Abstract
The emergence of antimicrobial-resistant bacteria poses a significant global health challenge, with ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) playing a major role in multidrug-resistant infections. While traditionally associated with hospital settings, these bacteria have increasingly been detected in wildlife, suggesting a complex web of transmission between human, animal, and environmental reservoirs. Wildlife may act as both sentinels and reservoirs for resistant pathogens, contributing to their persistence and dissemination across ecosystems. This review explores the presence of ESKAPE bacteria in wild animals, examining their clonal lineages, resistance profiles, and virulence traits. Understanding how these pathogens circulate in natural environments is crucial for designing effective strategies to mitigate antimicrobial resistance. By adopting a One Health perspective—integrating human, animal, and environmental health—efforts to control ESKAPE bacteria can extend beyond clinical interventions to broader ecological and public health frameworks. Addressing this issue requires comprehensive surveillance, responsible antibiotic use, and policies aimed at reducing environmental contamination, ultimately safeguarding both biodiversity and global health.
1. Introduction
The emergence and spread of antimicrobial-resistant bacteria is a growing global threat, with the ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) at the forefront. These pathogens are recognized for their ability to evade standard treatments, leading to difficult-to-manage infections, particularly in hospital settings where immunocompromised patients are most vulnerable [1,2]. Their name, “ESKAPE”, reflects their capacity to “escape” the effects of antibiotics through various mechanisms, including the rapid acquisition and dissemination of resistance genes and an array of virulence factors that enhance their ability to cause disease [1,3]. To contrapose the global menace posed by antimicrobial resistance, the World Health Organization (WHO) has assigned the pertinent bacterial pathogens into three categories of risk: critical, high, and medium priority. Remarkably, some constituents of the ESKAPE group are classified as extreme priority in relation to the threat that accompanies their infection such as carbapenem-resistant Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacteriaceae [4]. The existence of these bacteria within wildlife underscores the necessity of knowing how these bacteria circulate within a One Health approach.
Initially considered a primarily clinical issue, ESKAPE bacteria have increasingly been detected outside hospital environments, appearing in agriculture, the community, and even natural ecosystems [5]. This wider distribution points to a complex web of transmission involving humans, animals, and the environment [5,6]. In this context, wild animals have emerged as unexpected reservoirs for ESKAPE pathogens [5,6]. As they interact with diverse habitats, from urban landscapes to remote wilderness areas, wildlife can become exposed to contamination from human activities, such as agricultural runoff, untreated sewage, and waste from healthcare facilities [7]. This ongoing environmental exposure suggests that resistance is not confined to anthropogenic settings but is also maintained and potentially amplified through natural reservoirs, creating a cycle of transmission that connects diverse ecosystems [7,8].
This review examines the presence of ESKAPE bacteria in wildlife and explores their role in the broader ecology of antimicrobial resistance. By investigating the clonal lineages, resistance profiles, and virulence traits of these pathogens in wild animals, we can gain insights into how resistance spreads and persists across different settings. Understanding these dynamics is essential for formulating strategies that extend beyond traditional clinical approaches to embrace a One Health perspective that recognizes the interconnectedness of human, animal, and environmental health. While previous reviews have explored antimicrobial resistance in wildlife, most have focused on specific bacterial groups or individual species. This review uniquely examines the ESKAPE group as a whole, assessing their presence in diverse wildlife hosts, their resistance profiles, and their potential role in resistance transmission. By integrating data across multiple bacterial species and ecological contexts, this work provides a broader understanding of how ESKAPE pathogens circulate in natural environments and their implications for One Health. Additionally, this review identifies gaps in the literature, highlighting areas where further research is needed to fully understand the role of wildlife in the ecology of antimicrobial resistance.
2. ESKAPE Bacteria
The ESKAPE group of bacteria—comprising Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species—has gained notoriety in recent years as a leading cause of hospital-acquired infections worldwide (Figure 1). These pathogens are not only adept at evading the human immune system but also display a remarkable capacity to “escape” the effects of antibiotics, making them key drivers of antimicrobial resistance (AMR) [8,9]. Collectively, they are responsible for a significant proportion of difficult-to-treat infections, ranging from pneumonia and bloodstream infections to urinary tract and wound infections, particularly in immunocompromised patients or those undergoing invasive medical procedures [3,9].
Figure 1.
ESKAPE bacterial group.
The threat posed by ESKAPE bacteria is twofold. On the one hand, their ability to rapidly acquire and share resistance genes—often through mobile genetic elements like plasmids, transposons, and integrons—means that even once-effective antibiotics can quickly lose their efficacy [2,10,11]. On the other hand, many of these bacteria possess an array of virulence factors, including toxins, enzymes, and biofilm-forming capabilities, which allow them to colonize host tissues, evade immune responses, and persist on medical devices such as catheters and ventilators [3,12]. This dual ability to resist treatment and cause severe disease complicates infection control efforts and has led to a growing reliance on last-resort antibiotics, which are themselves increasingly compromised [13]. Drugs such as carbapenems and colistin, once considered the final line of defense, are becoming less effective due to the spread of resistant strains like carbapenem-resistant Klebsiella pneumoniae and colistin-resistant Acinetobacter baumannii [11,14].
Originally recognized as a major issue within hospital settings, the reach of ESKAPE pathogens extends beyond clinical environments. These bacteria have been detected in community settings, agricultural contexts, and natural ecosystems, underscoring the interconnected nature of AMR [5,14,15]. The detection of multidrug-resistant ESKAPE strains in livestock, wildlife, and even water sources suggests that the problem is not confined to hospitals but is also driven by environmental factors and the widespread use of antibiotics in both human and veterinary medicine [5,16]. This highlights the need for a One Health approach that addresses antimicrobial resistance as a shared challenge across human, animal, and environmental health.
Efforts to combat ESKAPE bacteria require a comprehensive strategy that goes beyond the development of new antibiotics. It involves improved infection prevention and control measures, responsible antibiotic use in both healthcare and agriculture, and proactive surveillance to track emerging resistance trends [2,17]. Understanding the mechanisms through which these pathogens spread and persist in different settings is critical for developing sustainable solutions to curb their impact. As the global community grapples with the growing crisis of AMR, ESKAPE bacteria remain at the forefront of this struggle, serving as both a symbol of the challenges ahead and a call to action for coordinated, multi-disciplinary efforts.
2.1. Staphylococcus aureus
S. aureus is a versatile and opportunistic pathogen known for its ability to colonize a variety of hosts, from humans to animals, and its remarkable adaptability to different environments [18,19]. As a Gram-positive bacterium, it is commonly found on the skin and mucous membranes, where it often exists as a harmless commensal [19]. However, under certain conditions, such as breaches in the skin barrier or immunosuppression, S. aureus can shift from a benign colonizer to a formidable pathogen, causing a range of infections from superficial skin lesions to life-threatening conditions like sepsis, endocarditis, and pneumonia [20]. S. aureus success as a pathogen can be attributed to its extensive arsenal of virulence factors, which include surface proteins that facilitate adhesion to host tissues, enzymes that promote invasion and tissue destruction, and toxins that disrupt the immune response [21,22,23]. Among these, the production of staphylococcal enterotoxins, hemolysins, and leukocidins plays a significant role in the severity of infections [24,25]. Additionally, S. aureus is adept at forming biofilms, particularly on medical devices, which not only helps it evade the host’s immune system but also significantly complicates treatment efforts [26,27]. The emergence of methicillin-resistant Staphylococcus aureus (MRSA) in the mid-20th century marked a significant turning point, transforming a once-treatable pathogen into a global public health threat. Today, MRSA is a major cause of hospital-associated (HA-MRSA) and community-associated (CA-MRSA) infections worldwide [28,29]. The ability of S. aureus to harbor resistance genes, such as mecA and mecC, which confer resistance to nearly all β-lactam antibiotics, underscores its resilience in the face of modern medicine [30,31]. The discovery of mecC-positive MRSA in both livestock and wildlife also highlights the broader environmental and zoonotic dimensions of S. aureus epidemiology [32]. The transmission dynamics of S. aureus are complex and multifactorial, involving not just person-to-person spread but also environmental reservoirs and animal hosts. Livestock-associated MRSA (LA-MRSA), particularly strains linked to clonal complex 398 (CC398), has demonstrated that farm animals can act as reservoirs for resistant strains, which can then spread to humans through direct contact or environmental contamination [33,34,35]. The detection of various S. aureus lineages in wildlife further expands this narrative, suggesting that these bacteria ecology extends well beyond human-associated settings [32]. Efforts to control S. aureus infections thus require a multifaceted approach, encompassing not only clinical interventions but also public health strategies that address the environmental and veterinary aspects of its transmission [36,37]. Understanding the interplay between human, animal, and environmental reservoirs is crucial for developing effective measures to combat this adaptable pathogen.
2.2. Enterobacteriaceae
Enterobacteriaceae are a diverse family of Gram-negative bacteria that inhabit a range of environments, including soil, water, and the gastrointestinal tracts of animals and humans [38]. While many members of this family play a role as commensals or environmental organisms, some, such as Escherichia coli and Klebsiella pneumoniae, can become opportunistic pathogens under certain conditions [39,40]. These bacteria are not only a significant cause of community and hospital-acquired infections, but they are also at the forefront of the global issue of antimicrobial resistance [41,42].
Escherichia coli, for instance, is a common inhabitant of the human gut, where it generally coexists harmlessly with the host [43]. However, certain strains have acquired virulence factors that enable them to cause a spectrum of infections, ranging from uncomplicated urinary tract infections (UTIs) to severe conditions like septicemia and meningitis [44,45]. Pathogenic E. coli strains are broadly classified into pathotypes based on their virulence mechanisms, with extraintestinal pathogenic E. coli (ExPEC) being a major contributor to UTIs and bloodstream infections [46]. The increasing prevalence of multidrug-resistant E. coli, especially those producing extended-spectrum β-lactamases (ESBLs) such as blaCTX-M, poses a significant challenge for treatment, as these enzymes confer resistance to many β-lactam antibiotics, including third-generation cephalosporins [47]. Moreover, the spread of plasmid-mediated colistin resistance (mcr genes) raises concerns about the diminishing efficacy of last-resort treatments [48].
Klebsiella pneumoniae is another key member of the Enterobacteriaceae family that has emerged as a major public health threat, particularly in healthcare settings. Known for its characteristic thick polysaccharide capsule, which enhances its virulence and helps it evade the host immune system, K. pneumoniae is a leading cause of hospital-acquired pneumonia, bloodstream infections, and urinary tract infections [49,50]. The rise in carbapenem-resistant K. pneumoniae has added to the urgency of controlling its spread, as carbapenems are often the last line of defense against MDR Gram-negative infections [51]. The resistance is frequently mediated by carbapenemase enzymes, such as KPC (Klebsiella pneumoniae carbapenemase), NDM (New Delhi metallo-β-lactamase), and OXA-48, which can be transferred between bacteria through mobile genetic elements, accelerating the dissemination of resistance [52,53].
Both E. coli and K. pneumoniae have demonstrated remarkable genetic plasticity, enabling them to adapt to different hosts and environments. This adaptability is facilitated by horizontal gene transfer (HGT), which allows for the acquisition of resistance genes and virulence factors from other bacteria [54]. The detection of MDR Enterobacteriaceae in animals, food, and the environment highlights the One Health aspect of AMR, where human, animal, and environmental health are interconnected [38,55]. Livestock, wildlife, and even companion animals can harbor and transmit resistant strains, potentially contributing to the spread of AMR through direct contact or environmental contamination [56,57].
Enterobacter species, although less commonly discussed than E. coli and Klebsiella, are significant contributors to the burden of hospital-acquired infections and antimicrobial resistance. These bacteria are naturally found in the environment, including water and soil, but can become opportunistic pathogens in vulnerable patients, causing respiratory, urinary, and bloodstream infections [58,59]. The clinical relevance of Enterobacter has been heightened by the emergence of multidrug-resistant (MDR) strains, often due to the production of AmpC β-lactamases and, more recently, carbapenemases. The ability of Enterobacter to upregulate these enzymes in response to antibiotic exposure makes treatment particularly challenging, often requiring the use of last-resort antibiotics such as carbapenems or colistin. Moreover, their capacity for acquiring resistance genes via horizontal gene transfer contributes to the broader dissemination of antimicrobial resistance across bacterial communities, highlighting the importance of monitoring and controlling Enterobacter as part of comprehensive AMR strategies [60,61].
Addressing the threat posed by Enterobacteriaceae, particularly E. coli and K. pneumoniae, requires a comprehensive approach that extends beyond the clinical setting. Strategies must include rigorous infection control measures in healthcare facilities, prudent antibiotic use in both human medicine and agriculture, and environmental monitoring to track the spread of resistance genes [62,63,64]. Understanding the intricate ecology of these bacteria, including their role in natural reservoirs, will be crucial for developing effective interventions to curb the tide of antimicrobial resistance.
2.3. Acinetobacter baumannii
Acinetobacter baumannii is a Gram-negative bacterium that has gained prominence as a formidable opportunistic pathogen, especially in healthcare settings [65]. Its natural habitat includes soil and water, where it can persist under harsh conditions, but it is in hospitals where A. baumannii truly thrives, causing a range of infections such as ventilator-associated pneumonia, bloodstream infections, and wound infections in critically ill patients [66,67]. This adaptability is largely due to its remarkable ability to survive on various surfaces for extended periods, making it a notorious cause of hospital-acquired infections [68].
The pathogenicity of A. baumannii is bolstered by a suite of virulence factors that allow it to evade the host’s immune response and adapt to different environments [69,70]. These include biofilm formation, which enhances its survival on medical devices, and an outer membrane that resists desiccation and antiseptics [71,72]. Furthermore, the bacteria’s capacity for acquiring and integrating foreign DNA facilitates the rapid spread of resistance genes. This is particularly concerning with the emergence of carbapenem-resistant A. baumannii (CRAB), which has severely limited treatment options. Carbapenem resistance is often mediated by the production of carbapenem-hydrolyzing enzymes known as oxacillinases (OXA), such as blaOXA-23, blaOXA-24/40, and blaOXA-58 [73,74]. The presence of these genes on mobile genetic elements like plasmids and transposons accelerates their spread within and between bacterial populations.
What makes A. baumannii especially challenging to control is its ability to rapidly develop resistance not just to carbapenems, but to multiple classes of antibiotics, including aminoglycosides, fluoroquinolones, and even polymyxins like colistin, which are considered last-resort treatments [75,76,77]. The emergence of colistin-resistant strains is alarming, as it leaves clinicians with very few therapeutic options [78,79]. Resistance mechanisms in A. baumannii often involve efflux pumps, enzymatic degradation, and modifications to target sites, reflecting the bacteria genetic versatility [80,81].
Beyond hospitals, the detection of A. baumannii in community settings and natural environments is increasingly reported, raising questions about the pathways through which this pathogen disseminates. Studies have found A. baumannii in diverse ecological niches, including wildlife, livestock, and aquatic environments [56,82,83]. These findings suggest that A. baumannii can persist outside clinical settings, potentially using environmental reservoirs to maintain a foothold from which it can re-enter healthcare facilities. Therefore, understanding their ecology, including the non-clinical reservoirs that may contribute to their persistence and transmission, will be crucial in formulating effective control measures. As such, A. baumannii remain a stark reminder of the challenges posed by antimicrobial resistance and the need for a coordinated One Health approach to tackle this global issue.
2.4. Pseudomonas aeruginosa
P. aeruginosa is a Gram-negative bacterium that stands out for its adaptability and resilience across a wide range of environments, from soil and water to the human body. While it is typically found in natural settings such as rivers, lakes, and soil, it is also a notorious opportunistic pathogen, particularly in healthcare settings. It poses a significant threat to immunocompromised patients, where it can cause severe infections, including pneumonia, bloodstream infections, urinary tract infections, and chronic lung infections in individuals with cystic fibrosis [84,85,86].
The success of P. aeruginosa as a pathogen is largely due to its diverse arsenal of virulence factors and sophisticated regulatory networks that allow it to sense and adapt to different conditions [84,85]. These include the ability to form biofilms, which are complex communities of bacteria encased in a protective matrix that adhere to surfaces like medical devices and tissues. Biofilm formation not only helps P. aeruginosa evade the host immune response but also confers significant tolerance to antibiotics, complicating treatment. Moreover, the bacterium produces a variety of toxins and enzymes, such as elastases, exotoxins (e.g., ExoU and ExoS), and proteases, which contribute to tissue damage and impair host defenses [87,88,89].
Compounding its virulence is P. aeruginosa’s impressive ability to develop resistance to multiple classes of antibiotics [90]. This multidrug resistance (MDR) is driven by several mechanisms, including the overexpression of efflux pumps that expel antibiotics from the cell, enzymatic degradation of antibiotic molecules, and mutations in target sites that reduce drug binding. The emergence of carbapenem-resistant P. aeruginosa (CRPA) has become a global concern, as carbapenems are often used as last-resort antibiotics for treating MDR bacterial infections [91]. Resistance is frequently mediated by the production of carbapenemases, such as metallo-β-lactamases (e.g., VIM and IMP), and loss of porins that decrease antibiotic uptake [17,92,93].
