Clarifying the Taxonomic Relationships of Tulipa iliensis and T. thianschanica Based on Multiple Evidences of Phenotypic, Karyotype, Molecular, and Chloroplast Genomes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Measurement of Major Morphological Traits
2.3. Measurements of Karyotype Parameters
2.4. Phylogenetic Analysis of ITSs
2.5. Phylogenetic Tree Mapping of the Chloroplast Genome
3. Results
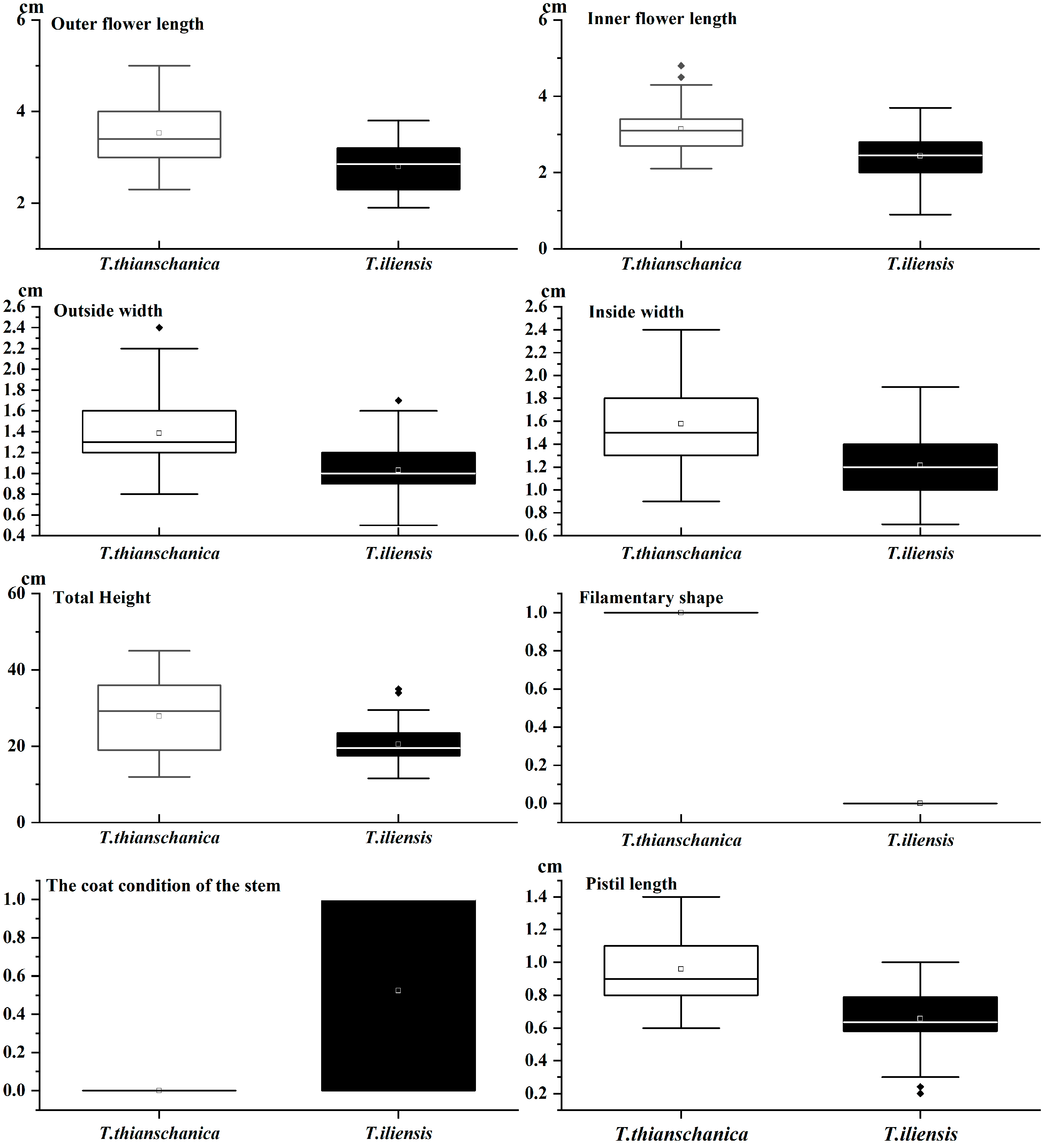
3.1. Morphological Comparison
3.2. Karyotyping
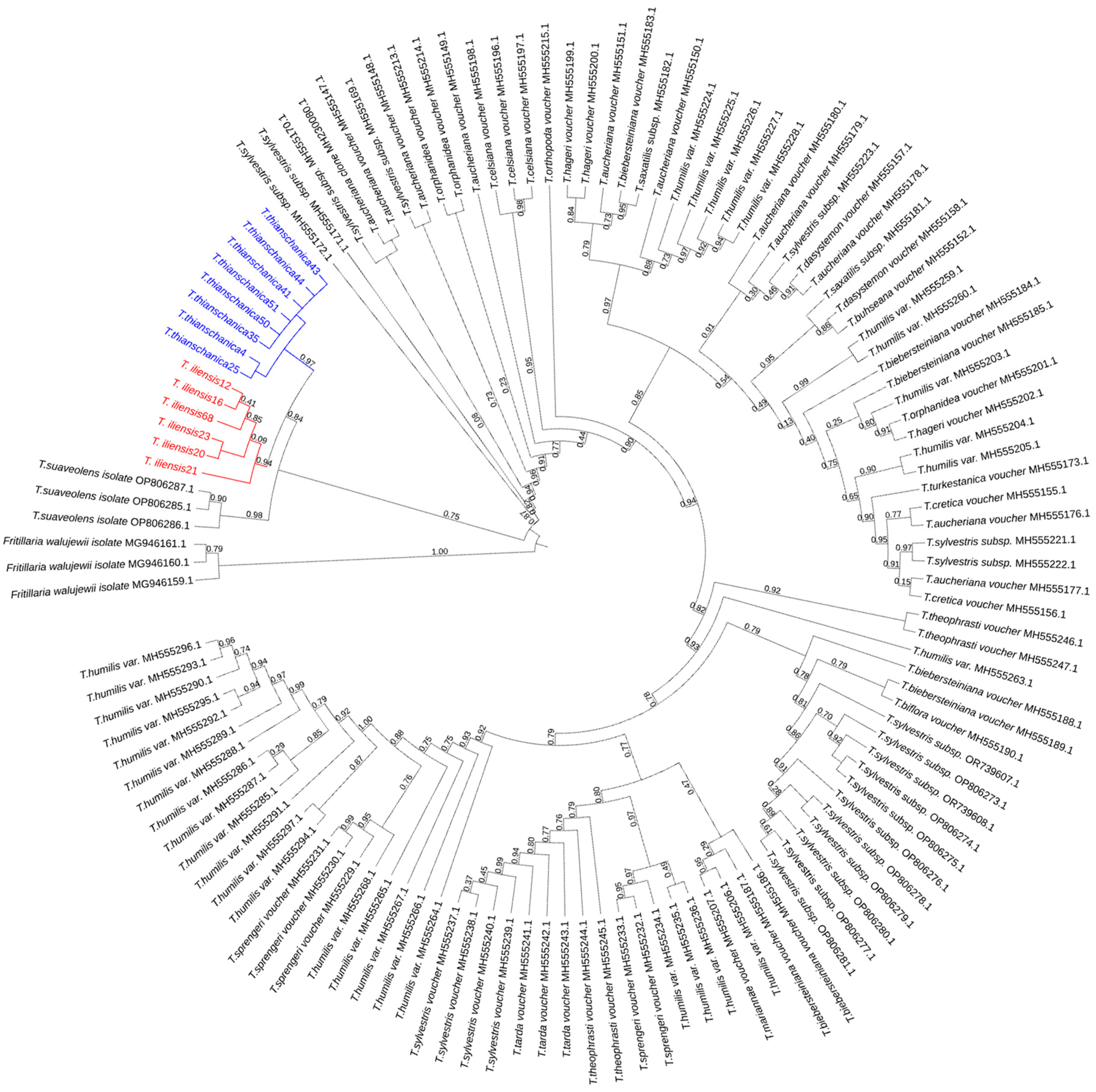
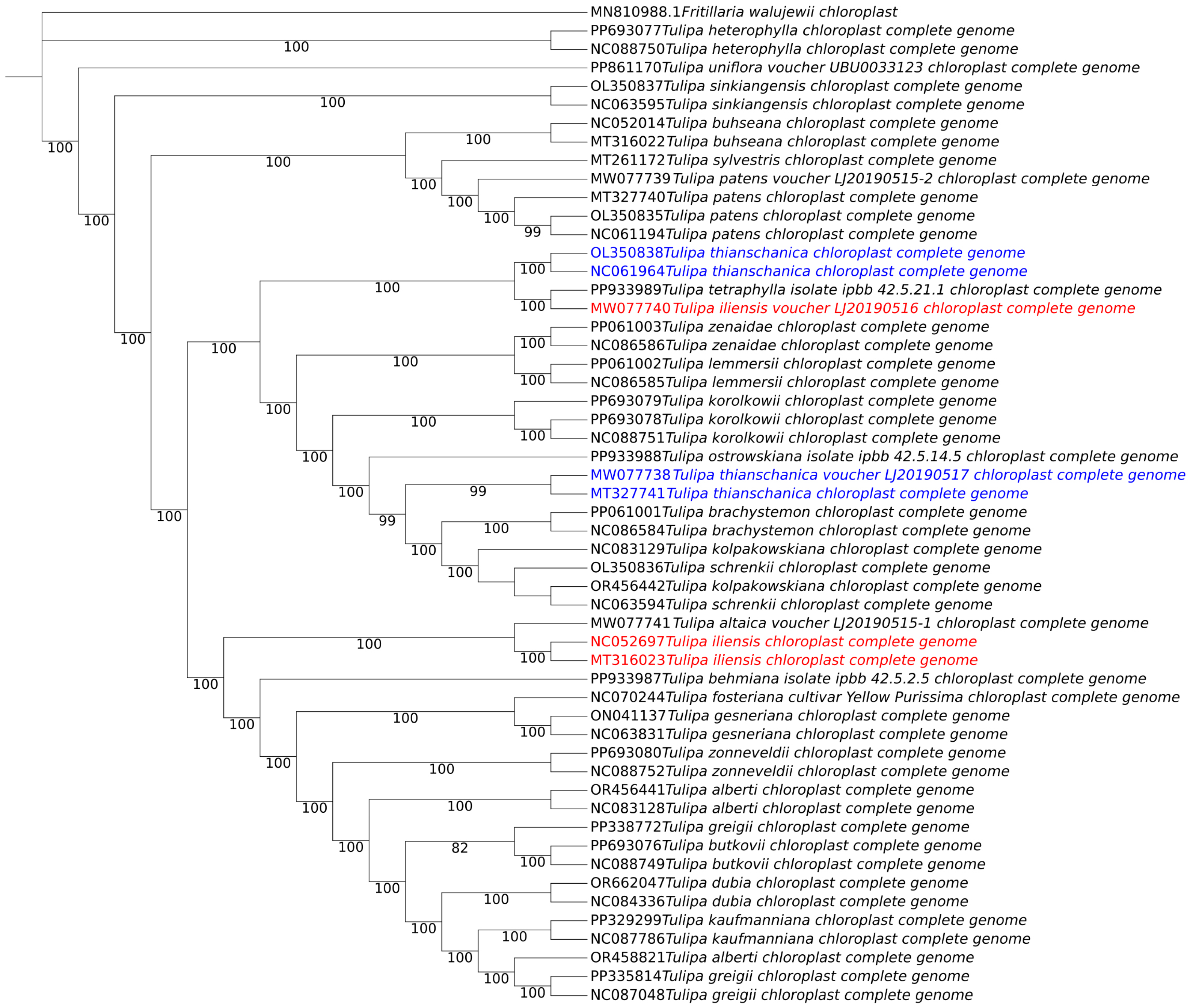
3.3. Molecular Phylogeny
4. Discussion
5. Conclusions
6. Taxonomic Treatment
- Tulipa iliensis Regel in Gartenfl. 28: 162, 1879; Vved. in F1. URSS 4: 347. 1935, Hall, Gen. Tulipa 136. 1940; Atlas of higher plants in China 5: 445, t. 7719. 1976. Flora of China 14: 91, 1980; Desert Flora of China 1: 221, 1985.
- 2.
