | 136 | Fgss | J_OR440875 | 198 | Fgss | J_LC796848 |
| 13 | F. asiaticum | C_MH448754 | 75 | F. asiaticum | C_KY283915 | 137 | Fgss | J_OR440874 | 199 | Fgss | J_LC796847 |
| 14 | F. asiaticum | C_MH448753 | 76 | F. asiaticum | C_KY283912 | 138 | Fgss | J_OR440873 | 200 | Fgss | J_OR529761 |
| 15 | F. asiaticum | C_MH448752 | 77 | F. asiaticum | C_KY283907 | 139 | Fgss | J_OR440872 | 201 | Fgss | J_OR528697 |
| 16 | F. asiaticum | C_MH448751 | 78 | F. asiaticum | C_KY283905 | 140 | Fgss | J_OR440871 | 202 | Fgss | J_OR528696 |
| 17 | F. asiaticum | C_MH448750 | 79 | F. asiaticum | C_KY283903 | 141 | Fgss | J_OR440870 | 203 | Fgss | J_OR528695 |
| 18 | F. asiaticum | C_MH448749 | 80 | F. asiaticum | C_KY283895 | 142 | Fgss | J_OR440869 | 204 | Fgss | J_OR528694 |
| 19 | F. asiaticum | C_MH448748 | 81 | F. asiaticum | C_KY283886 | 143 | Fgss | J_OR440868 | 205 | Fgss | J_OR528693 |
| 20 | F. asiaticum | C_KY466790 | 82 | F. asiaticum | C_KY283888 | 144 | Fgss | J_OR440867 | 206 | Fgss | J_OR424554 |
| 21 | F. asiaticum | C_KY466787 | 83 | F. asiaticum | C_KY283885 | 145 | Fgss | J_OR440866 | 207 | Fgss | J_OR424551 |
| 22 | F. asiaticum | C_KY466786 | 84 | F. asiaticum | C_KY283879 | 146 | Fgss | J_OR440865 | 208 | Fgss | J_OR424549 |
| 23 | F. asiaticum | C_KY466785 | 85 | F. asiaticum | C_KY283877 | 147 | Fgss | J_OR440864 | 209 | Fgss | J_OR424548 |
| 24 | F. asiaticum | C_KY466784 | 86 | F. asiaticum | C_KY283876 | 148 | Fgss | J_OR440863 | 210 | Fgss | J_OR424546 |
| 25 | F. asiaticum | C_KY466782 | 87 | F. asiaticum | C_KY283875 | 149 | Fgss | J_OR440862 | 211 | Fgss | J_OR424541 |
| 26 | F. asiaticum | C_KY466781 | 88 | F. asiaticum | C_KY283874 | 150 | Fgss | J_OR440861 | 212 | Fgss | J_OR424540 |
| 27 | F. asiaticum | C_KY466778 | 89 | F. asiaticum | C_KY283862 | 151 | Fgss | J_OR440860 | 213 | Fgss | J_OQ925578 |
| 28 | F. asiaticum | C_KY466777 | 90 | F. asiaticum | C_KY283867 | 152 | Fgss | J_OR440859 | 214 | Fgss | J_OQ925577 |
| 29 | F. asiaticum | C_KY466776 | 91 | F. asiaticum | C_KY283861 | 153 | Fgss | J_OR440858 | 215 | F. meridionale | K_PP034521 |
| 30 | F. asiaticum | C_KY466775 | 92 | F. asiaticum | C_KX702562 | 154 | Fgss | J_OR440857 | 216 | F. meridionale | K_MW233092 |
| 31 | F. asiaticum | C_KY466771 | 93 | F. asiaticum | C_KX702559 | 155 | Fgss | J_OR440856 | 217 | F. meridionale | K_MG838991 |
| 32 | F. asiaticum | C_KY466770 | 94 | F. asiaticum | C_DQ295124 | 156 | Fgss | J_OR440855 | 218 | F. meridionale | K_MG838988 |
| 33 | F. asiaticum | C_KY466769 | 95 | F. asiaticum | C_DQ295123 | 157 | Fgss | J_OR440854 | 219 | F. meridionale | K_MG838955 |
| 34 | F. asiaticum | C_KY466768 | 96 | F. asiaticum | C_HQ214263 | 158 | Fgss | J_OR440853 | 220 | F. meridionale | K_MG838949 |
| 35 | F. asiaticum | C_KY466767 | 97 | F. austroamericanum | D_MW233095 | 159 | Fgss | J_OR440852 | 221 | F. meridionale | K_MG838948 |
| 36 | F. asiaticum | C_KY466766 | 98 | F. boothii | E_OP245207 | 160 | Fgss | J_OR440851 | 222 | F. meridionale | K_MG838947 |
| 37 | F. asiaticum | C_KY466765 | 99 | F. boothii | E_OP245206 | 161 | Fgss | J_OR440850 | 223 | F. meridionale | K_MH448800 |
| 38 | F. asiaticum | C_KY466764 | 100 | F. boothii | E_OP245205 | 162 | Fgss | J_OR440849 | 224 | F. meridionale | K_MH448801 |
| 39 | F. asiaticum | C_KY466763 | 101 | F. boothii | E_OP245204 | 163 | Fgss | J_OR440848 | 225 | F. meridionale | K_MH448802 |
| 40 | F. asiaticum | C_KY466762 | 102 | F. boothii | E_PP035520 | 164 | Fgss | J_OR440847 | 226 | F. meridionale | K_MH448803 |
| 41 | F. asiaticum | C_KY466761 | 103 | F. boothii | E_PP035519 | 165 | Fgss | J_OR440846 | 227 | F. meridionale | K_MH448796 |
| 42 | F. asiaticum | C_KY466760 | 104 | F. boothii | E_ON601969 | 166 | Fgss | J_OR440845 | 228 | F. meridionale | K_MH448797 |
| 43 | F. asiaticum | C_KY466759 | 105 | F. boothii | E_ON601968 | 167 | Fgss | J_OR440844 | 229 | F. meridionale | K_MH448798 |