Beyond healthcare settings, P. aeruginosa is increasingly found in community and environmental contexts, including water sources, soil, and even in association with wildlife [94,95,96]. In aquatic environments, for example, P. aeruginosa can survive and persist, raising concerns about its role in the natural spread of resistance genes, which can be transferred to other bacteria via horizontal gene transfer [85,97].
To combat P. aeruginosa, a multifaceted approach is essential, combining the development of new therapeutic options with robust infection prevention measures, particularly in hospitals and long-term care facilities. Efforts to better understand its ecology, including the role of environmental reservoirs in sustaining and spreading resistant strains, are crucial for developing comprehensive strategies to curb the impact of this adaptable pathogen.
2.5. Enterococcus spp.
Enterococcus species, particularly Enterococcus faecalis and Enterococcus faecium, are important members of the gut microbiota in humans and animals, playing a largely commensal role [98]. However, they are also notorious as opportunistic pathogens capable of causing a variety of infections, including urinary tract infections, endocarditis, bacteremia, and wound infections [99]. These bacteria are well adapted to survive in harsh conditions, such as high salinity, a wide range of temperatures, and the presence of bile, which contributes to their persistence not only in the gut but also in hospital environments [98,100].
The shift from benign gut inhabitants to formidable pathogens is often associated with the acquisition of virulence factors that enhance their ability to colonize host tissues, evade immune defenses, and cause disease [98]. Virulence factors commonly found in pathogenic Enterococcus strains include surface adhesins like the enterococcal surface protein (esp), enzymes such as gelatinase (gelE) and hyaluronidase (hyl), and biofilm-forming capabilities that promote persistent infections, particularly on medical devices such as catheters and prosthetic valves [101,102].
Compounding their pathogenic potential is Enterococcus’s remarkable ability to develop resistance to a wide range of antibiotics, which is a major concern in healthcare settings. Enterococcus faecium, in particular, has emerged as a leading cause of hospital-acquired infections due to its high levels of resistance to multiple antibiotics, including aminoglycosides, β-lactams, and glycopeptides such as vancomycin [103,104]. The rise in vancomycin-resistant enterococci (VRE) poses a significant therapeutic challenge, as vancomycin is often used as a last-resort antibiotic for treating multidrug-resistant infections [105]. Resistance to vancomycin is typically mediated by the vanA and vanB gene clusters, which alter the bacterial cell wall target, reducing the efficacy of the antibiotic [106].
The ability of Enterococcus to acquire resistance genes through horizontal gene transfer is facilitated by the presence of mobile genetic elements like plasmids and transposons [107]. These elements can carry multiple resistance genes, allowing the bacteria to adapt rapidly to antibiotic pressures. The spread of resistance is further exacerbated by the environmental resilience of Enterococcus, which can survive in contaminated surfaces, water sources, and even in agricultural settings where antibiotics are used in livestock production [63,108]. This adaptability raises concerns about the dissemination of resistant Enterococcus strains from non-clinical settings to human populations, blurring the boundaries between hospital-acquired and community-associated infections.
In addition to healthcare environments, Enterococcus species have been increasingly detected in wildlife, livestock, and food products, highlighting their ecological versatility and the broader One Health implications [109,110,111]. The presence of VRE and multidrug-resistant strains in animals and the environment points to the interconnectedness of antimicrobial resistance across different sectors. Efforts to control the spread of resistant Enterococcus must therefore consider not only clinical interventions but also measures to reduce antibiotic use in agriculture and improve environmental management practices. Understanding the ecology and transmission dynamics of Enterococcus will be crucial in developing strategies to mitigate its impact as a pathogen and limit the spread of antimicrobial resistance.
3. ESKAPE Bacteria in Wildlife
In recent years, it has become increasingly clear that the environment plays a crucial role in the ecology of pathogenic and antibiotic-resistant bacteria [112]. Wild animals, as they interact with habitats ranging from remote forests to urban and agricultural areas, are frequently exposed to contamination sources such as industrial effluents, hospital waste, and veterinary products [113,114]. This exposure means that wildlife can serve as unexpected reservoirs for pathogens of public health concern, including those within the ESKAPE group. These bacteria, known for their ability to evade antimicrobial treatments, are found not only in hospitals and farms but also among wildlife and in natural environments [115]. The detection of resistant strains in animals inhabiting natural ecosystems challenges the notion that resistance is solely a problem of anthropogenic settings, suggesting a continuous cycle of transmission between humans, domestic animals, wildlife, and the natural environment. Previous studies examined the resistome and the microbiome of migratory birds through metagenomic techniques and studied the resistance in Escherichia coli of wild birds [116,117]. Still, this review differs from the literature by presenting a more holistic approach to ESKAPE pathogens in the domain of wildlife and their clonally related lineages, resistance strategies, and possible dispersal mechanisms. This effort, which spans multiple species of bacteria and taxa of hosts, enhances the understanding of the spread of antimicrobial resistance in the world’s ecosystems. This review provides a unique contribution to the field by consolidating data on ESKAPE pathogens in wildlife, offering a comparative perspective across different bacterial species and host taxa. Unlike previous studies that primarily address antimicrobial resistance trends in specific bacterial groups, our work synthesizes findings on all six ESKAPE pathogens, allowing for a more integrated understanding of their ecological and epidemiological significance. The identification of clonal lineages, resistance patterns, and transmission risks in wildlife underscores the importance of expanding surveillance efforts beyond clinical and agricultural settings. Future research should focus on genomic and functional studies to further elucidate the role of wildlife in AMR dissemination.
3.1. Staphylococcus aureus and MRSA in Wild Animals
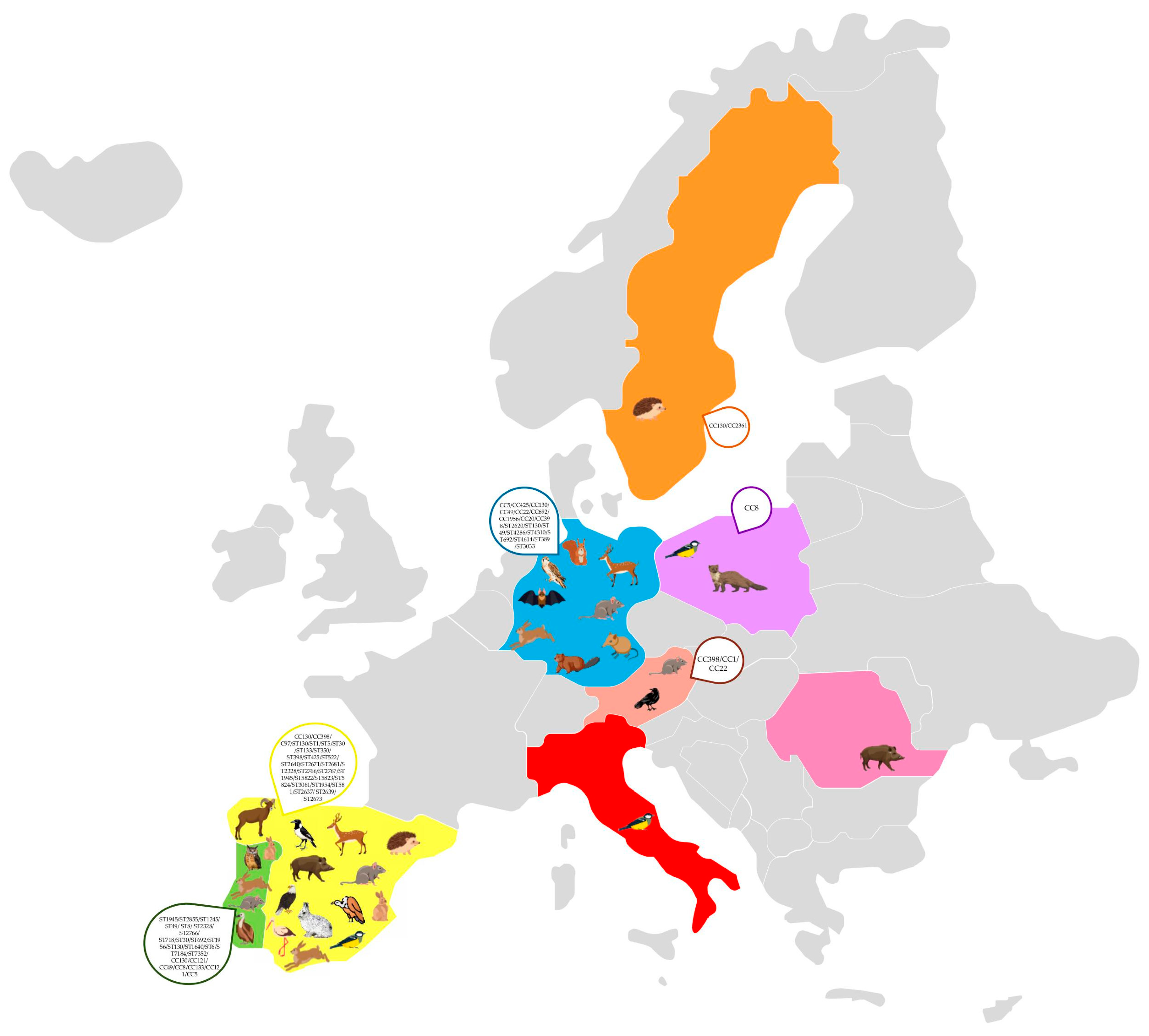
The detection of S. aureus, including methicillin-resistant S. aureus (MRSA), in wild animals across Europe highlights the potential role of wildlife as reservoirs and disseminators of antimicrobial resistance (Figure 2) [32]. Various studies have identified multiple clonal lineages, indicating a complex epidemiology with implications for both veterinary and human health (Table 1) [16,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141]. The most frequently reported clonal complex in wildlife is CC130, particularly in association with the mecC gene, which confers methicillin resistance. This clonal complex has been detected in numerous animal species, including rabbits, storks, hedgehogs, hares, rodents, and birds of prey, suggesting a widespread presence in different habitats [121,123,132,138,140,142]. In Spain and Portugal, for example, studies found mecC-positive MRSA belonging to ST130/CC130 in red deer, wild boars, and wild rodents, indicating that these animals may act as reservoirs for mecC-MRSA [118,137]. Similarly, in Sweden and Germany, mecC-carrying CC130 isolates have been found in a variety of wildlife, such as hedgehogs and rats, highlighting the geographical spread of this lineage across Northern and Central Europe [127,140].
Figure 2.
Molecular characteristics of Staphylococcus aureus clones in Europe.

Table 1.
Animal species, location, and genetic lineages of S. aureus isolated from European wild animals.
Besides CC130, other clonal lineages have been detected in wild animals, demonstrating a diverse population structure of S. aureus in wildlife. For instance, CC425 has been identified in red deer in Germany [129]. Additionally, rare lineages such as ST2855 (CC121) and ST49 (CC49) have been reported in wild rodents and hares in Portugal, indicating the existence of localized clonal variants [122,137]. The resistance profiles of S. aureus isolates from wild animals exhibit variability, with many isolates displaying resistance primarily to β-lactam antibiotics due to the presence of the mecA or mecC genes. For instance, mecC-positive MRSA isolates frequently show resistance to penicillin and cefoxitin, while remaining susceptible to other antibiotic classes, reflecting the typical resistance pattern associated with mecC-carrying strains. In contrast, some isolates from wildlife, particularly those associated with CC398, exhibit broader resistance profiles, including resistance to tetracycline, erythromycin, and clindamycin, which may be linked to antibiotic use in livestock [120,127,132,135,139]. Several studies have reported the transmission of MRSA from animals to humans [143,144,145], with particular emphasis on mecC-MRSA, a strain predominantly found in animals but increasingly detected in human infections. These findings suggest the potential for zoonotic spillover, reinforcing the need for surveillance to understand transmission pathways and public health implications.
In certain cases, multidrug-resistant MRSA strains have been identified. For example, wild birds in Portugal and red squirrels in Germany harbored isolates carrying both mecA and genes such as ermB (macrolide resistance) tetK (tetracycline resistance) and catpC221 (chloramphenicol resistance), indicating exposure to diverse antimicrobial agents [128,129,133]. The pathogenic potential of S. aureus isolates from wildlife is underscored by the detection of multiple virulence genes. Common virulence factors include genes encoding for enterotoxins (seg, seh), leukocidins (lukM/lukF-P83), and hemolysins (hla, hlb). For example, isolates from red deer in Spain have been found to carry the seg and seh genes, which encode staphylococcal enterotoxins implicated in food poisoning [118]. Similarly, strains isolated from red squirrels exhibited the presence of lukM/lukF-P83, which is known to enhance virulence in ruminants [129].
In addition to classical virulence genes, some wildlife-associated S. aureus strains possess unique adaptations. For instance, etd2, encoding an exfoliative toxin variant, has been found in strains from storks in Spain and owls in Portugal, suggesting the ability to colonize or infect avian hosts [123,128]. Additionally, some MRSA isolates from hedgehogs and rodents lacked certain immune evasion cluster (IEC) genes commonly found in human-adapted strains, potentially reflecting adaptations to the wildlife host [118,138].
3.2. Enterobacteriaceae in Wild Animals
The detection of Enterobacteriaceae in wild animals, especially antimicrobial-resistant strains, has significant implications for public health. These bacteria, which include pathogens such as E. coli and K. pneumoniae, are known for their ability to acquire and disseminate resistance genes, making them important indicators of environmental antimicrobial resistance. Studies across Europe have documented a diverse range of clonal lineages, resistance mechanisms, and virulence traits in Enterobacteriaceae isolated from wildlife, highlighting their role as potential reservoirs and sentinels for the spread of multidrug-resistant (MDR) bacteria [16,115,124,134,146,147,148,149,150,151,152,153,154,155,156,157,158,159] (Table 2). Various clonal lineages of E. coli have been reported in wild animals across Europe, with some lineages known for their relevance in human and animal infections. High-risk clones such as ST38, ST131, ST744, and ST410 have been identified in multiple studies [154,159]. For instance, ST38 and ST131, which are frequently associated with extraintestinal pathogenic E. coli (ExPEC) infections in humans, were found in gulls and other wild birds in Germany and Spain [154,159]. The presence of these lineages in wildlife suggests that wild birds may play a role in the environmental dissemination of ExPEC-related strains.

Table 2.
Animal species, location, and genetic lineages of Enterobacteriaceae isolated from European wild animals.
Similarly, ST744 and ST410, often linked to resistance to extended-spectrum β-lactamases (ESBLs) and carbapenemases, have been detected in wildlife species such as wild birds in Spain, indicating the wide distribution of these resistant clones [159]. The occurrence of diverse clonal lineages in various wildlife species suggests that these animals encounter different sources of contamination, including human sewage, agricultural runoff, and other environmental reservoirs.
Klebsiella pneumoniae in wild animals has also shown a concerning pattern of clonal diversity. High-risk clones such as ST11, ST307, and ST35, which are associated with healthcare-related infections in humans, have been isolated from wild animals in Spain and Italy [152,159]. For example, ST11, a notorious clone linked to carbapenem resistance, was found in wild birds, suggesting possible exposure to contaminated water bodies or food sources [159]. Additionally, other clones, such as ST290 in Germany and ST5670 in Italy, have been reported in various wild mammals and birds, further indicating the potential role of wildlife in the persistence and spread of these lineages [152,154]. The resistance profiles of Enterobacteriaceae isolated from wild animals exhibit a wide range of antimicrobial resistance, often mirroring the patterns observed in clinical and agricultural settings. Many isolates possess genes encoding for ESBLs, such as blaCTX-M, blaTEM, and blaSHV, which confer resistance to third-generation cephalosporins [124,146,147,153,154,156]. For example, studies in Germany have identified E. coli isolates from wild birds carrying blaCTX-M-15 and blaCTX-M-55, two of the most prevalent ESBL genes worldwide [154]. These findings suggest that wild birds may act as vectors for the dissemination of ESBL-producing strains across different geographic regions.
In addition to ESBLs, carbapenem-resistant Enterobacteriaceae (CRE) have also been detected in wildlife. E. coli-carrying carbapenemase genes such as blaVIM-1 and blaNDM-1 have been reported in aquatic birds and rodents in France, Greece, and Ukraine [115,146,155,159]. The detection of these resistance genes in wildlife is particularly concerning, as carbapenems are considered last-resort antibiotics for treating multidrug-resistant infections. The presence of CRE in wildlife habitats may be linked to the contamination of water bodies with untreated sewage or agricultural runoff.
Resistance to colistin, a last-resort antibiotic for treating multidrug-resistant Enterobacteriaceae, has also been documented. The mobile colistin resistance gene mcr-4 was found in E. coli isolates from deer and apennine chamois in Italy, indicating the spread of plasmid-mediated colistin resistance in natural environments [156]. This highlights the potential for wildlife to serve as reservoirs for colistin-resistant bacteria, which can pose a significant challenge to public health.
K. pneumoniae isolates from wild animals have shown multidrug resistance, including resistance to β-lactams, aminoglycosides, fluoroquinolones, and tetracyclines. For instance, isolates from gulls and raptors in Spain harbored genes such as blaKPC-3, blaOXA, and blaCTX-M-15, indicating the presence of highly resistant clones [153,154,159]. The detection of carbapenemase-producing K. pneumoniae in wild birds suggests that these animals could facilitate the environmental spread of carbapenem-resistant strains.