- Tulipa thianschanica Regel in Act. Hort. Petrop. 6: 503. 1880; et 8: t. 5. 1884; Vved. in F1. URSS 4: 349. 1935; ФJI.Кaзax. 2: 208. 1958; Flora of China 14: 91, 1980.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Peruzzi, L. A new infrafamilial taxonomic setting for Liliaceae, with a key to genera and tribes. Plant Biosyst. 2016, 150, 1341–1347. [Google Scholar] [CrossRef]
- Christenhusz, M.J.M.; Govaerts, R.; David, J.C.; Hall, T.; Borland, K.; Roberts, P.S.; Tuomisto, A.; Buerki, S.; Chase, M.W.; Fay, M.F. Tiptoe through the tulips—Cultural history, molecular phylogenetics and classification of Tulipa (Liliaceae). Bot. J. Linn. Soc. 2013, 172, 280–328. [Google Scholar] [CrossRef]
- Zonneveld, B.J.M. The systematic value of nuclear genome size for “all” species of Tulipa L. (Liliaceae). Plant Syst. Evol. 2009, 281, 217–245. [Google Scholar] [CrossRef]
- Asatulloev, T.; Dekhkonov, D.; Yusupov, Z.; Tojiboeva, U.; Cai, L.; Tojibaev, K.; Sun, W. Ecoregional and Phytogeographical Insights into the Distribution of Tulipa in the ’Nature Imperiled’ Area of Central Asia for Effective Conservation. Diversity 2023, 15, 1195. [Google Scholar] [CrossRef]
- Editorial Board of Xinjiang Flora. Xinjiang Flora M; Xinjiang Science Technology and Hygiene Press: Urumqi, China, 1992; pp. 495–504. [Google Scholar]
- Xing, G.M.; Qu, L.W.; Zhang, Y.Q.; Xue, L.; Su, J.W.; Lei, J.J. Collection and evaluation of wild tulip (Tulipa spp.) resources in China. Genet. Resour. Crop Evol. 2017, 64, 641–652. [Google Scholar] [CrossRef]
- Kang, X.S.; Zhang, Y.Z.; Yang, W.K.; Xiong, Q.; Wang, L.; Su, Z.H. Horticultural Value and Conservation Utilization of Wild Tulips in Xinjiang. J. Anhui Agric. Sci. 2018, 46, 39–41. [Google Scholar] [CrossRef]
- Qin, D.; Liu, W.; Tian, J.; Ju, X. Potentially Suitable Area and Change Trends of Tulipa iliensis under Climate Change. Phyton Int. J. Exp. Bot. 2024, 93, 981–1005. [Google Scholar]
- Qu, L.W.; Lei, J.J.; Zhang, Y.Q.; Xing, G.M.; Su, J.W. Research Status, Existing Problems, and Development Strategies of Tulipa in China. J. North. Hortic. 2016, 10, 188–194. [Google Scholar]
- Wang, Y.H.; Wang, Y.G.; Deng, C.H.; Cui, D.F. Current Status and Conservation Strategies of National Key Protected Wild Plants in Xinjiang. J. Xinjiang For. 2021, 9–14. [Google Scholar]
- Nazemi Ardakani, S.; Rahimi, R.; Mehrabian, A.; Mostafavi, H.; Kiadaliri, H. Ecological modeling of climate change effects on priority species of the Liliaceae family in Iran. Int. J. Environ. Sci. Technol. 2025, 22, 6435–6450. [Google Scholar] [CrossRef]
- Heidarian, M. Identification of a different ultrastructure type in Tulipa julia K. Koch. Palynology 2025, 2457462. [Google Scholar] [CrossRef]
- Wang, Y.R. Biological Characteristics and Resource Evaluation of Tulipa Plants in Xinjiang. Master’s Thesis, Xinjiang Agricultural University, Urumqi, China, 2001. [Google Scholar]
- Tan, D.Y. Systematic Study of Tulipa (sensu lato) in China. Ph.D. Thesis, Graduate University of Chinese Academy of Sciences, Beijing, China, 2005. [Google Scholar]
- Li, J.; Price, M.; Su, D.-M.; Zhang, Z.; Yu, Y.; Xie, D.-F.; Zhou, S.-D.; He, X.-J.; Gao, X.-F. Phylogeny and Comparative Analysis for the Plastid Genomes of Five Tulipa (Liliaceae). Biomed Res. Int. 2021, 2021, 6648429. [Google Scholar] [CrossRef]
- Li, X.R. Taxonomic Study of Tulipa Plants in China. Master’s Thesis, Xinjiang Agricultural University, Urumqi, China, 2003. [Google Scholar]
- Schindel, D.E.; Miller, S.E. DNA barcoding a useful tool for taxonomists. Nature 2005, 435, 17. [Google Scholar] [CrossRef]
- Davron, D.; Temur, A.; Umida, T.; Sari, I.; Komiljon, T.S. Suitable habitat prediction with a huge set of variables on some Central Asian tulips. J. Asia-Pac. Biodivers. 2023, 16, 75–82. [Google Scholar] [CrossRef]
- Qin, D.W.; Liu, W.Q.; Tian, J.T.; Ju, X.T. Construction of a cpDNA-PCR System and Genetic Diversity Analysis in Tulipa iliensis. Zhejiang Agric. J. 2025, 37, 78. Available online: https://kns.cnki.net/kcms/detail/33.1151.S.20241225.1314.004.html (accessed on 28 February 2025).