| 44 | F. asiaticum | C_KY466757 | 106 | F. boothii | E_ON601967 | 168 | Fgss | J_OR440843 | 230 | F. meridionale | K_MH448799 |
| 45 | F. asiaticum | C_KY466758 | 107 | F. boothii | E_MW233088 | 169 | Fgss | J_OR440842 | 231 | F. meridionale | K_KY466783 |
| 46 | F. asiaticum | C_KY466756 | 108 | F. boothii | E_KY794904 | 170 | Fgss | J_OR440841 | 232 | F. meridionale | K_KY466779 |
| 47 | F. asiaticum | C_KY466755 | 109 | F. brasilicum | F_MW233104 | 171 | Fgss | J_OR440840 | 233 | F. meridionale | K_KY466774 |
| 48 | F. asiaticum | C_KY466754 | 110 | F. cortaderiae | G_MW233098 | 172 | Fgss | J_OR440839 | 234 | F. meridionale | K_KY466773 |
| 49 | F. asiaticum | C_KY466752 | 111 | F. cortaderiae | G_MT193123 | 173 | Fgss | J_OR440838 | 235 | F. meridionale | K_KY466772 |
| 50 | F. asiaticum | C_KY466751 | 112 | F. cortaderiae | G_MT193124 | 174 | Fgss | J_OR440837 | 236 | F. meridionale | K_KY466735 |
| 51 | F. asiaticum | C_KY466749 | 113 | F. culmorum | H_OQ876777 | 175 | Fgss | J_OR440836 | 237 | F. meridionale | K_KY466733 |
| 52 | F. asiaticum | C_KY466747 | 114 | F. gerlachii | I_MW233118 | 176 | Fgss | J_OR440835 | 238 | F. meridionale | K_KY466732 |
| 53 | F. asiaticum | C_KY466746 | 115 | Fgss | J_OR440896 | 177 | Fgss | J_OR440834 | 239 | F. meridionale | K_KY466731 |
| 54 | F. asiaticum | C_KY466744 | 116 | Fgss | J_OR440895 | 178 | Fgss | J_OR440833 | 240 | F. meridionale | K_KY466718 |
| 55 | F. asiaticum | C_KY466743 | 117 | Fgss | J_OR440894 | 179 | Fgss | J_OR440832 | 241 | F. meridionale | K_HQ214262 |
| 56 | F. asiaticum | C_KY466740 | 118 | Fgss | J_OR440893 | 180 | Fgss | J_OR440831 | 242 | F. mesoamericanum | L_MW233083 |
| 57 | F. asiaticum | C_KY466741 | 119 | Fgss | J_OR440892 | 181 | Fgss | J_OR440830 | 243 | F. nepalense | M_MW233135 |
| 58 | F. asiaticum | C_KY466739 | 120 | Fgss | J_OR440891 | 182 | Fgss | J_OR440829 | 244 | F. solani | N_MG183712 |
| 59 | F. asiaticum | C_KY466738 | 121 | Fgss | J_OR440890 | 183 | Fgss | J_OR440828 | 245 | F. ussurianum | O_MW233125 |
| 60 | F. asiaticum | C_KY466737 | 122 | Fgss | J_OR440889 | 184 | Fgss | J_OR440826 | 246 | F. vorosii | P_MW233119 |
| 61 | F. asiaticum | C_KY466736 | 123 | Fgss | J_OR440888 | 185 | Fgss | J_OR440827 | 247 | F. vorosii | P_KY586243 |
| 62 | F. asiaticum | C_KY466734 | 124 | Fgss | J_OR440887 | 186 | Fgss | J_OR440825 | 248 | F. vorosii | P_MF974401 |
| Set | Parsimony Informative Sites | Singleton Sites | Constant Sites | Gap/Ambiguity | Chi2 Test | Base Frequencies (A/C/G/T) |
|---|---|---|---|---|---|---|
| Set I | 246 | 120 | 405 | 18.13% | p < 0.05, df = 3 | 0.220/0.301/0.220/0.259 |
| Set II | 36 | 158 | 554 | 20.38% | p < 0.05, df = 3 | 0.25/0.25/0.25/0.25 |
| Set III | 293 | 124 | 344 | 19.73% | p < 0.05, df = 3 | 0.25/0.25/0.25/0.25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yörük, E.; Yli-Mattila, T. Translation Elongation Factor 1-Alpha Sequencing Provides Reliable Tool for Identification of Fusarium graminearum Species Complex Members. Diversity 2024, 16, 481. https://doi.org/10.3390/d16080481
Yörük E, Yli-Mattila T. Translation Elongation Factor 1-Alpha Sequencing Provides Reliable Tool for Identification of Fusarium graminearum Species Complex Members. Diversity. 2024; 16(8):481. https://doi.org/10.3390/d16080481
Chicago/Turabian StyleYörük, Emre, and Tapani Yli-Mattila. 2024. "Translation Elongation Factor 1-Alpha Sequencing Provides Reliable Tool for Identification of Fusarium graminearum Species Complex Members" Diversity 16, no. 8: 481. https://doi.org/10.3390/d16080481
APA StyleYörük, E., & Yli-Mattila, T. (2024). Translation Elongation Factor 1-Alpha Sequencing Provides Reliable Tool for Identification of Fusarium graminearum Species Complex Members. Diversity, 16(8), 481. https://doi.org/10.3390/d16080481