The pathogenic potential of Enterobacteriaceae from wild animals is indicated by the presence of numerous virulence factors. In E. coli, common virulence genes include ompT, iss, papC, and cva/cvi, which are associated with ExPEC strains capable of causing urinary tract infections and sepsis in humans [160,161]. For instance, E. coli isolates from birds in Poland were found to carry combinations of these genes, suggesting that these strains could pose a zoonotic risk [147].
In K. pneumoniae, virulence factors such as siderophore genes (ybt, iro, iuc), which enhance iron acquisition, and capsular polysaccharide genes (rmpA, rmpA2), associated with hypervirulence, were detected in isolates from gulls and other birds [152]. These virulence traits, coupled with antibiotic resistance, indicate that wildlife-associated K. pneumoniae may have a significant pathogenic potential, similar to the strains found in human clinical settings. Numerous studies provide evidence of the transmission of Enterobacteriaceae from animals to humans, particularly in pets and food-producing animals, where close contact or the food chain facilitates dissemination [162,163,164,165]. However, transmission from wild animals to humans is less evident and much more challenging to trace. Despite this, it can still occur through direct contact with wildlife or indirectly via environmental contamination, such as through water sources, soil, or food products exposed to resistant strains.
3.3. A. baumannii in Wild Animals
The presence of A. baumannii in wild animals is a growing concern due to its potential role as a reservoir for multidrug-resistant strains that pose a significant risk to both veterinary and human health. However, studies of A. baumannii in wildlife are still very scarce [134,148,150,153,166,167,168] (Table 3). This opportunistic pathogen is known for causing healthcare-associated infections, and its detection in wildlife suggests that environmental transmission pathways could contribute to the spread of resistance genes [169,170]. Studies across Europe have identified various clonal lineages, resistance mechanisms, and virulence traits in A. baumannii isolated from different wild animal species.

Table 3.
Animal species, location, and genetic lineages of Acinetobacter baumanni isolated from European wild animals.
The detection of A. baumannii in wildlife has been reported in several European countries, with varying clonal lineages indicating a broad genetic diversity [134,148,150,153,166,167,168]. However, high-risk clones such as ST2, ST25, and ST78, which are often associated with hospital outbreaks, were not detected in wild animals in those studies. A. baumannii clones ST1447 and ST836, which have been identified in geese in Germany, were first reported in Brazilian hospitals and in sewage water from a poultry slaughterhouse, respectively [166,171,172]. These findings highlight the potential for wildlife to harbor distinct A. baumanni istrains not commonly found in clinical settings, possibly reflecting local adaptation or exposure to environmental sources of contamination. In Southern Europe, particularly in Italy, Spain, and Portugal, A. baumannii has been isolated from various wild animals, including turtles, raptors, and wild mammals, suggesting that wildlife in Mediterranean areas may act as a reservoir for specific A. baumannii genotypes, some of which may possess environmental origins [134,148,150,153]. The detection of these strains across different animal species and geographical regions points to the potential for cross-species transmission or shared environmental sources, such as water bodies or contaminated soils.
The antimicrobial resistance profiles of A. baumannii isolated from wild animals often indicate multidrug resistance (MDR), with resistance to β-lactams, aminoglycosides, fluoroquinolones, and tetracyclines being frequently observed. A significant concern is the detection of carbapenem-resistant A. baumannii (CRAB), as carbapenems are considered last-resort antibiotics for treating MDR bacterial infections [173,174]. Carbapenem resistance in wildlife-associated A. baumannii is often mediated by the presence of blaOXA genes, which encode carbapenem-hydrolyzing class D β-lactamases [166,167,168]. For instance, the blaOXA-314 gene was detected in isolates from geese in Germany and from geese and white storks in Poland, indicating a geographically widespread distribution of these resistance genes in wildlife. The presence of these resistance genes suggests exposure to contaminated environments where antibiotics may be present, such as agricultural lands, wastewater discharge areas, or locations impacted by human activity. A. baumannii is primarily known as a human pathogen associated with healthcare infections, but increasing evidence suggests its presence in animals, raising concerns about zoonotic transmission. Human-to-animal transmission has been documented, particularly in pets such as dogs and cats, where strains genetically similar to human clinical isolates have been identified. Animal-to-human transmission is less well understood but has been reported in livestock, where A. baumannii has been isolated from cattle and pigs, with potential transmission routes through direct contact or environmental contamination [175,176,177,178].
3.4. P. aeruginosa in Wild Animals
Studies in P. aeruginosa isolated from wild animals are also very scarce [95,124,134,136,148,179,180]. P. aeruginosa is a versatile opportunistic pathogen found in various environments and is a significant concern due to its ability to acquire multidrug resistance [181,182]. Although it is primarily associated with hospital-acquired infections, its presence in wildlife suggests that natural environments could serve as reservoirs or transmission pathways for resistant strains. Several studies suggest that P. aeruginosa can be transmitted between animals and humans, either through direct contact with infected animals or indirectly via contaminated environments, water, or food. While most cases in humans are linked to healthcare settings, evidence indicates zoonotic and reverse zoonotic transmission. For example, a study documented the transmission of a carbapenem-resistant P. aeruginosa strain (ST233, VIM-2 producer) between a dog, its owner, and their household environment. Additionally, poultry has been identified as a potential reservoir, with strains capable of infecting humans through foodborne exposure [96,183,184,185]. These findings highlight the need for surveillance and a One Health approach to monitor the spread of P. aeruginosa across human, animal, and environmental interfaces. Studies across Europe have reported various clonal lineages, resistance patterns, and virulence traits in P. aeruginosa isolates from wild animals, indicating a potentially significant role in the ecology of antimicrobial resistance (Table 4) [95,124,134,136,148,179,180].

Table 4.
Animal species, location, and genetic lineages of P. aeruginosa isolated from European wild animals.
Among the collected studies, only two documented diverse clonal lineages of P. aeruginosa in wild animals. The globally prevalent high-risk clone ST274 and its associated clonal complex (CC274) are primarily linked to hospital environments and are frequently found colonizing cystic fibrosis patients across different regions [186]. Nevertheless, this high-risk clone has been identified in wild raptors in Spain, suggesting a possible spillover from human-impacted environments to wildlife. [179,180]. Additionally, novel lineages have been identified in wild animals, including ST1711 and ST2252 in Spain, which have not been commonly associated with human infections. P. aeruginosa ST1711 has been reported in non-clinical samples, while ST2252 has been identified in both non-clinical samples and among healthy animals also in Spain [179,187]. In some cases, the isolates showed a high degree of genetic diversity, indicating the presence of both well-known and novel clonal types circulating among various animal species. Geographically, P. aeruginosa has been reported in a wide range of habitats, including wetlands, forests, and coastal areas, where animals such as wild boars, turtles, and birds of prey reside [94,188]. The widespread detection across different ecosystems suggests that wildlife could play a role in the environmental dissemination of P. aeruginosa.
The antimicrobial resistance profiles of P. aeruginosa isolated from wild animals often reveal significant multidrug resistance, including resistance to β-lactams, aminoglycosides, fluoroquinolones, and carbapenems. Resistance to carbapenems, such as imipenem (IMI) and doripenem (DOR), is particularly concerning, as these antibiotics are used as last-resort treatments for serious P. aeruginosa infections [189,190]. Carbapenem resistance mechanisms in wildlife-associated isolates are often related to the production of β-lactamases, such as VIM, IMP, OXA, and GES metallo-β-lactamases, as well as alterations in porin channels and efflux pump overexpression. For instance, carbapenem-resistant P. aeruginosa isolates from red deer in Portugal were found to carry blaOXA-486 [180]. Resistance to other antibiotics, including fluoroquinolones (e.g., ciprofloxacin), aminoglycosides (e.g., gentamicin), and tetracyclines, has also been observed. In birds of prey from Spain and wild boars from Italy, P. aeruginosa isolates exhibited high resistance rates to aminoglycosides and fluoroquinolones, possibly due to exposure to contaminated environments where these antimicrobials are present, such as agricultural runoff or water bodies affected by human waste [134,191].
The pathogenic potential of P. aeruginosa isolates from wild animals is underscored by the detection of various virulence factors, which contribute to the bacterium’s ability to cause disease in humans and animals. Only one study reported the presence of virulence genes among P. aeruginosa from wild boars in Spain [179]. Virulence genes identified include those encoding exotoxins (exoS, exoY, exoT), elastases (lasB), and alkaline proteases (aprA), which are associated with tissue invasion and immune evasion [179]. The presence of quorum sensing systems, which regulate the expression of virulence factors, further indicates the pathogenic potential of these isolates. Genes like lasI and rhlI involved in quorum sensing were found in isolates from healthy animals, suggesting that these bacteria may coordinate virulence gene expression to adapt to different hosts or environmental conditions.
3.5. E. faecium in Wild Animals
Enterococcus spp., particularly Enterococcus faecium and Enterococcus faecalis, are commensal bacteria of the gastrointestinal tract in various hosts but can also act as opportunistic pathogens causing severe infections in humans and animals [98,192]. The detection of multidrug-resistant Enterococcus in wild animals suggests that wildlife may serve as reservoirs or vectors for the dissemination of resistance genes and virulent strains. Studies across Europe have identified diverse clonal lineages, resistance mechanisms, and virulence factors in Enterococcus isolates from wildlife, highlighting their significance in the One Health context (Table 5) [16,124,125,134,149,156,193,194,195,196,197,198,199,200,201,202,203,204].

Table 5.
Animal species, location, and genetic lineages of Enterococcus faecium isolated from European wild animals.
The genetic diversity of Enterococcus [109] isolates from wild animals is considerable, with various clonal lineages commonly associated with human and veterinary infections. In Europe, CC17, a high-risk lineage associated with hospital-acquired infections, has been frequently identified in wild birds and mammals, indicating that these animals could act as reservoirs for lineages typically associated with human clinical settings [193,200]. In addition to CC17, other clonal lineages such as ST121, ST32, and ST448 have been reported in wild animals. For instance, ST121 and ST32 were detected in wild birds in Germany and Poland, suggesting potential transmission or environmental contamination linked to agricultural activities or human waste [203,204]. ST448, associated with multidrug resistance, was found in wild partridges in Portugal, indicating that wildlife in various ecological niches can harbor diverse Enterococcus lineages.
Geographically, Enterococcus isolates from wildlife have been reported across a wide range of habitats, from forested areas and rural landscapes to urban environments. Studies have documented the presence of Enterococcus in species such as birds, rodents, foxes, and wild boars across countries including Spain, Italy, Poland, Slovakia, and the Czech Republic. This widespread distribution indicates that wild animals living in different environments are exposed to factors that promote the acquisition and spread of Enterococcus strains [200,205].
The antimicrobial resistance patterns of Enterococcus isolates from wild animals often reflect significant multidrug resistance, with resistance to antibiotics such as glycopeptides (e.g., vancomycin), aminoglycosides (e.g., gentamicin), tetracyclines, macrolides (e.g., erythromycin), and β-lactams (e.g., ampicillin). The detection of vancomycin-resistant enterococci (VRE) in wildlife is of particular concern, as vancomycin is a last-resort antibiotic for treating MDR Enterococcus infections [206]. Vancomycin resistance in Enterococcus is usually mediated by the presence of vanA or vanB gene clusters, which encode enzymes that alter the bacterial cell wall to reduce vancomycin binding [207]. Several studies have reported the presence of van genes in E. faecium isolates from wild birds and mammals in Poland, Portugal, and Slovakia [125,200,202]. In addition to glycopeptide resistance, Enterococcus isolates from wild animals often exhibit resistance to other antibiotics. Resistance genes such as ermB (macrolides), tetM and tetL (tetracyclines), aac(6′)-I and ant(6)-Ia (aminoglycosides), and pbp5 (β-lactams) have been commonly detected. For example, Red foxes in Latvia harbored Enterococcus strains resistant to macrolides, tetracyclines, and lincosamidses, and rodents in Bulgaria carried E. faecium showing resistance to ciprofloxacin, aminoglicosides, macrolides, and trimethoprim-sulfamethoxazole [149,197]. The presence of these resistances suggests that wildlife is exposed to environments contaminated with antibiotic residues, which may select for resistance. The detection of multidrug resistance, including combinations of vancomycin resistance and resistance to other antibiotic classes, poses a significant risk for the transfer of resistance genes across different reservoirs, including humans, animals, and the environment.
Virulence factors in Enterococcus species contribute to their ability to cause infections in humans and animals. These factors include genes encoding for surface adhesins, biofilm formation, and extracellular enzymes [208,209]. In wildlife-associated isolates, common virulence genes include esp (enterococcal surface protein), gelE (gelatinase), asa1 (aggregation substance), efaA (endocarditis antigen), and hyl (hyaluronidase). The presence of the esp gene, which is associated with biofilm formation and colonization of host tissues, has been frequently reported in E. faecium isolates from wild animals, including wild birds in Portugal and several wild mammals in Italy [195,200,210]. The ability to form biofilms enhances the persistence of these bacteria in the host and environment, potentially facilitating transmission and colonization of new hosts [208,209]. The detection of gelE and asa1 in Enterococcus isolates from wildlife further indicates a potential for pathogenicity. For example, several studies reported the presence of gelE and asa1 in E. faecalis isolates from Apennine chamois and deer in Italy, as well as several animal species in Poland [156,194,199], suggesting that these strains could pose a risk for causing opportunistic infections. Additionally, the presence of hyl, found in some VRE isolates from wild birds in Portugal, suggests an enhanced ability to degrade host tissues and evade immune responses [200]. The genetic adaptability of Enterococcus is often facilitated by horizontal gene transfer (HGT), which allows for the acquisition of antibiotic resistance genes and virulence factors. Mobile genetic elements such as plasmids, transposons, and integrative conjugative elements play a key role in the dissemination of resistance genes across different bacterial populations [211,212,213]. In wildlife-associated Enterococcus, several studies have documented the presence of mobile genetic elements carrying resistance genes. For example, the Tn1546 transposon, which carries the vanA gene cluster, has been found in VRE isolates from wild birds in Portugal [200]. Similarly, conjugative plasmids carrying multiple resistance genes, including ermB, aac(6′)-I, and tetM, have been identified in E. faecium and E. faecalis from wild mammals in Poland and the Czech Republic [125,204]. The detection of these elements highlights the potential for wildlife to contribute to the spread of resistance genes through HGT. Evidence of human-to-animal and animal-to-human transmission has been reported, particularly in companion animals and livestock [199,214,215,216]. Pets, such as dogs and cats, have been found to carry strains genetically similar to those isolated from hospitalized patients, suggesting possible bidirectional transmission. In livestock, E. faecium has been detected in farm animals, particularly in those exposed to antibiotics, raising concerns about its potential spread to humans through direct contact or the food chain. The increasing detection of VRE in non-human hosts highlights the importance of surveillance and a One Health approach to mitigate its spread across human, animal, and environmental interfaces.
4. Implications for One Health
The presence of ESKAPE bacteria in wild animals has critical implications for public health, veterinary medicine, and ecosystem integrity. These bacteria, known for their role in hospital-acquired infections and multidrug resistance, are increasingly found in wildlife, raising concerns about the broader environmental dynamics of antimicrobial resistance and the potential risks posed to humans and domestic animals. The detection of diverse resistant and virulent strains across various wildlife species and geographical locations underscores the interconnectedness of human, animal, and environmental health, emphasizing the need for a One Health approach to combatting AMR [16].
Wild animals are often exposed to contaminated environments, which can act as reservoirs for resistant bacteria and antimicrobial residues [217]. Habitats near urban centers, agricultural areas, or water bodies impacted by human activities frequently harbor high levels of contaminants, including antibiotics and resistant bacteria shed by humans or livestock [217,218]. For example, surface waters polluted with untreated sewage, agricultural runoff, or waste from pharmaceutical production can serve as hotspots for the spread of resistant bacteria across different species [219]. Wild birds, rodents, and aquatic mammals that frequent these areas can acquire and carry ESKAPE bacteria, effectively acting as vectors that facilitate the movement of resistance genes across ecological boundaries. The evidence of ESKAPE pathogens, such as carbapenem-resistant Acinetobacter baumannii and ESBL-producing Enterobacteriaceae, in wildlife highlights the permeability of barriers between natural and human-impacted environments [56,217]. The presence of bacteria in wild animals suggests a bidirectional flow of resistance genes between wildlife and anthropogenic sources. While some strains found in wildlife may be of environmental origin, others share clonal lineages with human clinical isolates, indicating possible spillover from healthcare settings or animal husbandry practices. Such findings suggest that natural habitats are not isolated reservoirs but rather part of a complex web of transmission pathways that include human, animal, and environmental interfaces [220,221].
Wild animals can serve as sentinels for monitoring the spread and emergence of antimicrobial resistance. Species that live in close proximity to human activity, such as birds of prey, scavengers, and small mammals, are particularly useful indicators of environmental AMR levels because of their exposure to diverse sources of contamination [128]. The detection of methicillin-resistant Staphylococcus aureus (MRSA) in hedgehogs and other mammals, for instance, indicates the widespread nature of mecC-mediated resistance beyond clinical settings. The persistence of MRSA in natural populations suggests that resistant strains can thrive outside of traditional reservoirs, further complicating efforts to contain AMR. In the case of carbapenemase-producing Klebsiella pneumoniae and Pseudomonas aeruginosa, their detection in gulls and other migratory birds provides evidence of the potential for long-distance transmission of resistant bacteria. Migratory species can disseminate AMR genes across continents, contributing to the global spread of resistance [222,223]. The identification of high-risk clones in wildlife emphasizes the relevance of including wild animal populations in AMR surveillance programs. These programs should not only focus on the pathogens commonly associated with human health but also account for the broader environmental and ecological drivers of resistance.