- Luan, Q.F.; Ouyang, T.; Jiang, Y.C.; Wang, C.X. RAPD Analysis of Wild and Cultivated Tulipa in Xinjiang. J. Jiangxi Agric. Univ. 2008, 30, 656–660. [Google Scholar] [CrossRef]
- Ju, X.T.; Pan, A.Q.; Jiang, F.J.; Tang, N.; Hou, Z.Q. ISSR Analysis of Genetic Diversity in Tulipa Germplasm Resources. Genom. Appl. Biol. 2019, 38, 3667–3674. [Google Scholar] [CrossRef]
- Shimada, H.; Sugiura, M. Fine-Structural Features of the Chloroplast Genome—Comparison of the Sequenced Chloroplast Genomes. Nucleic Acids Res. 1991, 19, 983–995. [Google Scholar] [CrossRef]
- Xing, G.M.; Zhang, Y.Q.; Zhang, H.H.; Wu, T.Y.; Lu, J.J.; Tian, Z.Z.; Qu, L.W. Identification and Evaluation of Wild Tulipa Resources in China. In Proceedings of the 16th Annual Conference of China Bulb and Perennial Flower, Beijing, China, 20 August 2022. [Google Scholar] [CrossRef]
- Song, Q.; Su, Y.R.; Wang, T.M.; Xu, J.; Liang, L.X.; Dong, H.Y.; Tai, T.L. Comparative Study of Chloroplast Genomes of Spiraea × media and Spiraea pubescens in the Forest-Grassland Ecotone of Inner Mongolia. J. Beijing For. Univ. 2024, 46, 103–114. [Google Scholar] [CrossRef]
- Zhang, X.; Bauman, N.; Brown, R.; Richardson, T.H.; Akella, S.; Hann, E.; Morey, R.; Smith, D.R. The mitochondrial and chloroplast genomes of the green alga Haematococcus are made up of nearly identical repetitive sequences. Curr. Biol. 2019, 29, R736–R737. [Google Scholar] [CrossRef]
- Zhao, K.H.; Yuan, F.; Zhao, F.Y.; Lan, X.Z. DNA Barcode Identification of Rhodiola Plants Based on ITS2 and matK Sequences. J. Plateau Agric. 2022, 6, 462–468. [Google Scholar] [CrossRef]
- Xia, Y.M.; Yin, J.; Zeng, L.S.; Wang, Y.G.; Zhan, R.T.; Lin, Y.; Zhang, G.F. Interspecific Identification of Hoya Plants Based on ITS2 Sequences. J. Chin. Med. Mater. 2023, 46, 603–609. [Google Scholar] [CrossRef]
- Zhao, Y. SPSS Implementation of Independent Two-Sample t-Test. J. Stat. Decis. 2021, 37, 44–47. [Google Scholar] [CrossRef]
- Seifert, E. Origin Pro 9.1: Scientific Data Analysis and Graphing Software-Software Review. J. Chem. Inf. Model. 2014, 54, 1552. [Google Scholar] [CrossRef]
- Liu, G.X.; Lan, Y.; Qu, L.W.; Zhao, Y.L.; Xin, H.Y.; Xi, M.L. Analyzing the genetic relationships in Tulipa based on karyotypes and 5S rDNA sequences. Sci. Hortic. 2022, 302, 111178. [Google Scholar] [CrossRef]
- Paesold, S.; Borchardt, D.; Schmidt, T.; Dechyeva, D. A sugar beet (Beta vulgaris L.) reference FISH karyotype for chromosome and chromosome-arm identification, integration of genetic linkage groups and analysis of major repeat family distribution. Plant J. 2012, 72, 600–611. [Google Scholar] [CrossRef]
- Fan, Y.; Hao, L.; Zhu, W.; Teixeira da Silva, J.A.; Yu, X. Chromosome karyotype analysis of some Fritillaria species. Maejo Int. J. Sci. Technol. 2022, 16, 223–237. [Google Scholar]
- Saensouk, P.; Saensouk, S.; Senavongse, R. Cytogenetic Studies of Six Species in Family Araceae from Thailand. Caryologia 2022, 75, 5–13. [Google Scholar] [CrossRef]
- Levan, A.; Fredga, K.; Sandberg, A.A. Nomenclature for Centromeric Position on Chromosomes. Hered.-Genet. Ark. 1964, 52, 201–220. [Google Scholar] [CrossRef]