The spread of AMR is not limited to the movement of resistant bacterial strains but is also driven by horizontal gene transfer, facilitated by mobile genetic elements such as plasmids, transposons, and integrons [224,225]. Wildlife-associated ESKAPE bacteria often harbor resistance genes on mobile elements, which can be transferred across different bacterial species and genera. The detection of plasmid-mediated colistin resistance (mcr genes) in Enterobacteriaceae from deer and wild boars, as well as the presence of carbapenemase genes (blaOXA, blaNDM) in Acinetobacter baumannii from birds and mammals, suggests that natural environments provide a setting for the exchange of genetic material that enhances bacterial adaptability. The role of wildlife as reservoirs for resistance genes extends beyond individual bacterial species; it involves a network of genetic exchanges that can have significant consequences for public health. In particular, the combination of multidrug resistance and virulence factors in wildlife-associated strains raises concerns about the potential emergence of highly pathogenic, resistant bacteria [118,226]. The presence of conjugative plasmids carrying multiple resistance genes, such as ermB, aac(6′)-Ii, vanA, and tetM, in Enterococcus spp. from wild mammals, points to the complexity of the resistance gene pool circulating in natural environments. This gene pool is not static but dynamic, continuously shaped by selective pressures such as antibiotic use, ecological interactions, and environmental factors. The risk levels associated with antimicrobial resistance in wildlife vary depending on factors such as bacterial species, resistance mechanisms, and ecological interactions. High-risk resistance genes, such as those encoding carbapenemases (blaKPC, blaNDM, blaOXA), vancomycin resistance (vanA, vanB), and extended-spectrum β-lactamases (blaCTX-M), have been identified in ESKAPE bacteria isolated from wild animals. These genes are particularly concerning due to their association with multidrug-resistant infections in humans. However, the mere presence of a resistance gene does not always indicate immediate clinical relevance, as some may remain unexpressed under natural conditions. The expression of antimicrobial resistance genes can be influenced by environmental stressors, antibiotic exposure, and genetic regulation mechanisms. Silent resistance genes may still play a critical role in AMR dissemination, as they can be activated under selective pressure or transferred to other bacteria via horizontal gene transfer. This highlights the need for further studies evaluating not only the genetic presence of resistance determinants but also their phenotypic expression and potential impact on human and animal health.
The detection of ESKAPE bacteria in wildlife has direct implications for zoonotic transmission and human health risks. While the majority of studies focus on livestock and domestic animals as potential sources of resistant pathogens, wildlife represents a less controlled, yet equally significant, reservoir. The movement of wild animals into human-dominated landscapes, whether through natural migration, habitat encroachment, or attraction to urban food sources, increases the likelihood of direct or indirect transmission of resistant bacteria to humans and domestic animals [227,228]. For instance, scavenging species like foxes, raccoons, and vultures, which feed on waste or carcasses, can act as vectors for resistant bacteria that originate from agricultural or urban settings [128]. The risk extends to the transmission of virulent strains with enhanced pathogenicity. Wildlife-associated isolates of P. aeruginosa, for example, often carry virulence genes such as exoS and lasB, which can increase their capacity to cause severe infections in immunocompromised hosts [229]. Likewise, E. faecium strains from wild animals that carry biofilm-associated genes may have an enhanced ability to colonize medical devices or persist in hospital environments [210]. The combination of antimicrobial resistance and virulence traits in ESKAPE pathogens from wildlife thus represents a dual threat, complicating infection control and treatment strategies.
Addressing the implications of ESKAPE bacteria in wild animals requires an integrated One Health approach that combines efforts from human medicine, veterinary science, and environmental management [223,230]. Surveillance of antimicrobial resistance should not be confined to clinical settings but should include a broader range of ecosystems, recognizing the interconnectedness of different AMR reservoirs. Such integrated surveillance would enable earlier detection of emerging resistance patterns, providing critical data for guiding policy decisions and mitigating the spread of AMR. Efforts to reduce environmental contamination with antibiotics and resistant bacteria are also essential. This includes better management of agricultural practices to limit the use of antibiotics in livestock, improving wastewater treatment to prevent the release of resistant bacteria into natural water bodies, and implementing regulations on the disposal of pharmaceutical waste [231,232]. Wildlife rehabilitation centers, zoos, and conservation programs should also incorporate AMR monitoring to ensure that released animals do not inadvertently contribute to the spread of resistance. Lastly, public awareness and education about the risks of AMR should extend beyond healthcare and agriculture to include the role of wildlife and natural ecosystems. Understanding that antimicrobial resistance is not confined to hospitals or farms but is a broader environmental issue can help garner support for policies aimed at addressing AMR on a global scale. The challenge of ESKAPE bacteria in wild animals exemplifies the need for a comprehensive and multidisciplinary approach to effectively tackle antimicrobial resistance in the One Health framework. Finding ESKAPE pathogens in wildlife poses a risk for their potential spread to humans, whether by direct contact with animals or through contaminated environments. Wildlife can serve as reservoirs, aiding in the maintenance and dissemination of these bacteria in natural ecosystems from where they may eventually be transmitted to people via food, water, or other vector species. This reiterates the need for an integrated multisectoral approach that incorporates monitoring and evaluation activities regarding the health of humans, animals, and the environment. In addition, integrating results with the WHO list of prioritized pathogens associated with antimicrobial resistance makes it possible to evaluate risks and produce comprehensive solutions with increased precision. Although earlier studies contributed useful information regarding the resistome of certain wildlife species like migratory birds, this review places ESKAPE pathogens in a wider context. By integrating information from different host species and resistance patterns, our study demonstrates that the circulation of antimicrobial resistance is more complex than clinical and agricultural environments. This point reinforces the need for more comprehensive surveillance systems that cover various wildlife reservoirs and their relationships with human and ecosystem health.
5. Conclusions
The detection of ESKAPE bacteria in wildlife underlines the complexity of antimicrobial resistance as a global issue that transcends healthcare settings. The evidence of diverse resistant strains across different animal species and geographic regions highlights the interconnected nature of human, animal, and environmental health. Wild animals, whether as reservoirs, sentinels, or vectors, play a significant role in the ecology of antimicrobial resistance. They are not only exposed to resistance genes through environmental contamination but can also contribute to their dissemination across ecological boundaries, potentially spreading resistance to human populations.
The findings suggest that controlling ESKAPE pathogens requires a comprehensive and multidisciplinary approach. Surveillance efforts must expand beyond hospitals to encompass broader environmental contexts, including wildlife and natural ecosystems. The inclusion of wild animals in AMR monitoring programs can provide valuable insights into emerging resistance patterns and help guide targeted interventions. This approach, combined with strategies to reduce environmental contamination, such as better management of antibiotic use in agriculture and improvements in wastewater treatment, can help to mitigate the spread of resistance.
Ultimately, addressing the challenges posed by ESKAPE bacteria in wildlife is not just about protecting animal health; it is about safeguarding public health and ensuring the sustainability of ecosystems. The One Health approach emphasizes that tackling antimicrobial resistance must involve coordinated efforts across human medicine, veterinary science, and environmental management. As our understanding of the dynamics of resistance continues to evolve, it becomes increasingly clear that efforts to curb the tide of antimicrobial resistance must consider the full spectrum of reservoirs and transmission pathways that extend well beyond the walls of hospitals. This review highlights the importance of evaluating AMR risk levels by considering both genetic detection and functional expression of resistance genes. While some antimicrobial resistance genes remain unexpressed in wildlife isolates, they may still pose a future threat due to their potential activation under selective pressure. Future research should focus on transcriptomic and proteomic analyses to determine gene expression patterns in different environmental contexts. Implementing a One Health surveillance approach that integrates genomic and phenotypic resistance data is essential for predicting and mitigating the spread of AMR across ecological boundaries.
Author Contributions
Conceptualization, V.S.; methodology, V.S.; validation, J.E.P., P.P. and G.I.; formal analysis, M.C. and J.E.P.; investigation, V.S. and S.A.; data curation, V.S. and S.A.; writing—original draft preparation, V.S.; writing—review and editing, V.S.; supervision, M.C., P.P. and G.I. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the projects UI/00772 and LA/P/0059/2020 funded by the Portuguese Foundation for Science and Technology (FCT). This work received financial support from FCT/MCTES (UIDB/00772/2020, Doi:10.54499/UIDB/00772/2020, UIDB/50006/2020 DOI 10.54499/UIDB/50006/2020, LA/P/0008/2020 DOI 10.54499/LA/P/0008/2020, UIDP/50006/2020 DOI 10.54499/UIDP/50006/2020).
Institutional Review Board Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Panda, S.K.; Buroni, S.; Swain, S.S.; Bonacorsi, A.; da Fonseca Amorim, E.A.; Kulshrestha, M.; Da Silva, L.C.N.; Tiwari, V. Recent advances to combat ESKAPE pathogens with special reference to essential oils. Front. Microbiol. 2022, 13, 1029098. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.B.; Nolan, A.C.; Zeden, M.S. How can we escape the ESKAPEs: Antimicrobial resistance mechanisms and what lies ahead? PLoS Pathog. 2024, 20, e1012270. [Google Scholar] [CrossRef] [PubMed]
- Venkateswaran, P.; Vasudevan, S.; David, H.; Shaktivel, A.; Shanmugam, K.; Neelakantan, P.; Solomon, A.P. Revisiting ESKAPE Pathogens: Virulence, resistance, and combating strategies focusing on quorum sensing. Front. Cell. Infect. Microbiol. 2023, 13, 1159798. [Google Scholar] [CrossRef]
- WHO World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Aguilar-Salazar, A.; Martínez-Vázquez, A.V.; Aguilera-Arreola, G.; de Jesus de Luna-Santillana, E.; Cruz-Hernández, M.A.; Escobedo-Bonilla, C.M.; Lara-Ramírez, E.; Sánchez-Sánchez, M.; Guerrero, A.; Rivera, G.; et al. Prevalence of ESKAPE Bacteria in Surface Water and Wastewater Sources: Multidrug Resistance and Molecular Characterization, an Updated Review. Water 2023, 15, 3200. [Google Scholar] [CrossRef]
- Khasapane, N.G.; Nkhebenyane, S.J.; Lekota, K.; Thekisoe, O.; Ramatla, T. “One Health” Perspective on Prevalence of ESKAPE Pathogens in Africa: A Systematic Review and Meta-Analysis. Pathogens 2024, 13, 787. [Google Scholar] [CrossRef]
- Xia, Y.; Zhang, M.; Tsang, D.C.W.; Geng, N.; Lu, D.; Zhu, L.; Igalavithana, A.D.; Dissanayake, P.D.; Rinklebe, J.; Yang, X.; et al. Recent advances in control technologies for non-point source pollution with nitrogen and phosphorous from agricultural runoff: Current practices and future prospects. Appl. Biol. Chem. 2020, 63, 8. [Google Scholar] [CrossRef]
- Akhter, S.; Bhat, M.A.; Ahmed, S.; Siddiqui, W.A. Antibiotic residue contamination in the aquatic environment, sources and associated potential health risks. Environ. Geochem. Health 2024, 46, 387. [Google Scholar] [CrossRef]
- Bereanu, A.-S.; Bereanu, R.; Mohor, C.; Vintilă, B.I.; Codru, I.R.; Olteanu, C.; Sava, M. Prevalence of Infections and Antimicrobial Resistance of ESKAPE Group Bacteria Isolated from Patients Admitted to the Intensive Care Unit of a County Emergency Hospital in Romania. Antibiotics 2024, 13, 400. [Google Scholar] [CrossRef]
- Vrancianu, C.O.; Pelcaru, C.F.; Alistar, A.; Gheorghe, I.; Marutescu, L.; Popa, M.; Czobor, I.; Gradisteanu, G.; Dobre, E.G.; Chifiriuc, M.C. Escaping from ESKAPE. Clinical significance and antibiotic resistance mechanisms in acinetobacter baumannii: A review. Biointerface Res. Appl. Chem. 2021, 11, 8190–8203. [Google Scholar]
- Ranganathan, S.; Nagarajan, H.; Busi, S.; Ampasala, D.R.; Lee, J.-K. Mechanistic Understanding of Antibiotic Resistance in ESKAPE Pathogens. In ESKAPE Pathogens: Detection, Mechanisms and Treatment Strategies; Busi, S., Prasad, R., Eds.; Springer Nature: Singapore, 2024; pp. 79–118. ISBN 978-981-99-8799-3. [Google Scholar]
- Mishra, A.; Aggarwal, A.; Khan, F. Medical Device-Associated Infections Caused by Biofilm-Forming Microbial Pathogens and Controlling Strategies. Antibiotics 2024, 13, 623. [Google Scholar] [CrossRef]
- Artini, M.; Papa, R.; Vrenna, G.; Trecca, M.; Paris, I.; D’Angelo, C.; Tutino, M.L.; Parrilli, E.; Selan, L. Antarctic marine bacteria as a source of anti-biofilm molecules to combat ESKAPE pathogens. Antibiotics 2023, 12, 1556. [Google Scholar] [CrossRef] [PubMed]
- Ravi, K.; Singh, B. ESKAPE: Navigating the Global Battlefield for Antimicrobial Resistance and Defense in Hospitals. Bacteria 2024, 3, 76–98. [Google Scholar] [CrossRef]
- Miller, W.R.; Arias, C.A. ESKAPE pathogens: Antimicrobial resistance, epidemiology, clinical impact and therapeutics. Nat. Rev. Microbiol. 2024, 22, 598–616. [Google Scholar] [CrossRef] [PubMed]
- Russo, T.P.; Minichino, A.; Gargiulo, A.; Varriale, L.; Borrelli, L.; Pace, A.; Santaniello, A.; Pompameo, M.; Fioretti, A.; Dipineto, L. Prevalence and phenotypic antimicrobial resistance among ESKAPE bacteria and Enterobacterales strains in wild birds. Antibiotics 2022, 11, 1825. [Google Scholar] [CrossRef]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Correia, S.; Silva, V.; García-Díez, J.; Teixeira, P.; Pimenta, K.; Pereira, J.E.; Oliveira, S.; Rocha, J.; Manaia, C.M.; Igrejas, G.; et al. One Health Approach Reveals the Absence of Methicillin-Resistant Staphylococcus aureus in Autochthonous Cattle and Their Environments. Front. Microbiol. 2019, 10, 2735. [Google Scholar] [CrossRef]
- Silva, V.; Correia, E.; Pereira, J.E.; González-Machado, C.; Capita, R.; Alonso-Calleja, C.; Igrejas, G.; Poeta, P. Biofilm Formation of Staphylococcus aureus from Pets, Livestock, and Wild Animals: Relationship with Clonal Lineages and Antimicrobial Resistance. Antibiotics 2022, 11, 772. [Google Scholar] [CrossRef]
- Silva, V.; Ribeiro, J.; Teixeira, P.; Pinto, P.; Vieira-Pinto, M.; Poeta, P.; Caniça, M.; Igrejas, G. Genetic Complexity of CC5 Staphylococcus aureus Isolates Associated with Sternal Bursitis in Chickens: Antimicrobial Resistance, Virulence, Plasmids, and Biofilm Formation. Pathogens 2024, 13, 519. [Google Scholar] [CrossRef]
- Waryah, C.B.; Gogoi-Tiwari, J.; Wells, K.; Eto, K.Y.; Masoumi, E.; Costantino, P.; Kotiw, M.; Mukkur, T. Diversity of Virulence Factors Associated with West Australian Methicillin-Sensitive Staphylococcus aureus Isolates of Human Origin. Biomed Res. Int. 2016, 2016, 8651918. [Google Scholar] [CrossRef]
- Tam, K.; Torres, V.J. Staphylococcus aureus secreted toxins and extracellular enzymes. Microbiol. Spectr. 2019, 7, 10–1128. [Google Scholar] [CrossRef]
- Feng, Y.; Chen, C.-J.; Su, L.-H.; Hu, S.; Yu, J.; Chiu, C.-H. Evolution and pathogenesis of Staphylococcus aureus: Lessons learned from genotyping and comparative genomics. FEMS Microbiol. Rev. 2008, 32, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Divyakolu, S.; Chikkala, R.; Ratnakar, K.S.; Sritharan, V. Hemolysins of Staphylococcus aureus—An update on their biology, role in pathogenesis and as targets for anti-virulence therapy. Adv. Infect. Dis. 2019, 9, 80–104. [Google Scholar]
- Kong, C.; Neoh, H.M.; Nathan, S. Targeting Staphylococcus aureus toxins: A potential form of anti-virulence therapy. Toxins 2016, 8, 72. [Google Scholar] [CrossRef]
- Sedarat, Z.; Taylor-Robinson, A.W. Biofilm formation by pathogenic bacteria: Applying a Staphylococcus aureus model to appraise potential targets for therapeutic intervention. Pathogens 2022, 11, 388. [Google Scholar] [CrossRef] [PubMed]
- Kranjec, C.; Morales Angeles, D.; Torrissen Mårli, M.; Fernández, L.; García, P.; Kjos, M.; Diep, D.B. Staphylococcal biofilms: Challenges and novel therapeutic perspectives. Antibiotics 2021, 10, 131. [Google Scholar] [CrossRef]
- Tsouklidis, N.; Kumar, R.; Heindl, S.E.; Soni, R.; Khan, S. Understanding the fight against resistance: Hospital-acquired methicillin-resistant Staphylococcus aureus vs. community-acquired methicillin-resistant Staphylococcus aureus. Cureus 2020, 12, e8867. [Google Scholar] [CrossRef]
- Lynch, J.P.; Zhanel, G.G. Escalation of antimicrobial resistance among MRSA part 1: Focus on global spread. Expert Rev. Anti. Infect. Ther. 2023, 21, 99–113. [Google Scholar] [CrossRef]
- Gopikrishnan, M.; Haryini, S. Emerging strategies and therapeutic innovations for combating drug resistance in Staphylococcus aureus strains: A comprehensive review. J. Basic Microbiol. 2024, 64, 2300579. [Google Scholar] [CrossRef]
- Shah, S.N.; Bhat, M.A.; Bhat, M.A.; Jan, A.T. Antimicrobial Resistance: An Overview. In Nanotechnology Based Strategies for Combating Antimicrobial Resistance; Springer: Singapore, 2024; pp. 1–44. [Google Scholar] [CrossRef]
- Silva, V.; Capelo, J.L.; Igrejas, G.; Poeta, P. Molecular Epidemiology of Staphylococcus aureus Lineages in Wild Animals in Europe: A Review. Antibiotics 2020, 9, 122. [Google Scholar] [CrossRef]
- Silva, V.; Monteiro, A.; Pereira, J.E.; Maltez, L.; Igrejas, G.; Poeta, P. MRSA in Humans, Pets and Livestock in Portugal: Where We Came from and Where We Are Going. Pathogens 2022, 11, 1110. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, P.; Wu, J.; Chen, S.; Jin, Y.; Long, J.; Duan, G.; Yang, H. Transmission of livestock-associated methicillin-resistant Staphylococcus aureus between animals, environment, and humans in the farm. Environ. Sci. Pollut. Res. 2023, 30, 86521–86539. [Google Scholar] [CrossRef] [PubMed]
- Crespo-Piazuelo, D.; Lawlor, P.G. Livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) prevalence in humans in close contact with animals and measures to reduce on-farm colonisation. Ir. Vet. J. 2021, 74, 21. [Google Scholar] [CrossRef]
- Dalton, K.R.; Rock, C.; Carroll, K.C.; Davis, M.F. One Health in hospitals: How understanding the dynamics of people, animals, and the hospital built-environment can be used to better inform interventions for antimicrobial-resistant gram-positive infections. Antimicrob. Resist. Infect. Control 2020, 9, 78. [Google Scholar] [CrossRef]
- Wang, S.-H.; Kebede, S.; Abate, E.; Amir, A.; Calderon, E.; Hoet, A.E.; Ikram, A.; LeJeune, J.T.; Mekuria, Z.; Suzuki, S. Emergence and dissemination of antimicrobial resistance at the interface of humans, animals, and the environment. In Modernizing Global Health Security to Prevent, Detect, and Respond; Elsevier: Amsterdam, The Netherlands, 2024; pp. 113–136. [Google Scholar]
- Taggar, G.; Attiq Rheman, M.; Boerlin, P.; Diarra, M.S. Molecular epidemiology of carbapenemases in Enterobacteriales from humans, animals, food and the environment. Antibiotics 2020, 9, 693. [Google Scholar] [CrossRef]
- Hu, Y.; Anes, J.; Devineau, S.; Fanning, S. Klebsiella pneumoniae: Prevalence, reservoirs, antimicrobial resistance, pathogenicity, and infection: A hitherto unrecognized zoonotic bacterium. Foodborne Pathog. Dis. 2021, 18, 63–84. [Google Scholar] [CrossRef]
- Sanz-García, F.; Gil-Gil, T.; Laborda, P.; Ochoa-Sánchez, L.E.; Martínez, J.L.; Hernando-Amado, S. Coming from the Wild: Multidrug Resistant Opportunistic Pathogens Presenting a Primary, Not Human-Linked, Environmental Habitat. Int. J. Mol. Sci. 2021, 22, 8080. [Google Scholar] [CrossRef]
- Abban, M.K.; Ayerakwa, E.A.; Mosi, L.; Isawumi, A. The burden of hospital acquired infections and antimicrobial resistance. Heliyon 2023, 9, e20561. [Google Scholar] [CrossRef]
- Lynch III, J.P.; Clark, N.M.; Zhanel, G.G. Escalating antimicrobial resistance among Enterobacteriaceae: Focus on carbapenemases. Expert Opin. Pharmacother. 2021, 22, 1455–1474. [Google Scholar] [CrossRef]
- Foster-Nyarko, E.; Pallen, M.J. The microbial ecology of Escherichia coli in the vertebrate gut. FEMS Microbiol. Rev. 2022, 46, fuac008. [Google Scholar] [CrossRef]
- Johnson, T.J.; Logue, C.M.; Johnson, J.R.; Kuskowski, M.A.; Sherwood, J.S.; Barnes, H.J.; DebRoy, C.; Wannemuehler, Y.M.; Obata-Yasuoka, M.; Spanjaard, L.; et al. Associations between multidrug resistance, plasmid content, and virulence potential among extraintestinal pathogenic and commensal Escherichia coli from humans and poultry. Foodborne Pathog. Dis. 2012, 9, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Xicohtencatl-Cortes, J.; Ochoa, S.A.; Cruz-Córdova, A.; Flores-Oropeza, M.A.; Hernández-Castro, R. New Strategies for the Prevention of Urinary Tract Infections by Uropathogenic Escherichia coli. In Urinary Tract Infections-New Insights; IntechOpen: London, UK, 2022; ISBN 1837682127. [Google Scholar]
- Santos, A.C.M.; Santos-Neto, J.F.; Trovão, L.O.; Romano, R.F.T.; Silva, R.M.; Gomes, T.A.T. Characterization of unconventional pathogenic Escherichia coli isolated from bloodstream infection: Virulence beyond the opportunism. Braz. J. Microbiol. 2023, 54, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Husna, A.; Rahman, M.M.; Badruzzaman, A.T.M.; Sikder, M.H.; Islam, M.R.; Rahman, M.T.; Alam, J.; Ashour, H.M. Extended-spectrum β-lactamases (ESBL): Challenges and opportunities. Biomedicines 2023, 11, 2937. [Google Scholar] [CrossRef]
- Mondal, A.H.; Khare, K.; Saxena, P.; Debnath, P.; Mukhopadhyay, K.; Yadav, D. A Review on Colistin Resistance: An Antibiotic of Last Resort. Microorganisms 2024, 12, 772. [Google Scholar] [CrossRef]
- Xu, L.; Wang, M.; Yuan, J.; Wang, H.; Li, M.; Zhang, F.; Tian, Y.; Yang, J.; Wang, J.; Li, B. The KbvR regulator contributes to capsule production, outer membrane protein biosynthesis, antiphagocytosis, and virulence in Klebsiella pneumoniae. Infect. Immun. 2021, 89, e00016-21. [Google Scholar] [CrossRef]
- Zhang, G.; Zhang, M.; Sun, F.; Zhou, J.; Wang, Y.; Zhu, D.; Chen, Z.; Chen, Q.; Chang, Q.; Liu, H. Epidemiology, mortality and risk factors for patients with K. pneumoniae bloodstream infections: Clinical impact of carbapenem resistance in a tertiary university teaching hospital of Beijing. J. Infect. Public Health 2020, 13, 1710–1714. [Google Scholar] [CrossRef]
- Karampatakis, T.; Tsergouli, K.; Behzadi, P. Carbapenem-resistant Klebsiella pneumoniae: Virulence factors, molecular epidemiology and latest updates in treatment options. Antibiotics 2023, 12, 234. [Google Scholar] [CrossRef]
- Boyd, S.E.; Holmes, A.; Peck, R.; Livermore, D.M.; Hope, W. OXA-48-like β-lactamases: Global epidemiology, treatment options, and development pipeline. Antimicrob. Agents Chemother. 2022, 66, e00216-22. [Google Scholar] [CrossRef]
- Qamar, M.U.; Ejaz, H.; Walsh, T.R.; Shah, A.A.; Al Farraj, D.A.; Alkufeidy, R.M.; Alkubaisi, N.A.; Saleem, S.; Jahan, S. Clonal relatedness and plasmid profiling of extensively drug-resistant New Delhi metallo-β-lactamase-producing Klebsiella pneumoniae clinical isolates. Future Microbiol. 2021, 16, 229–239. [Google Scholar] [CrossRef]
- Arnold, B.J.; Huang, I.-T.; Hanage, W.P. Horizontal gene transfer and adaptive evolution in bacteria. Nat. Rev. Microbiol. 2022, 20, 206–218. [Google Scholar] [CrossRef]
- Oyenuga, N.; Cobo-Díaz, J.F.; Alvarez-Ordóñez, A.; Alexa, E.-A. Overview of Antimicrobial Resistant ESKAPEE Pathogens in Food Sources and Their Implications from a One Health Perspective. Microorganisms 2024, 12, 2084. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Castillo, F.Y.; Guerrero-Barrera, A.L.; Avelar-González, F.J. An overview of carbapenem-resistant organisms from food-producing animals, seafood, aquaculture, companion animals, and wildlife. Front. Vet. Sci. 2023, 10, 1158588. [Google Scholar] [CrossRef] [PubMed]
- Kabali, E.; Pandey, G.S.; Munyeme, M.; Kapila, P.; Mukubesa, A.N.; Ndebe, J.; Muma, J.B.; Mubita, C.; Muleya, W.; Muonga, E.M. Identification of Escherichia coli and Related Enterobacteriaceae and Examination of Their Phenotypic Antimicrobial Resistance Patterns: A Pilot Study at A Wildlife–Livestock Interface in Lusaka, Zambia. Antibiotics 2021, 10, 238. [Google Scholar] [CrossRef] [PubMed]
- Anju, V.T.; Siddhardha, B.; Dyavaiah, M. Enterobacter infections and antimicrobial drug resistance. Model Org. Microb. Pathog. Biofilm Form. Antimicrob. Drug Discov. 2020, 175–194. [Google Scholar] [CrossRef]
- Salimiyan Rizi, K.; Ghazvini, K.; Farsiani, H. Clinical and pathogenesis overview of Enterobacter infections. Rev. Clin. Med. 2020, 6, 146–154. [Google Scholar]
- Fu, S.; Wang, Q.; Wang, R.; Zhang, Y.; Lan, R.; He, F.; Yang, Q. Horizontal transfer of antibiotic resistance genes within the bacterial communities in aquacultural environment. Sci. Total Environ. 2022, 820, 153286. [Google Scholar] [CrossRef]
- Leverstein-van Hall, M.A.; Box, A.T.A.; Blok, H.E.M.; Paauw, A.; Fluit, A.C.; Verhoef, J. Evidence of Extensive Interspecies Transfer of Integron-Mediated Antimicrobial Resistance Genes among Multidrug-Resistant Enterobacteriaceae in a Clinical Setting. J. Infect. Dis. 2002, 186, 49–56. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Burns, K.; Rodríguez Baño, J.; Borg, M.; Daikos, G.; Dumpis, U.; Lucet, J.C.; Moro, M.L.; Tacconelli, E.; Simonsen, G.S.; et al. Infection prevention and control measures and tools for the prevention of entry of carbapenem-resistant Enterobacteriaceae into healthcare settings: Guidance from the European Centre for Disease Prevention and Control. Antimicrob. Resist. Infect. Control 2017, 6, 113. [Google Scholar] [CrossRef]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef]
- Partridge, S.R. Resistance mechanisms in Enterobacteriaceae. Pathology 2015, 47, 276–284. [Google Scholar] [CrossRef]
- Ahuatzin-Flores, O.E.; Torres, E.; Chávez-Bravo, E. Acinetobacter baumannii, a Multidrug-Resistant Opportunistic Pathogen in New Habitats: A Systematic Review. Microorganisms 2024, 12, 644. [Google Scholar] [CrossRef] [PubMed]
- Doughari, H.J.; Ndakidemi, P.A.; Human, I.S.; Benade, S. The Ecology, Biology and Pathogenesis of Acinetobacter spp.: An Overview. Microbes Environ. 2011, 26, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Corcione, S.; Longo, B.M.; Scabini, S.; Pivetta, E.; Curtoni, A.; Shbaklo, N.; Costa, C.; De Rosa, F.G. Risk factors for mortality in Acinetobacter baumannii bloodstream infections and development of a predictive mortality model. J. Glob. Antimicrob. Resist. 2024, 38, 317–326. [Google Scholar] [CrossRef]
- Bravo, Z.; Orruño, M.; Navascues, T.; Ogayar, E.; Ramos-Vivas, J.; Kaberdin, V.R.; Arana, I. Analysis of Acinetobacter baumannii survival in liquid media and on solid matrices as well as effect of disinfectants. J. Hosp. Infect. 2019, 103, e42–e52. [Google Scholar] [CrossRef]
- Chapartegui-González, I.; Lázaro-Díez, M.; Bravo, Z.; Navas, J.; Icardo, J.M.; Ramos-Vivas, J. Acinetobacter baumannii maintains its virulence after long-time starvation. PLoS ONE 2018, 13, e0201961. [Google Scholar] [CrossRef]
- Geisinger, E.; Mortman, N.J.; Vargas-Cuebas, G.; Tai, A.K.; Isberg, R.R. A global regulatory system links virulence and antibiotic resistance to envelope homeostasis in Acinetobacter baumannii. PLOS Pathog. 2018, 14, e1007030. [Google Scholar] [CrossRef]