- Goto, M. 8. Image Processing Using ImageJ. Nihon Hoshasen Gijutsu Gakkai Zasshi 2019, 75, 688–692. [Google Scholar] [CrossRef]
- Peruzzi, L.; Eroglu, H.E. Karyotype asymmetry: Again, how to measure and what to measure? Comp. Cytogenet. 2013, 7, 1. [Google Scholar] [CrossRef]
- Peruzzi, L.; Altinordu, F. A proposal for a multivariate quantitative approach to infer karyological relationships among taxa. Comp. Cytogenet. 2014, 8, 337–349. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL-W—Improving the Sensitivity of Progressive Multiple Sequence Alignment Through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Lord, P.W.; Selley, J.N.; Attwood, T.K. CINEMA-MX: A modular multiple alignment editor. Bioinformatics 2002, 18, 1402–1403. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002, 2, 2.3. [Google Scholar] [CrossRef]
- Felsenstein, J. Phylogenies and the Comparative Method. Am. Nat. 1985, 125, 1–15. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Zhou, T.; Xu, K.; Zhao, F.; Liu, W.; Li, L.; Hua, Z.; Zhou, X. itol.toolkit accelerates working with iTOL (Interactive Tree of Life) by an automated generation of annotation files. Bioinformatics 2023, 39, btad339. [Google Scholar] [CrossRef]
- Katoh, K.; Toh, H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics (Oxf. Engl.) 2010, 26, 1899–1900. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT: Iterative refinement and additional methods. Methods Mol. Biol. 2014, 1079, 131–146. [Google Scholar] [CrossRef]
- Livingston, K. Conservation genetics. Case histories from nature—Avise, JC, Hamrick, JL. Science 1996, 272, 364. [Google Scholar] [CrossRef]
- Nasab, F.K.; Mehrabian, A.R. A Taxonomic Revision of Two Species Complexes Belonging to the Haplotricha Subsection in the Genus Onosma (Boraginaceae): A Realistic Approach to Plant Diversity. Diversity 2022, 14, 671. [Google Scholar] [CrossRef]
- Szlachetko, D.L.; Dudek, M.; Naczk, A.; Kolanowska, M. Taxonomy and Biogeography of Andinia-Complex (Orchidaceae). Diversity 2022, 14, 372. [Google Scholar] [CrossRef]
- Song, K.; Zhang, J.; Yan, E.; Wang, X.; Song, Y. EcoFloVCS: An eco-physiognomic-floristic vegetation classification system. Appl. Veg. Sci. 2024, 27, e70005. [Google Scholar] [CrossRef]
- de Almeida, R.F.; de Morais, I.L.; Alves-Silva, T.; Antonio-Domingues, H.; Pellegrini, M.O.O. A new classification system and taxonomic synopsis for Malpighiaceae (Malpighiales, Rosids) based on molecular phylogenetics, morphology, palynology, and chemistry. Phytokeys 2024, 242, 69–138. [Google Scholar] [CrossRef]
- Alotaibi, N.M.; Kenyon, E.J.; Bertelli, C.M.; Al-Qthanin, R.N.; Mead, J.; Parry, M.; Bull, J.C. Environment predicts seagrass genotype, phenotype, and associated biodiversity in a temperate ecosystem. Front. Plant Sci. 2022, 13, 887474. [Google Scholar] [CrossRef]
- Mueller, C.; Junker, R.R. Chemical phenotype as important and dynamic niche dimension of plants. New Phytol. 2022, 234, 1168–1174. [Google Scholar] [CrossRef]