- Pompilio, A.; Scribano, D.; Sarshar, M.; Di Bonaventura, G.; Palamara, A.T.; Ambrosi, C. Gram-Negative Bacteria Holding Together in a Biofilm: The Acinetobacter baumannii Way. Microorganisms 2021, 9, 1353. [Google Scholar] [CrossRef]
- Smitran, A.; Lukovic, B.; Bozic, L.J.; Jelic, D.; Jovicevic, M.; Kabic, J.; Kekic, D.; Ranin, J.; Opavski, N.; Gajic, I. Carbapenem-Resistant Acinetobacter baumannii: Biofilm-Associated Genes, Biofilm-Eradication Potential of Disinfectants, and Biofilm-Inhibitory Effects of Selenium Nanoparticles. Microorganisms 2023, 11, 171. [Google Scholar] [CrossRef]
- Castanheira, M.; Mendes, R.E.; Gales, A.C. Global Epidemiology and Mechanisms of Resistance of Acinetobacter baumannii-calcoaceticus Complex. Clin. Infect. Dis. 2023, 76, S166–S178. [Google Scholar] [CrossRef]
- Thacharodi, A.; Vithlani, A.; Hassan, S.; Alqahtani, A.; Pugazhendhi, A. Carbapenem-resistant Acinetobacter baumannii raises global alarm for new antibiotic regimens. iScience 2024, 27, 111367. [Google Scholar] [CrossRef]
- Mohamed, R.A.E.; Moustafa, N.M.; Mahmoud, F.M.; Elsaadawy, Y.S.; Aziz, H.S.A.; Gaber, S.A.B.; Hussin, A.M.; Seadawy, M.G. Whole-genome sequencing of two multidrug-resistant acinetobacter baumannii strains isolated from a neonatal intensive care unit in Egypt: A prospective cross-sectional study. BMC Microbiol. 2024, 24, 362. [Google Scholar] [CrossRef] [PubMed]
- Bonomo, R.A.; Szabo, D. Mechanisms of Multidrug Resistance in Acinetobacter Species and Pseudomonas aeruginosa. Clin. Infect. Dis. 2006, 43, S49–S56. [Google Scholar] [CrossRef]
- Kyriakidis, I.; Vasileiou, E.; Pana, Z.D.; Tragiannidis, A. Acinetobacter baumannii Antibiotic Resistance Mechanisms. Pathogens 2021, 10, 373. [Google Scholar] [CrossRef]
- Novović, K.; Jovčić, B. Colistin Resistance in Acinetobacter baumannii: Molecular Mechanisms and Epidemiology. Antibiotics 2023, 12, 516. [Google Scholar] [CrossRef]
- Moffatt, J.H.; Harper, M.; Harrison, P.; Hale, J.D.; Vinogradov, E.; Seemann, T.; Henry, R.; Crane, B.; St Michael, F.; Cox, A.D.; et al. Colistin Resistance in Acinetobacter baumannii Is Mediated by Complete Loss of Lipopolysaccharide Production. Antimicrob. Agents Chemother. 2010, 54, 4971–4977. [Google Scholar] [CrossRef]
- Shi, J.; Cheng, J.; Liu, S.; Zhu, Y.; Zhu, M. Acinetobacter baumannii: An evolving and cunning opponent. Front. Microbiol. 2024, 15, 1332108. [Google Scholar] [CrossRef]
- Hernández-Durán, M.; Colín-Castro, C.A.; Fernández-Rodríguez, D.; Delgado, G.; Morales-Espinosa, R.; Martínez-Zavaleta, M.G.; Shekhar, C.; Ortíz-Álvarez, J.; García-Contreras, R.; Franco-Cendejas, R.; et al. Inside-out, antimicrobial resistance mediated by efflux pumps in clinical strains of Acinetobacter baumannii isolated from burn wound infections. Braz. J. Microbiol. 2024, 55, 3629–3641. [Google Scholar] [CrossRef]
- Leu-Burke, G.; Edwards, E.; Jones, H. Zoonotic transmission risk of Acinetobacter baumannii from Alaskan wildlife. Am. J. Clin. Pathol. 2023, 160, S83. [Google Scholar] [CrossRef]
- Gheorghe-Barbu, I.; Dragomir, R.-I.; Gradisteanu Pircalabioru, G.; Surleac, M.; Dinu, I.A.; Gaboreanu, M.D.; Czobor Barbu, I. Tracing Acinetobacter baumannii’s Journey from Hospitals to Aquatic Ecosystems. Microorganisms 2024, 12, 1703. [Google Scholar] [CrossRef]
- Reynolds, D.; Kollef, M. The Epidemiology and Pathogenesis and Treatment of Pseudomonas aeruginosa Infections: An Update. Drugs 2021, 81, 2117–2131. [Google Scholar] [CrossRef]
- Moradali, M.F.; Ghods, S.; Rehm, B.H.A. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Xiao, W.; Zhou, C.; Pu, Q.; Deng, X.; Lan, L.; Liang, H.; Song, X.; Wu, M. Pseudomonas aeruginosa: Pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct. Target. Ther. 2022, 7, 199. [Google Scholar] [CrossRef] [PubMed]
- Georgescu, M.; Gheorghe, I.; Curutiu, C.; Lazar, V.; Bleotu, C.; Chifiriuc, M.-C. Virulence and resistance features of Pseudomonas aeruginosa strains isolated from chronic leg ulcers. BMC Infect. Dis. 2016, 16, 92. [Google Scholar] [CrossRef] [PubMed]
- Kamali, E.; Jamali, A.; Ardebili, A.; Ezadi, F.; Mohebbi, A. Evaluation of antimicrobial resistance, biofilm forming potential, and the presence of biofilm-related genes among clinical isolates of Pseudomonas aeruginosa. BMC Res. Notes 2020, 13, 27. [Google Scholar] [CrossRef]
- Sawa, T.; Shimizu, M.; Moriyama, K.; Wiener-Kronish, J.P. Association between Pseudomonas aeruginosa type III secretion, antibiotic resistance, and clinical outcome: A review. Crit. Care 2014, 18, 668. [Google Scholar] [CrossRef]
- Behzadi, P.; Gajdács, M.; Pallós, P.; Ónodi, B.; Stájer, A.; Matusovits, D.; Kárpáti, K.; Burián, K.; Battah, B.; Ferrari, M.; et al. Relationship between Biofilm-Formation, Phenotypic Virulence Factors and Antibiotic Resistance in Environmental Pseudomonas aeruginosa. Pathogens 2022, 11, 1015. [Google Scholar] [CrossRef]
- Saravanan, M.; Belete, M.A.; Arockiaraj, J. Carbapenem-resistant Pseudomonas aeruginosa in intensive care units increase mortality as an emerging global threat. Int. J. Surg. 2023, 109, 1034–1036. [Google Scholar] [CrossRef]
- Yoon, E.-J.; Jeong, S.H. Mobile Carbapenemase Genes in Pseudomonas aeruginosa. Front. Microbiol. 2021, 12, 614058. [Google Scholar] [CrossRef]
- Halat, D.H.; Moubareck, C.A. The intriguing carbapenemases of Pseudomonas aeruginosa: Current status, genetic profile, and global epidemiology. Yale J. Biol. Med 2022, 95, 507–515. [Google Scholar]
- Henriot, C.P.; Martak, D.; Genet, S.; Bornette, G.; Hocquet, D. Origin, fluxes, and reservoirs of Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa in aquatic ecosystems of a French floodplain. Sci. Total Environ. 2022, 834, 155353. [Google Scholar] [CrossRef]
- GC Rodrigues, J.; Nair, H.P.; O’Kane, C.; Walker, C.A. Prevalence of multidrug resistance in Pseudomonas spp. isolated from wild bird feces in an urban aquatic environment. Ecol. Evol. 2021, 11, 14303–14311. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Ghany, W.A. Pseudomonas aeruginosa infection of avian origin: Zoonosis and one health implications. Vet. World 2021, 14, 2155. [Google Scholar] [CrossRef] [PubMed]
- Devarajan, N.; Köhler, T.; Sivalingam, P.; van Delden, C.; Mulaji, C.K.; Mpiana, P.T.; Ibelings, B.W.; Poté, J. Antibiotic resistant Pseudomonas spp. in the aquatic environment: A prevalence study under tropical and temperate climate conditions. Water Res. 2017, 115, 256–265. [Google Scholar] [CrossRef]
- Krawczyk, B.; Wityk, P.; Gałęcka, M.; Michalik, M. The many faces of Enterococcus spp.—Commensal, probiotic and opportunistic pathogen. Microorganisms 2021, 9, 1900. [Google Scholar] [CrossRef]
- Bhardwaj, S.B. Enterococci: An important nosocomial pathogen. In Pathogenic Bacteria; IntechOpen: London, UK, 2019. [Google Scholar]
- Singhal, N.; Maurya, A.K.; Mohanty, S.; Kumar, M.; Virdi, J.S. Evaluation of Bile Salt Hydrolases, Cholesterol-Lowering Capabilities, and Probiotic Potential of Enterococcus faecium Isolated from Rhizosphere. Front. Microbiol. 2019, 10, 1567. [Google Scholar] [CrossRef]
- Bin-Asif, H.; Ali, S.A. The genus Enterococcus and its associated virulent factors. In Microorganisms; IntechOpen: London, UK, 2019; pp. 109–130. [Google Scholar]
- Li, P.; Yin, R.; Cheng, J.; Lin, J. Bacterial biofilm formation on biomaterials and approaches to its treatment and prevention. Int. J. Mol. Sci. 2023, 24, 11680. [Google Scholar] [CrossRef]
- Brinkwirth, S.; Ayobami, O.; Eckmanns, T.; Markwart, R. Hospital-acquired infections caused by enterococci: A systematic review and meta-analysis, WHO European Region, 1 January 2010 to 4 February 2020. Eurosurveillance 2021, 26, 2001628. [Google Scholar] [CrossRef]
- Gao, W.; Howden, B.P.; Stinear, T.P. Evolution of virulence in Enterococcus faecium, a hospital-adapted opportunistic pathogen. Curr. Opin. Microbiol. 2018, 41, 76–82. [Google Scholar] [CrossRef]
- Miller, W.R.; Murray, B.E.; Rice, L.B.; Arias, C.A. Vancomycin-resistant enterococci: Therapeutic challenges in the 21st century. Infect. Dis. Clin. 2016, 30, 415–439. [Google Scholar]
- Stogios, P.J.; Savchenko, A. Molecular mechanisms of vancomycin resistance. Protein Sci. 2020, 29, 654–669. [Google Scholar] [CrossRef]
- Mikalsen, T.; Pedersen, T.; Willems, R.; Coque, T.M.; Werner, G.; Sadowy, E.; van Schaik, W.; Jensen, L.B.; Sundsfjord, A.; Hegstad, K. Investigating the mobilome in clinically important lineages of Enterococcus faecium and Enterococcus faecalis. BMC Genom. 2015, 16, 282. [Google Scholar]
- Cho, S.; Jackson, C.R.; Frye, J.G. The prevalence and antimicrobial resistance phenotypes of Salmonella, Escherichia coli and Enterococcus sp. in surface water. Lett. Appl. Microbiol. 2020, 71, 3–25. [Google Scholar] [CrossRef] [PubMed]
- Nowakiewicz, A.; Zięba, P.; Gnat, S.; Trościańczyk, A.; Osińska, M.; Łagowski, D.; Kosior-Korzecka, U.; Puzio, I. A significant number of multi-drug resistant Enterococcus faecalis in wildlife animals; long-term consequences and new or known reservoirs of resistance? Sci. Total Environ. 2020, 705, 135830. [Google Scholar] [CrossRef] [PubMed]
- Nüesch-Inderbinen, M.; Heyvaert, L.; Treier, A.; Zurfluh, K.; Cernela, N.; Biggel, M.; Stephan, R. High occurrence of Enterococcus faecalis, Enterococcus faecium, and Vagococcus lutrae harbouring oxazolidinone resistance genes in raw meat-based diets for companion animals–a public health issue, Switzerland, September 2018 to May 2020. Eurosurveillance 2023, 28, 2200496. [Google Scholar] [CrossRef]
- Amuasi, G.R.; Dsani, E.; Owusu-Nyantakyi, C.; Owusu, F.A.; Mohktar, Q.; Nilsson, P.; Adu, B.; Hendriksen, R.S.; Egyir, B. Enterococcus species: Insights into antimicrobial resistance and whole-genome features of isolates recovered from livestock and raw meat in Ghana. Front. Microbiol. 2023, 14, 1254896. [Google Scholar] [CrossRef]
- Larsson, D.G.; Flach, C.-F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Soulsbury, C.D.; White, P.C.L. Human–wildlife interactions in urban areas: A review of conflicts, benefits and opportunities. Wildl. Res. 2015, 42, 541–553. [Google Scholar] [CrossRef]
- Khan, A.H.; Aziz, H.A.; Khan, N.A.; Hasan, M.A.; Ahmed, S.; Farooqi, I.H.; Dhingra, A.; Vambol, V.; Changani, F.; Yousefi, M.; et al. Impact, disease outbreak and the eco-hazards associated with pharmaceutical residues: A Critical review. Int. J. Environ. Sci. Technol. 2022, 19, 677–688. [Google Scholar] [CrossRef]
- Bonardi, S.; Pitino, R. Carbapenemase-producing bacteria in food-producing animals, wildlife and environment: A challenge for human health. Ital. J. Food Saf. 2019, 8, 77–92. [Google Scholar] [CrossRef]
- Cao, J.; Hu, Y.; Liu, F.; Wang, Y.; Bi, Y.; Lv, N.; Li, J.; Zhu, B.; Gao, G.F. Metagenomic analysis reveals the microbiome and resistome in migratory birds. Microbiome 2020, 8, 26. [Google Scholar] [CrossRef]
- Ahmed, N.A.; Gulhan, T. Determination of antibiotic resistance patterns and genotypes of Escherichia coli isolated from wild birds. Microbiome 2024, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Abdullahi, I.N.; Fernández-Fernández, R.; Juárez-Fernández, G.; Martínez-Álvarez, S.; Eguizábal, P.; Zarazaga, M.; Lozano, C.; Torres, C. Wild animals are reservoirs and sentinels of Staphylococcus aureus and MRSA clones: A problem with “One Health” concern. Antibiotics 2021, 10, 1556. [Google Scholar] [CrossRef] [PubMed]
- Sousa, M.; Silva, V.; Silva, A.; Silva, N.; Ribeiro, J.; Tejedor-Junco, M.T.; Capita, R.; Chenouf, N.S.; Alonso-Calleja, C.; Rodrigues, T.M.; et al. Staphylococci among Wild European Rabbits from the Azores: A Potential Zoonotic Issue? J. Food Prot. 2020, 83, 1110–1114. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Grúa, E.; Pérez-Fuentes, S.; Viana, D.; Cardells, J.; Lizana, V.; Aguiló, J.; Selva, L.; Corpa, J.M. Marked presence of methicillin-resistant staphylococcus aureus in wild lagomorphs in Valencia, Spain. Animals 2020, 10, 1109. [Google Scholar] [CrossRef]
- Loncaric, I.; Kübber-Heiss, A.; Posautz, A.; Stalder, G.L.; Hoffmann, D.; Rosengarten, R.; Walzer, C. Characterization of methicillin-resistant Staphylococcus spp. carrying the mecC gene, isolated from wildlife. J. Antimicrob. Chemother. 2013, 68, 2222–2225. [Google Scholar] [CrossRef]
- Silva, V.; Pereira, J.E.; Maltez, L.; Ferreira, E.; Manageiro, V.; Caniça, M.; Capelo, J.L.; Igrejas, G.; Poeta, P. Diversity of methicillin-resistant staphylococci among wild Lepus granatensis: First detection of mecA-MRSA in hares. FEMS Microbiol. Ecol. 2019, 96, fiz204. [Google Scholar] [CrossRef]
- Gómez, P.; Lozano, C.; Camacho, M.C.; Lima-Barbero, J.-F.; Hernández, J.-M.; Zarazaga, M.; Höfle, Ú.; Torres, C. Detection of MRSA ST3061-t843-mecC and ST398-t011-mecA in white stork nestlings exposed to human residues. J. Antimicrob. Chemother. 2016, 71, 53–57. [Google Scholar] [CrossRef]
- Gambino, D.; Vicari, D.; Vitale, M.; Schirò, G.; Mira, F.; Giglia, M.L.; Riccardi, A.; Gentile, A.; Giardina, S.; Carrozzo, A.; et al. Study on Bacteria Isolates and Antimicrobial Resistance in Wildlife in Sicily, Southern Italy. Microorganisms 2021, 9, 203. [Google Scholar] [CrossRef]
- Kutkowska, J.; Turska-Szewczuk, A.; Kucharczyk, M.; Kucharczyk, H.; Zalewska, J.; Urbanik-Sypniewska, T. Methicillin-resistant Staphylococcus aureus and glycopeptide-resistant enterococci in fecal samples of birds from South-Eastern Poland. BMC Vet. Res. 2019, 15, 472. [Google Scholar] [CrossRef]
- Sánchez-Ortiz, E.; Blanco Gutiérrez, M.d.M.; Calvo-Fernandez, C.; Mencía-Gutiérrez, A.; Pastor Tiburón, N.; Alvarado Piqueras, A.; Pablos-Tanarro, A.; Martín-Maldonado, B. Addressing Challenges in Wildlife Rehabilitation: Antimicrobial-Resistant Bacteria from Wounds and Fractures in Wild Birds. Animals 2024, 14, 1151. [Google Scholar] [CrossRef]
- Raafat, D.; Mrochen, D.M.; Al’Sholui, F.; Heuser, E.; Ryll, R.; Pritchett-Corning, K.R.; Jacob, J.; Walther, B.; Matuschka, F.-R.; Richter, D.; et al. Molecular Epidemiology of Methicillin-Susceptible and Methicillin-Resistant Staphylococcus aureus in Wild, Captive and Laboratory Rats: Effect of Habitat on the Nasal S. aureus Population. Toxins 2020, 12, 80. [Google Scholar] [CrossRef] [PubMed]
- Silva, V.; Lopes, A.F.; Soeiro, V.; Caniça, M.; Manageiro, V.; Pereira, J.E.; Maltez, L.; Capelo, J.L.; Igrejas, G.; Poeta, P. Nocturnal Birds of Prey as Carriers of Staphylococcus aureus and Other Staphylococci: Diversity, Antimicrobial Resistance and Clonal Lineages. Antibiotics 2022, 11, 240. [Google Scholar] [CrossRef] [PubMed]