- Walker, C. Increases in plant phenotypic diversity in response to aridity and grazing. Nat. Plants 2024, 10, 1277. [Google Scholar] [CrossRef]
- Zhou, T.; Ning, K.; Zhang, W.X.; Chen, H.; Lu, X.Q.; Zhang, D.L.; El-Kassaby, Y.A.; Bian, J. Phenotypic variation of floral organs in flowering crabapples and its taxonomic significance. BMC Plant Biol. 2021, 21, 503. [Google Scholar] [CrossRef]
- Rosas-Guerrero, V.; Quesada, M.; Armbruster, W.S.; Pérez-Barrales, R.; Smith, S.D. Influence of Pollination Specialization and Breeding System on Floral Integration and Phenotypic Variation in Ipomoea. Evolution 2011, 65, 350–364. [Google Scholar] [CrossRef]
- Stebbins, G.L. Research on Evolution of Higher-Plants—Problems and Prospects. Can. J. Genet. Cytol. 1972, 14, 453–462. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Ratnasingham, S.; de Waard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B-Biol. Sci. 2003, 270, S96–S99. [Google Scholar] [CrossRef]
- Chen, Z.; Gao, L.; Wang, H.; Feng, S. Molecular Identification and Phylogenetic Analysis of Cymbidium Species (Orchidaceae) Based on the Potential DNA Barcodes matK, rbcL, psbA-trnH, and Internal Transcribed Spacer. Agronomy 2024, 14, 933. [Google Scholar] [CrossRef]
- Mano, H.; Boltenkov, E.V.; Marchuk, E.A.; Nakamura, K.; Yoichi, W. The complete chloroplast genome sequence of Hypecoum erectum L. (Papaveraceae). Mitochondrial DNA Part B Resour. 2024, 9, 1010–1014. [Google Scholar] [CrossRef]
- Zeng, X.; Zhao, W.; Deng, A.; Xiang, L.; Tang, X.; Li, H.; Yin, H.; Huang, R.; Xiao, Y.; Liu, Y.; et al. Characterization and phylogenetic analysis of the chloroplast genome of Diospyros aff. oleifera. Mitochondrial DNA Part B Resour. 2024, 9, 1467–1472. [Google Scholar] [CrossRef]
- Li, S.; Yuan, W.; He, S.; Bai, W.; Wu, H. The complete chloroplast genome and phylogenetic analysis of Gentiana arethusae Burkill (Gentianaceae) from China. Mitochondrial DNA Part B Resour. 2021, 6, 3132–3133. [Google Scholar] [CrossRef]
- Liu, W.Q.; Qin, D.W.; Shi, G.M.; Xu, T.L.; Ju, X.T. Codon Usage Bias Analysis of the Chloroplast Genome in Tulipa iliensis. J. Chin. Wild Plant Resour. 2024, 43, 39–45. [Google Scholar] [CrossRef]
- Fainelli, F.; Baldesi, G.; Pallanza, M.; Orsenigo, S. Extinct or Not? Confirming the “Extinct” Status of Hieracium tolstoii (Asteraceae) with Integrated Taxonomic Investigation. Diversity 2024, 16, 591. [Google Scholar] [CrossRef]
- Jian, X.; Wang, Y.L.; Li, Q.; Miao, Y.M. Plastid Phylogenetics, Biogeography, and Character Evolution of the Chinese Endemic Genus Sinojackia Hu. Diversity 2024, 16, 305. [Google Scholar] [CrossRef]
- Mifsud, S.; Lanfranco, S. An Updated Taxonomic Appraisal of Narcissus (Amaryllidaceae) in the Maltese Islands. Diversity 2024, 16, 397. [Google Scholar] [CrossRef]
- Wu, X.; Chen, J.; Cui, G. Proposals for updating the List of National Key Protected Wild Plants-Based on an analysis of existing conservation lists. Biodivers. Sci. 2023, 31, 22622. [Google Scholar]