- Feßler, A.T.; Thomas, P.; Mühldorfer, K.; Grobbel, M.; Brombach, J.; Eichhorn, I.; Monecke, S.; Ehricht, R.; Schwarz, S. Phenotypic and genotypic characteristics of Staphylococcus aureus isolates from zoo and wild animals. Vet. Microbiol. 2018, 218, 98–103. [Google Scholar] [CrossRef]
- Nowakiewicz, A.; Ziółkowska, G.; Zięba, P.; Gnat, S.; Wojtanowicz-Markiewicz, K.; Trościańczyk, A. Coagulase-positive Staphylococcus isolated from wildlife: Identification, molecular characterization and evaluation of resistance profiles with focus on a methicillin-resistant strain. Comp. Immunol. Microbiol. Infect. Dis. 2016, 44, 21–28. [Google Scholar] [CrossRef]
- Loncaric, I.; Stalder, G.L.; Mehinagic, K.; Rosengarten, R.; Hoelzl, F.; Knauer, F.; Walzer, C. Comparison of ESBL—And AmpC Producing Enterobacteriaceae and Methicillin-Resistant Staphylococcus aureus (MRSA) Isolated from Migratory and Resident Population of Rooks (Corvus frugilegus) in Austria. PLoS ONE 2014, 8, e84048. [Google Scholar] [CrossRef]
- Ruiz-Ripa, L.; Gómez, P.; Alonso, C.A.; Camacho, M.C.; de la Puente, J.; Fernández-Fernández, R.; Ramiro, Y.; Quevedo, M.A.; Blanco, J.M.; Zarazaga, M.; et al. Detection of MRSA of Lineages CC130-mecC and CC398-mecA and Staphylococcus delphini-lnu(A) in Magpies and Cinereous Vultures in Spain. Microb. Ecol. 2019, 78, 409–415. [Google Scholar] [CrossRef]
- Sousa, M.; Silva, N.; Igrejas, G.; Silva, F.; Sargo, R.; Alegria, N.; Benito, D.; Gómez, P.; Lozano, C.; Gómez-Sanz, E.; et al. Antimicrobial resistance determinants in Staphylococcus spp. recovered from birds of prey in Portugal. Vet. Microbiol. 2014, 171, 436–440. [Google Scholar] [CrossRef]
- Vidal, A.; Baldomà, L.; Molina-López, R.A.; Martin, M.; Darwich, L. Microbiological diagnosis and antimicrobial sensitivity profiles in diseased free-living raptors. Avian Pathol. 2017, 46, 442–450. [Google Scholar] [CrossRef]
- Porrero, M.C.; Mentaberre, G.; Sánchez, S.; Fernández-Llario, P.; Casas-Díaz, E.; Mateos, A.; Vidal, D.; Lavín, S.; Fernández-Garayzábal, J.-F.; Domínguez, L. Carriage of Staphylococcus aureus by Free-Living Wild Animals in Spain. Appl. Environ. Microbiol. 2014, 80, 4865–4870. [Google Scholar] [CrossRef]
- Pall, E.; Spînu, M.; ȘANDRU, C.D.; Duca, G.; Suătean, M.I.; Szafta, A.-A.; Diana, O.; Vasiu, A. A comparison of antibiotic resistance and multiple antibiotic resistance index in wild boars from covasna and Cluj counties. Sci. Work. Ser. C Vet. Med. 2022, 68, 94–99. [Google Scholar]
- Silva, V.; Gabriel, S.I.; Borrego, S.B.; Tejedor-Junco, M.T.; Manageiro, V.; Ferreira, E.; Reis, L.; Caniça, M.; Capelo, J.L.; Igrejas, G.; et al. Antimicrobial Resistance and Genetic Lineages of Staphylococcus aureus from Wild Rodents: First Report of mecC-Positive Methicillin-Resistant S. aureus (MRSA) in Portugal. Animals 2021, 11, 1537. [Google Scholar] [CrossRef] [PubMed]
- Mrochen, D.M.; Schulz, D.; Fischer, S.; Jeske, K.; El Gohary, H.; Reil, D.; Imholt, C.; Trübe, P.; Suchomel, J.; Tricaud, E.; et al. Wild rodents and shrews are natural hosts of Staphylococcus aureus. Int. J. Med. Microbiol. 2018, 308, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Desvars-Larrive, A.; Ruppitsch, W.; Lepuschitz, S.; Szostak, M.P.; Spergser, J.; Feßler, A.T.; Schwarz, S.; Monecke, S.; Ehricht, R.; Walzer, C.; et al. Urban brown rats (Rattus norvegicus) as possible source of multidrug-resistant Enterobacteriaceae and meticillin-resistant Staphylococcus spp., Vienna, Austria, 2016 and 2017. Euro Surveill. 2019, 24, 1900149. [Google Scholar] [CrossRef]
- Bengtsson, B.; Persson, L.; Ekström, K.; Unnerstad, H.E.; Uhlhorn, H.; Börjesson, S. High occurrence of mecC-MRSA in wild hedgehogs (Erinaceus europaeus) in Sweden. Vet. Microbiol. 2017, 207, 103–107. [Google Scholar] [CrossRef]
- Ruiz-Ripa, L.; Alcalá, L.; Simón, C.; Gómez, P.; Mama, O.M.; Rezusta, A.; Zarazaga, M.; Torres, C. Diversity of Staphylococcus aureus clones in wild mammals in Aragon, Spain, with detection of MRSA ST130-mecC in wild rabbits. J. Appl. Microbiol. 2019, 127, 284–291. [Google Scholar] [CrossRef]
- Gómez, P.; González-Barrio, D.; Benito, D.; García, J.T.; Viñuela, J.; Zarazaga, M.; Ruiz-Fons, F.; Torres, C. Detection of methicillin-resistant Staphylococcus aureus (MRSA) carrying the mecC gene in wild small mammals in Spain. J. Antimicrob. Chemother. 2014, 69, 2061–2064. [Google Scholar] [CrossRef]
- Petersen, A.; Stegger, M.; Heltberg, O.; Christensen, J.; Zeuthen, A.; Knudsen, L.K.; Urth, T.; Sorum, M.; Schouls, L.; Larsen, J.; et al. Epidemiology of methicillin-resistant Staphylococcus aureus carrying the novel mecC gene in Denmark corroborates a zoonotic reservoir with transmission to humans. Clin. Microbiol. Infect. 2013, 19, E16–E22. [Google Scholar] [CrossRef]
- Lindgren, A.-K.; Gustafsson, E.; Petersson, A.C.; Melander, E. Methicillin-resistant Staphylococcus aureus with mecC: A description of 45 human cases in southern Sweden. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 971–975. [Google Scholar] [CrossRef]
- Hussain, S.; Zeshan, B.; Arshad, R.; Kabir, S.; Ahmed, N. MRSA Clinical Isolates Harboring mecC Gene Imply Zoonotic Transmission to Humans and Colonization by Biofilm Formation. Pak. J. Zool. 2022, 55, 999–1002. [Google Scholar] [CrossRef]
- Athanasakopoulou, Z.; Diezel, C.; Braun, S.D.; Sofia, M.; Giannakopoulos, A.; Monecke, S.; Gary, D.; Krähmer, D.; Chatzopoulos, D.C.; Touloudi, A. Occurrence and characteristics of ESBL-and carbapenemase-producing Escherichia coli from wild and feral birds in Greece. Microorganisms 2022, 10, 1217. [Google Scholar] [CrossRef]
- Nowaczek, A.; Dec, M.; Stępień-Pyśniak, D.; Urban-Chmiel, R.; Marek, A.; Różański, P. Antibiotic resistance and virulence profiles of Escherichia coli strains isolated from wild birds in Poland. Pathogens 2021, 10, 1059. [Google Scholar] [CrossRef] [PubMed]
- Blasi, M.F.; Migliore, L.; Mattei, D.; Rotini, A.; Thaller, M.C.; Alduina, R. Antibiotic resistance of gram-negative bacteria from wild captured loggerhead sea turtles. Antibiotics 2020, 9, 162. [Google Scholar] [CrossRef] [PubMed]
- Ilieva, Y.; Zaharieva, M.M.; Dimitrova, L.; Kaleva, M.D.; Jordanova, J.; Dimitrova, M.; Beltcheva, M.; Aleksieva, I.; Georgiev, Y.; Manasiev, Y. Preliminary Data on Escherichia coli, Yersinia enterocolitica, and Other Bacteria, as Well as Absent African Swine Fever Virus in the Gut Microbiota of Wild Mice and Voles from Bulgaria. Microbiol. Res. 2023, 14, 1788–1819. [Google Scholar] [CrossRef]
- Foti, M.; Siclari, A.; Mascetti, A.; Fisichella, V. Study of the spread of antimicrobial-resistant Enterobacteriaceae from wild mammals in the National Park of Aspromonte (Calabria, Italy). Environ. Toxicol. Pharmacol. 2018, 63, 69–73. [Google Scholar] [CrossRef]
- Panwar, K.; Venu, G.; Satpathy, M.M.; Tanwar, A.; Dutta, S. Comparative evaluation of prevalence of extended spectrum beta lactamase genes in Enterobacteriaceae isolates from wild-life. Int. J. Veter. Sci. Anim. Husb. 2024, 9, 41–44. [Google Scholar] [CrossRef]
- Chiaverini, A.; Cornacchia, A.; Centorotola, G.; Tieri, E.E.; Sulli, N.; Del Matto, I.; Iannitto, G.; Petrone, D.; Petrini, A.; Pomilio, F. Phenotypic and genetic characterization of Klebsiella pneumoniae isolates from wild animals in Central Italy. Animals 2022, 12, 1347. [Google Scholar] [CrossRef]
- Dias, C.; Borges, A.; Oliveira, D.; Martinez-Murcia, A.; Saavedra, M.J.; Simões, M. Biofilms and antibiotic susceptibility of multidrug-resistant bacteria from wild animals. PeerJ 2018, 6, e4974. [Google Scholar] [CrossRef]
- Brendecke, J.; Homeier-Bachmann, T.; Schmitz Ornés, A.; Guenther, S.; Heiden, S.E.; Schwabe, M.; Eger, E.; Schaufler, K. Multidrug-resistant high-risk Escherichia coli and Klebsiella pneumoniae clonal lineages occur in black-headed gulls from two conservation islands in Germany. Antibiotics 2022, 11, 1357. [Google Scholar] [CrossRef]
- Vittecoq, M.; Laurens, C.; Brazier, L.; Durand, P.; Elguero, E.; Arnal, A.; Thomas, F.; Aberkane, S.; Renaud, N.; Prugnolle, F. VIM-1 carbapenemase-producing Escherichia coli in gulls from southern France. Ecol. Evol. 2017, 7, 1224–1232. [Google Scholar] [CrossRef]
- Smoglica, C.; Vergara, A.; Angelucci, S.; Festino, A.R.; Antonucci, A.; Marsilio, F.; Di Francesco, C.E. Antibiotic-resistant bacteria dissemination in the wildlife, livestock, and water of Maiella National Park, Italy. Animals 2023, 13, 432. [Google Scholar] [CrossRef]
- Musa, L.; Stefanetti, V.; Casagrande Proietti, P.; Grilli, G.; Gobbi, M.; Toppi, V.; Brustenga, L.; Magistrali, C.F.; Franciosini, M.P. Antimicrobial Susceptibility of Commensal E. coli Isolated from Wild Birds in Umbria (Central Italy). Animals 2023, 13, 1776. [Google Scholar] [CrossRef] [PubMed]
- Prandi, I.; Bellato, A.; Nebbia, P.; Stella, M.C.; Ala, U.; von Degerfeld, M.M.; Quaranta, G.; Robino, P. Antibiotic resistant Escherichia coli in wild birds hospitalised in a wildlife rescue centre. Comp. Immunol. Microbiol. Infect. Dis. 2023, 93, 101945. [Google Scholar] [CrossRef] [PubMed]
- Ahlstrom, C.A.; Woksepp, H.; Sandegren, L.; Mohsin, M.; Hasan, B.; Muzyka, D.; Hernandez, J.; Aguirre, F.; Tok, A.; Söderman, J. Genomically diverse carbapenem resistant Enterobacteriaceae from wild birds provide insight into global patterns of spatiotemporal dissemination. Sci. Total Environ. 2022, 824, 153632. [Google Scholar] [CrossRef]
- Mahmoud, E.; El-Kholi, S.A.A.M.; Rady, M.A.; El-Tarabili, R.M.; Hashem, M.A.; Elfeil, W.M.K. Molecular Characterization of Virulence Genes among MDR and XDR Avian Pathogenic E. coli. J. Adv. Vet. Res. 2023, 13, 2014–2018. [Google Scholar]
- Farzin, H.R.; Ghaniei, A.; Jamshidian Mojaver, M.; Amir, M. Investigation of the prevalence of virulence and antibiotic resistance genes in Escherichia coli isolated from ostriches and human urinary tract infections. Vet. Res. Biol. Prod. 2022, 35, 92–100. [Google Scholar]
- Zhang, X.-F.; Doi, Y.; Huang, X.; Li, H.-Y.; Zhong, L.-L.; Zeng, K.-J.; Zhang, Y.-F.; Patil, S.; Tian, G.-B. Possible transmission of mcr-1–harboring Escherichia coli between companion animals and human. Emerg. Infect. Dis. 2016, 22, 1679. [Google Scholar] [CrossRef]
- Schaufler, K.; Semmler, T.; Wieler, L.H.; Wöhrmann, M.; Baddam, R.; Ahmed, N.; Müller, K.; Kola, A.; Fruth, A.; Ewers, C.; et al. Clonal spread and interspecies transmission of clinically relevant ESBL-producing Escherichia coli of ST410—Another successful pandemic clone? FEMS Microbiol. Ecol. 2016, 92, fiv155. [Google Scholar] [CrossRef]
- Day, M.J.; Hopkins, K.L.; Wareham, D.W.; Toleman, M.A.; Elviss, N.; Randall, L.; Teale, C.; Cleary, P.; Wiuff, C.; Doumith, M.; et al. Extended-spectrum β-lactamase-producing Escherichia coli in human-derived and foodchain-derived samples from England, Wales, and Scotland: An epidemiological surveillance and typing study. Lancet Infect. Dis. 2019, 19, 1325–1335. [Google Scholar] [CrossRef]
- Dahms, C.; Hübner, N.-O.; Kossow, A.; Mellmann, A.; Dittmann, K.; Kramer, A. Occurrence of ESBL-producing Escherichia coli in livestock and farm workers in Mecklenburg-Western Pomerania, Germany. PLoS ONE 2015, 10, e0143326. [Google Scholar] [CrossRef]
- Hernández-González, I.L.; Castillo-Ramírez, S. Antibiotic-resistant Acinetobacter baumannii is a One Health problem. Lancet Microbe 2020, 1, e279. [Google Scholar] [CrossRef]
- Wilharm, G.; Skiebe, E.; Higgins, P.G.; Poppel, M.T.; Blaschke, U.; Leser, S.; Heider, C.; Heindorf, M.; Brauner, P.; Jäckel, U. Relatedness of wildlife and livestock avian isolates of the nosocomial pathogen Acinetobacter baumannii to lineages spread in hospitals worldwide. Environ. Microbiol. 2017, 19, 4349–4364. [Google Scholar] [CrossRef] [PubMed]
- Unger, F.; Eisenberg, T.; Prenger-Berninghoff, E.; Leidner, U.; Semmler, T.; Ewers, C. Imported pet reptiles and their “Blind passengers”—In-depth characterization of 80 Acinetobacter species isolates. Microorganisms 2022, 10, 893. [Google Scholar] [CrossRef]
- Teerawattanapong, N.; Panich, P.; Kulpokin, D.; Ranong, S.N.; Kongpakwattana, K.; Saksinanon, A.; Goh, B.-H.; Lee, L.-H.; Apisarnthanarak, A.; Chaiyakunapruk, N. A systematic review of the burden of multidrug-resistant healthcare-associated infections among intensive care unit patients in Southeast Asia: The rise of multidrug-resistant Acinetobacter baumannii. Infect. Control Hosp. Epidemiol. 2018, 39, 525–533. [Google Scholar] [CrossRef]
- Lv, Y.; Xiang, Q.; Jin, Y.Z.; Fang, Y.; Wu, Y.J.; Zeng, B.; Yu, H.; Cai, H.M.; Wei, Q.D.; Wang, C. Faucet aerators as a reservoir for Carbapenem-resistant Acinetobacter baumannii: A healthcare-associated infection outbreak in a neurosurgical intensive care unit. Antimicrob. Resist. Infect. Control 2019, 8, 205. [Google Scholar] [CrossRef]
- Camargo, C.H.; Cunha, M.P.V.; de Barcellos, T.A.F.; Bueno, M.S.; Bertani, A.M.d.J.; dos Santos, C.A.; Nagamori, F.O.; Takagi, E.H.; Chimara, E.; de Carvalho, E.; et al. Genomic and phenotypic characterisation of antimicrobial resistance in carbapenem-resistant Acinetobacter baumannii hyperendemic clones CC1, CC15, CC79 and CC25. Int. J. Antimicrob. Agents 2020, 56, 106195. [Google Scholar] [CrossRef]
- Savin, M.; Parcina, M.; Schmoger, S.; Kreyenschmidt, J.; Käsbohrer, A.; Hammerl, J.A. Draft genome sequences of Acinetobacter baumannii isolates recovered from sewage water from a poultry slaughterhouse in Germany. Microbiol. Resour. Announc. 2019, 8, 10–1128. [Google Scholar] [CrossRef]
- Verkaik, N.J.; Wielders, C.C.H.; den Boer, H.; Langerak, D.; Vogel, M.; Witteveen, S.; de Haan, A.; Bos, J.; van Westreenen, M.; Notermans, D.W. Antimicrobial susceptibility to last-resort antibiotics in carbapenemase-producing bacteria from Ukrainian patients. Microbiol. Spectr. 2024, 12, e01142-24. [Google Scholar] [CrossRef]
- Terreni, M.; Taccani, M.; Pregnolato, M. New antibiotics for multidrug-resistant bacterial strains: Latest research developments and future perspectives. Molecules 2021, 26, 2671. [Google Scholar] [CrossRef]
- Pailhoriès, H.; Belmonte, O.; Kempf, M.; Lemarié, C.; Cuziat, J.; Quinqueneau, C.; Ramont, C.; Joly-Guillou, M.-L.; Eveillard, M. Diversity of Acinetobacter baumannii strains isolated in humans, companion animals, and the environment in Reunion Island: An exploratory study. Int. J. Infect. Dis. 2015, 37, 64–69. [Google Scholar] [CrossRef]
- Eveillard, M.; Kempf, M.; Belmonte, O.; Pailhoriès, H.; Joly-Guillou, M.-L. Reservoirs of Acinetobacter baumannii outside the hospital and potential involvement in emerging human community-acquired infections. Int. J. Infect. Dis. 2013, 17, e802–e805. [Google Scholar] [CrossRef]
- Castillo-Ramírez, S. Zoonotic Acinetobacter baumannii: The need for genomic epidemiology in a One Health context. Lancet Microbe 2022, 3, e895–e896. [Google Scholar] [CrossRef] [PubMed]
- Linz, B.; Mukhtar, N.; Shabbir, M.Z.; Rivera, I.; Ivanov, Y.V.; Tahir, Z.; Yaqub, T.; Harvill, E.T. Virulent epidemic pneumonia in sheep caused by the human pathogen Acinetobacter baumannii. Front. Microbiol. 2018, 9, 2616. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Roldán, L.; Rojo-Bezares, B.; de Toro, M.; López, M.; Toledano, P.; Lozano, C.; Chichón, G.; Alvarez-Erviti, L.; Torres, C.; Sáenz, Y. Antimicrobial resistance and virulence of Pseudomonas spp. among healthy animals: Concern about exolysin ExlA detection. Sci. Rep. 2020, 10, 11667. [Google Scholar] [CrossRef] [PubMed]
- Torres, R.T.; Cunha, M.V.; Ferreira, H.; Fonseca, C.; Palmeira, J.D. A high-risk carbapenem-resistant Pseudomonas aeruginosa clone detected in red deer (Cervus elaphus) from Portugal. Sci. Total Environ. 2022, 829, 154699. [Google Scholar] [CrossRef]
- Laborda, P.; Sanz-García, F.; Hernando-Amado, S.; Martínez, J.L. Pseudomonas aeruginosa: An antibiotic resilient pathogen with environmental origin. Curr. Opin. Microbiol. 2021, 64, 125–132. [Google Scholar] [CrossRef]
- Jurado-Martín, I.; Sainz-Mejías, M.; McClean, S. Pseudomonas aeruginosa: An audacious pathogen with an adaptable arsenal of virulence factors. Int. J. Mol. Sci. 2021, 22, 3128. [Google Scholar] [CrossRef]
- Fernandes, M.R.; Sellera, F.P.; Moura, Q.; Carvalho, M.P.N.; Rosato, P.N.; Cerdeira, L.; Lincopan, N. Zooanthroponotic transmission of drug-resistant Pseudomonas aeruginosa, Brazil. Emerg. Infect. Dis. 2018, 24, 1160. [Google Scholar] [CrossRef]
- Gharieb, R.; Saad, M.; Khedr, M.; El Gohary, A.; Ibrahim, H. Occurrence, virulence, carbapenem resistance, susceptibility to disinfectants and public health hazard of Pseudomonas aeruginosa isolated from animals, humans and environment in intensive farms. J. Appl. Microbiol. 2022, 132, 256–267. [Google Scholar] [CrossRef]
- Abdullahi, I.N.; Mejri, S.; Okwume, C.C.; Lawal, N.A.; Olusegun, O.A.; Sallem, R.B.; Slama, K.B. Global epidemiology of high priority and pandemic Pseudomonas aeruginosa in pets, livestock, wild, and aquatic animals: A systematic review and meta-analysis. Lett. Appl. Microbiol. 2025, 78, ovaf028. [Google Scholar] [CrossRef]
- Chichón, G.; López, M.; de Toro, M.; Ruiz-Roldán, L.; Rojo-Bezares, B.; Sáenz, Y. Spread of Pseudomonas aeruginosa ST274 Clone in Different Niches: Resistome, Virulome, and Phylogenetic Relationship. Antibiotics 2023, 12, 1561. [Google Scholar] [CrossRef]
- López, M.; Rojo-Bezares, B.; Chichón, G.; Sáenz, Y. Resistance to Fluoroquinolones in Pseudomonas aeruginosa from Human, Animal, Food and Environmental Origin: The Role of CrpP and Mobilizable ICEs. Antibiotics 2022, 11, 1271. [Google Scholar] [CrossRef] [PubMed]
- Januário, A.P.; Afonso, C.N.; Mendes, S.; Rodrigues, M.J. Faecal indicator bacteria and Pseudomonas aeruginosa in marine coastal waters: Is there a relationship? Pathogens 2019, 9, 13. [Google Scholar] [CrossRef] [PubMed]
- Larcher, R.; Laffont-Lozes, P.; Roger, C.; Doncesco, R.; Groul-Viaud, C.; Martin, A.; Loubet, P.; Lavigne, J.-P.; Pantel, A.; Sotto, A. Last resort beta-lactam antibiotics for treatment of New-Delhi Metallo-Beta-Lactamase producing Enterobacterales and other Difficult-to-Treat Resistance in Gram-negative bacteria: A real-life study. Front. Cell. Infect. Microbiol. 2022, 12, 1048633. [Google Scholar] [CrossRef]
- Rossolini, G.M.; Mantengoli, E. Treatment and control of severe infections caused by multiresistant Pseudomonas aeruginosa. Clin. Microbiol. Infect. 2005, 11, 17–32. [Google Scholar] [CrossRef]
- Kerr, K.G.; Snelling, A.M. Pseudomonas aeruginosa: A formidable and ever-present adversary. J. Hosp. Infect. 2009, 73, 338–344. [Google Scholar] [CrossRef]
- Fiore, E.; Van Tyne, D.; Gilmore, M.S. Pathogenicity of Enterococci. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Stępień-Pyśniak, D.; Hauschild, T.; Dec, M.; Marek, A.; Urban-Chmiel, R. Clonal structure and antibiotic resistance of Enterococcus spp. from wild birds in Poland. Microb. Drug Resist. 2019, 25, 1227–1237. [Google Scholar] [CrossRef]
- Stępień-Pyśniak, D.; Hauschild, T.; Kosikowska, U.; Dec, M.; Urban-Chmiel, R. Biofilm formation capacity and presence of virulence factors among commensal Enterococcus spp. from wild birds. Sci. Rep. 2019, 9, 11204. [Google Scholar] [CrossRef]
- Dec, M.; Stępień-Pyśniak, D.; Gnat, S.; Fratini, F.; Urban-Chmiel, R.; Cerri, D.; Winiarczyk, S.; Turchi, B. Antibiotic susceptibility and virulence genes in Enterococcus isolates from wild mammals living in Tuscany, Italy. Microb. Drug Resist. 2020, 26, 505–519. [Google Scholar] [CrossRef]
- García, L.A.; Torres, C.; López, A.R.; Rodríguez, C.O.; Valencia, C.S. Antimicrobial resistance of Enterococcus species isolated from wild mammals in Aragón, Spain. J. Vet. Res. 2022, 66, 151. [Google Scholar] [CrossRef]
- Terentjeva, M.; Ķibilds, J.; Avsejenko, J.; Cīrulis, A.; Labecka, L.; Bērziņš, A. Antimicrobial Resistance in Enterococcus spp. Isolates from Red Foxes (Vulpes vulpes) in Latvia. Antibiotics 2024, 13, 114. [Google Scholar] [CrossRef] [PubMed]
- Trościańczyk, A.; Nowakiewicz, A.; Gnat, S.; Łagowski, D.; Osińska, M.; Chudzik-Rząd, B. Comparative study of multidrug-resistant Enterococcus faecium obtained from different hosts. J. Med. Microbiol. 2021, 70, 1340. [Google Scholar] [CrossRef] [PubMed]
- Trościańczyk, A.; Nowakiewicz, A.; Osińska, M.; Łagowski, D.; Gnat, S.; Chudzik-Rząd, B. Comparative characteristics of sequence types, genotypes and virulence of multidrug-resistant Enterococcus faecium isolated from various hosts in eastern Poland. Spread of clonal complex 17 in humans and animals. Res. Microbiol. 2022, 173, 103925. [Google Scholar] [CrossRef]
- Silva, V.; Igrejas, G.; Carvalho, I.; Peixoto, F.; Cardoso, L.; Pereira, E.; Campo, R.; Poeta, P. Genetic Characterization of vanA-Enterococcus faecium Isolates from Wild Red-Legged Partridges in Portugal. Microb. Drug Resist. 2017, 24, 89–94. [Google Scholar] [CrossRef]
- Cagnoli, G.; Bertelloni, F.; Interrante, P.; Ceccherelli, R.; Marzoni, M.; Ebani, V.V. Antimicrobial-resistant Enterococcus spp. in wild avifauna from central Italy. Antibiotics 2022, 11, 852. [Google Scholar] [CrossRef]
- Hamarova, L.; Kopcakova, A.; Kocianova-Adamcova, M.; Piknova, M.; Javorsky, P.; Pristas, P. Antimicrobial Resistance of Enterococci from Wild Animals in Slovakia. Polish J. Environ. Stud. 2021, 30, 2085–2091. [Google Scholar] [CrossRef]
- Kwit, R.; Zając, M.; Śmiałowska-Węglińska, A.; Skarżyńska, M.; Bomba, A.; Lalak, A.; Skrzypiec, E.; Wojdat, D.; Koza, W.; Mikos-Wojewoda, E. Prevalence of Enterococcus spp. and the whole-genome characteristics of Enterococcus faecium and Enterococcus faecalis strains isolated from free-living birds in Poland. Pathogens 2023, 12, 836. [Google Scholar] [CrossRef]
- Oravcová, V.; Peixe, L.; Coque, T.M.; Novais, C.; Francia, M.V.; Literák, I.; Freitas, A.R. Wild corvid birds colonized with vancomycin-resistant Enterococcus faecium of human origin harbor epidemic vanA plasmids. Environ. Int. 2018, 118, 125–133. [Google Scholar] [CrossRef]
- García-Solache, M.; Rice, L.B. The Enterococcus: A Model of Adaptability to Its Environment. Clin. Microbiol. Rev. 2019, 32, e00058-18. [Google Scholar] [CrossRef]
- Wada, Y.; Harun, A.; Yean, C.Y.; Abdul-Rahman, Z. Global Prevalence of Vancomycin-resistant Enterococcus in Wildlife: The First Meta-Analysis and Systematic Review. Int. J. Infect. Dis. 2022, 116, S9–S10. [Google Scholar] [CrossRef]
- Ahmed, M.O.; Baptiste, K.E. Vancomycin-resistant enterococci: A review of antimicrobial resistance mechanisms and perspectives of human and animal health. Microb. Drug Resist. 2018, 24, 590–606. [Google Scholar] [CrossRef] [PubMed]
- Șchiopu, P.; Toc, D.A.; Colosi, I.A.; Costache, C.; Ruospo, G.; Berar, G.; Gălbău, Ș.-G.; Ghilea, A.C.; Botan, A.; Pană, A.-G.; et al. An overview of the factors involved in biofilm production by the enterococcus genus. Int. J. Mol. Sci. 2023, 24, 11577. [Google Scholar] [CrossRef] [PubMed]
- Too, E.; Masila, E. The Interconnection between Virulence Factors, Biofilm Formation, and Horizontal Gene Transfer in Enterococcus: A Review. In Enterococcus—Unveiling the Emergence of a Potent Pathogen; IntechOpen: London, UK, 2024. [Google Scholar]
- Rahimi, N.; Poursina, F.; sadat Ghaziasgar, F.; Sepehrpor, S.; Hassanzadeh, A. Presence of virulence factor genes (gelE and esp) and biofilm formation in clinical Enterococcus faecalis and Enterococcus faecium isolated from urinary tract infection in Isfahan, Iran. Gene Rep. 2018, 13, 72–75. [Google Scholar] [CrossRef]
- Johnson, C.N.; Sheriff, E.K.; Duerkop, B.A.; Chatterjee, A. Let me upgrade you: Impact of mobile genetic elements on enterococcal adaptation and evolution. J. Bacteriol. 2021, 203, 10–1128. [Google Scholar] [CrossRef]
- Tokuda, M.; Shintani, M. Microbial evolution through horizontal gene transfer by mobile genetic elements. Microb. Biotechnol. 2024, 17, e14408. [Google Scholar] [CrossRef]
- Conwell, M.; Dooley, J.S.G.; Naughton, P.J. Enterococcal biofilm—A nidus for antibiotic resistance transfer? J. Appl. Microbiol. 2022, 132, 3444–3460. [Google Scholar] [CrossRef]
- Jahan, M.; Zhanel, G.G.; Sparling, R.; Holley, R.A. Horizontal transfer of antibiotic resistance from Enterococcus faecium of fermented meat origin to clinical isolates of E. faecium and Enterococcus faecalis. Int. J. Food Microbiol. 2015, 199, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Guardabassi, L. Zoonotic Transmission of Antimicrobial-Resistant Enterococci: A Threat to Public Health or an Overemphasized Risk? In Zoonoses: Infections Affecting Humans and Animals; Springer: Berlin/Heidelberg, Germany, 2023; pp. 1–33. [Google Scholar]
- Abat, C.; Huart, M.; Garcia, V.; Dubourg, G.; Raoult, D. Enterococcus faecalis urinary-tract infections: Do they have a zoonotic origin? J. Infect. 2016, 73, 305–313. [Google Scholar] [CrossRef]
- Dolejska, M. Antibiotic-resistant bacteria in wildlife. In Antibiotic Resistance in the Environment: A Worldwide Overview; Springer: Berlin/Heidelberg, Germany, 2020; pp. 19–70. [Google Scholar]
- Singh, A.; Pratap, S.G.; Raj, A. Occurrence and dissemination of antibiotics and antibiotic resistance in aquatic environment and its ecological implications: A review. Environ. Sci. Pollut. Res. 2024, 31, 47505–47529. [Google Scholar] [CrossRef]
- Silva, V.; Ferreira, E.; Manageiro, V.; Reis, L.; Tejedor-Junco, M.T.; Sampaio, A.; Capelo, J.L.; Caniça, M.; Igrejas, G.; Poeta, P. Distribution and Clonal Diversity of Staphylococcus aureus and Other Staphylococci in Surface Waters: Detection of ST425-t742 and ST130-t843 mecC-Positive MRSA Strains. Antibiotics 2021, 10, 1416. [Google Scholar] [CrossRef]
- Rees, E.M.; Minter, A.; Edmunds, W.J.; Lau, C.L.; Kucharski, A.J.; Lowe, R. Transmission modelling of environmentally persistent zoonotic diseases: A systematic review. Lancet Planet. Health 2021, 5, e466–e478. [Google Scholar] [CrossRef] [PubMed]
- Baquero, F.; Martinez, J.L.; Lanza, V.F.; Rodríguez-Beltrán, J.; Galán, J.C.; San Millán, A.; Cantón, R.; Coque, T.M. Evolutionary pathways and trajectories in antibiotic resistance. Clin. Microbiol. Rev. 2021, 34, e00050-19. [Google Scholar] [CrossRef] [PubMed]
- Motlhalamme, T.; Paul, L.; Singh, V. Environmental Reservoirs, Genomic Epidemiology, and Mobile Genetic Elements. In Antimicrobial Resistance: Factors to Findings: Omics and Systems Biology Approaches; Springer: Berlin/Heidelberg, Germany, 2024; pp. 239–273. [Google Scholar]
- Laborda, P.; Sanz-García, F.; Ochoa-Sánchez, L.E.; Gil-Gil, T.; Hernando-Amado, S.; Martínez, J.L. Wildlife and antibiotic resistance. Front. Cell. Infect. Microbiol. 2022, 12, 873989. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef]
- Liu, G.; Thomsen, L.E.; Olsen, J.E. Antimicrobial-induced horizontal transfer of antimicrobial resistance genes in bacteria: A mini-review. J. Antimicrob. Chemother. 2022, 77, 556–567. [Google Scholar] [CrossRef]
- Li, X.; Mowlaboccus, S.; Jackson, B.; Cai, C.; Coombs, G.W. Antimicrobial Resistance Among Clinically Significant Bacteria in Wildlife: An Overlooked One Health Concern. Int. J. Antimicrob. Agents 2024, 64, 107251. [Google Scholar] [CrossRef]
- Gwenzi, W.; Chaukura, N.; Muisa-Zikali, N.; Teta, C.; Musvuugwa, T.; Rzymski, P.; Abia, A.L.K. Insects, rodents, and pets as reservoirs, vectors, and sentinels of antimicrobial resistance. Antibiotics 2021, 10, 68. [Google Scholar] [CrossRef]
- Hassell, J.M. Ecological and Epidemiological Consequences of Rapid Urbanisation at Wildlife-Livestock-Human Interfaces; The University of Liverpool: Liverpool, UK, 2018; ISBN 9798662378728. [Google Scholar]
- Shams Eldeen, M.A.; Elsaid, R.E.; Salem, H.S.; Eisa, E.A.; Shalaby, R.E. LasB, ExoS and Nan1 genes as potential predictors of site-specific Pseudomonas aeruginosa pathogenicity in nosocomial isolates. Microbes Infect. Dis. 2024, 5, 770–780. [Google Scholar] [CrossRef]
- Aslam, B.; Khurshid, M.; Arshad, M.I.; Muzammil, S.; Rasool, M.; Yasmeen, N.; Shah, T.; Chaudhry, T.H.; Rasool, M.H.; Shahid, A. Antibiotic resistance: One health one world outlook. Front. Cell. Infect. Microbiol. 2021, 11, 771510. [Google Scholar] [CrossRef]
- He, Y.; Yuan, Q.; Mathieu, J.; Stadler, L.; Senehi, N.; Sun, R.; Alvarez, P.J.J. Antibiotic resistance genes from livestock waste: Occurrence, dissemination, and treatment. NPJ Clean Water 2020, 3, 4. [Google Scholar] [CrossRef]
- Robles-Jimenez, L.E.; Aranda-Aguirre, E.; Castelan-Ortega, O.A.; Shettino-Bermudez, B.S.; Ortiz-Salinas, R.; Miranda, M.; Li, X.; Angeles-Hernandez, J.C.; Vargas-Bello-Pérez, E.; Gonzalez-Ronquillo, M. Worldwide traceability of antibiotic residues from livestock in wastewater and soil: A systematic review. Animals 2021, 12, 60. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).