- Jiang, X.; Liu, W.J.; Zhu, Y.Z.; Cao, Y.T.; Yang, X.M.; Geng, Y.; Zhang, F.J.; Sun, R.Q.; Jia, R.W.; Yan, C.L.; et al. Impacts of Climate Changes on Geographic Distribution of Primula filchnerae, an Endangered Herb in China. Plants 2023, 12, 3561. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Wu, J.Y.; Zhang, H.; Lobora, A.; Hou, Y.L.; Wen, Y.L. Conflict Governance between Protected Areas and Surrounding Communities: Willingness and Behaviors of Communities-Empirical Evidence from Tanzania. Diversity 2024, 16, 278. [Google Scholar] [CrossRef]
- Perry, G.; Cox, R.D. Opportunities for Biodiversity Conservation via Urban Ecosystem Regeneration. Diversity 2024, 16, 131. [Google Scholar] [CrossRef]
- Wei, Z.; Lock, J. Flora of China; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2010. [Google Scholar]


| Pop. N. | Species | Location | Latitude | Longitude | Elevation |
|---|---|---|---|---|---|
| 1 | T. thianschanica | Qapqal Xibe Autonomous County S237 | 43.50° N | 81.12° E | 1737 m |
| 2 | T. thianschanica | Tokkuzkuzur County G577 | 43.37° N | 81.88° E | 1551 m |
| 3 | T. thianschanica, T. iliensis | Senmatas village, Kashagar town, Zhaosu County | 42.84° N | 81.08° E | 1713 m |
| 4 | T. iliensis | Baibird Lake, Urumqi City | 43.83° N | 87.44° E | 768 m |
| 5 | T. iliensis | Urumqi small green valley | 43.84° N | 87.50° E | 798 m |
| 6 | T. iliensis | Urumqi Yamalik Mountain | 43.80° N | 87.58° E | 863 m |
| 7 | T. iliensis | Alemale town, Xinyuan County | 43.60° N | 81.24° E | 1155 m |
| 8 | T. iliensis | Artificial forest of dolomite mine in Xinyuan County | 43.38° N | 83.56° E | 2697 m |
| 9 | T. iliensis | Yangpo of dolomite mine in Xinyuan County | 43.41° N | 83.58° E | 1255 m |
| 10 | T. iliensis | Emin county wild fruit forest scenic spot | 46.69° N | 84.72° E | 938 m |
| 11 | T. iliensis | Shawan City cattle circle ranch | 43.93° N | 85.70° E | 1181 m |
| 12 | T. iliensis | Yumin County Baluk Mountain scenic spot | 45.84° N | 82.56° E | 1673 m |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Wang, X.; Liu, H.; Liu, S.; Wei, Y. Clarifying the Taxonomic Relationships of Tulipa iliensis and T. thianschanica Based on Multiple Evidences of Phenotypic, Karyotype, Molecular, and Chloroplast Genomes. Diversity 2025, 17, 219. https://doi.org/10.3390/d17030219
Zhang H, Wang X, Liu H, Liu S, Wei Y. Clarifying the Taxonomic Relationships of Tulipa iliensis and T. thianschanica Based on Multiple Evidences of Phenotypic, Karyotype, Molecular, and Chloroplast Genomes. Diversity. 2025; 17(3):219. https://doi.org/10.3390/d17030219
Chicago/Turabian StyleZhang, Huimin, Xiyong Wang, Huawei Liu, Shiqing Liu, and Yan Wei. 2025. "Clarifying the Taxonomic Relationships of Tulipa iliensis and T. thianschanica Based on Multiple Evidences of Phenotypic, Karyotype, Molecular, and Chloroplast Genomes" Diversity 17, no. 3: 219. https://doi.org/10.3390/d17030219
APA StyleZhang, H., Wang, X., Liu, H., Liu, S., & Wei, Y. (2025). Clarifying the Taxonomic Relationships of Tulipa iliensis and T. thianschanica Based on Multiple Evidences of Phenotypic, Karyotype, Molecular, and Chloroplast Genomes. Diversity, 17(3), 219. https://doi.org/10.3390/d17030219






