The Genetic Diversity and Phylogeography of the Iberian Endemic Steppe Plant Moricandia moricandioides (Boiss.) Heywood, Inferred from ISSR, Plastid DNA, and ITS Sequences
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Strategy
2.2. DNA Isolation and ISSR Amplification
2.3. Nuclear and Plastid DNA Amplification and Sequencing
2.4. ISSR Analysis
2.5. Sequence Alignment and Analysis
3. Results
3.1. ISSR Genetic Diversity in Moricandia moricandioides
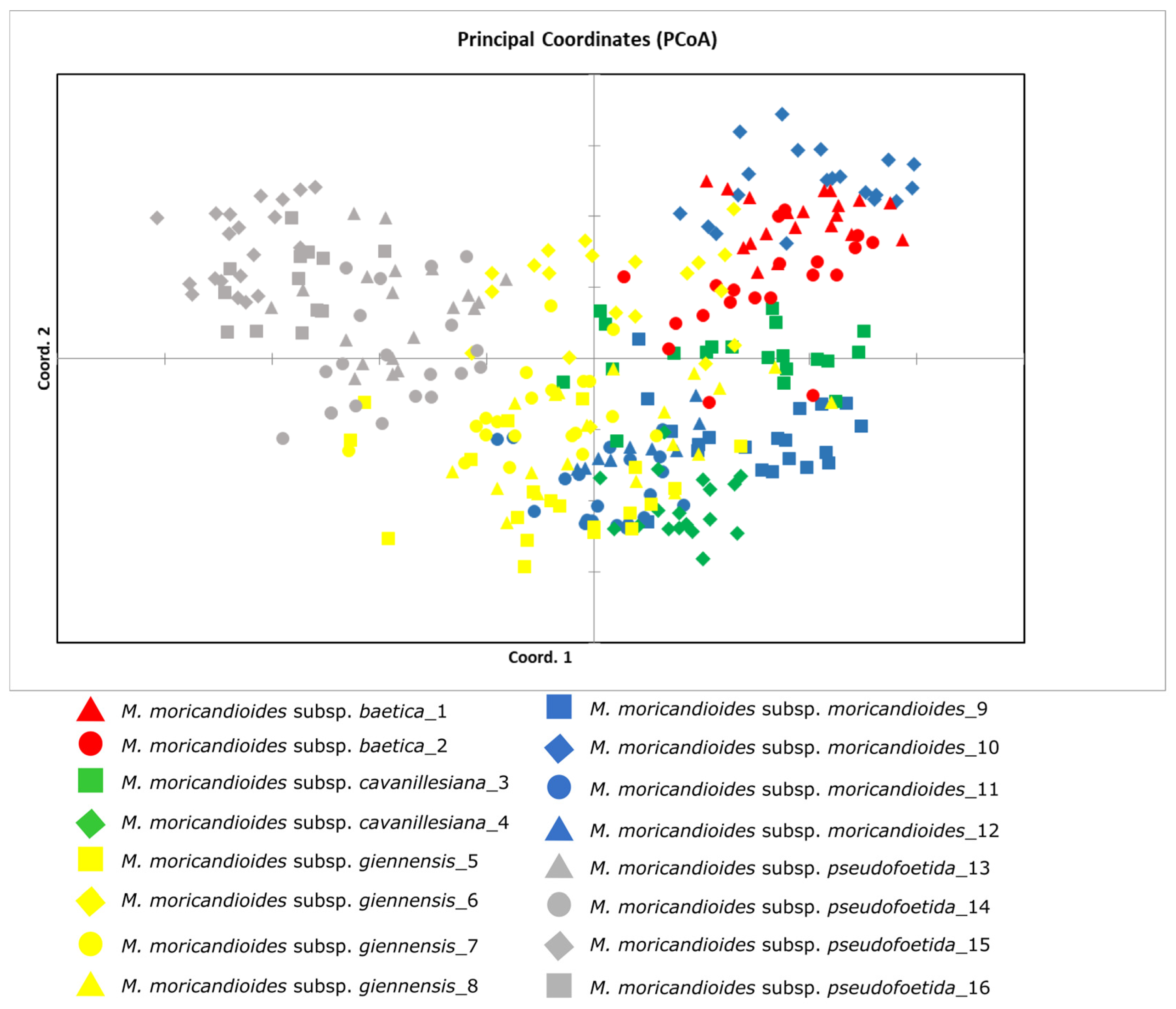
3.2. Nuclear DNA Variation, Geographical Distribution and Phylogenetic Analysis
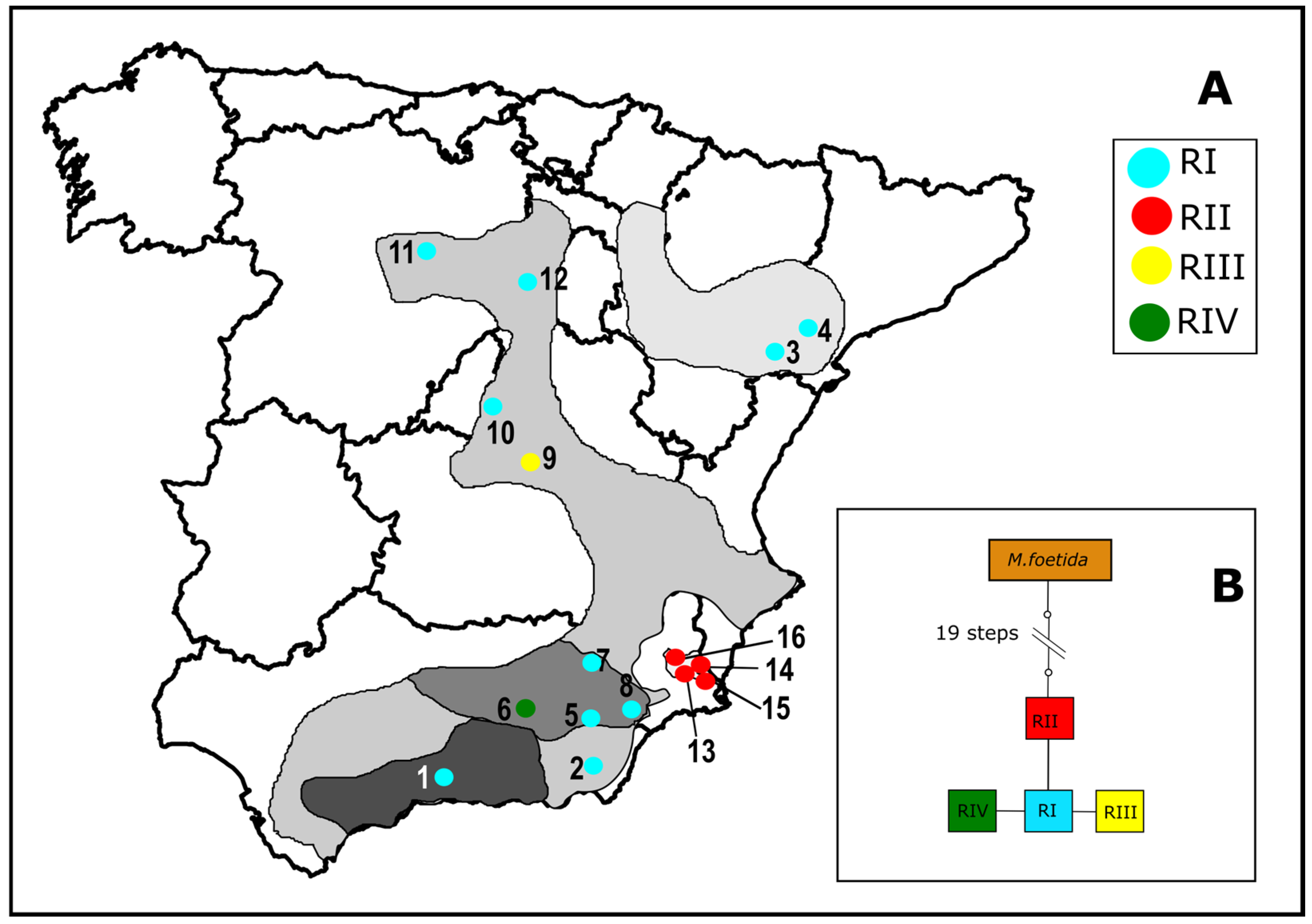
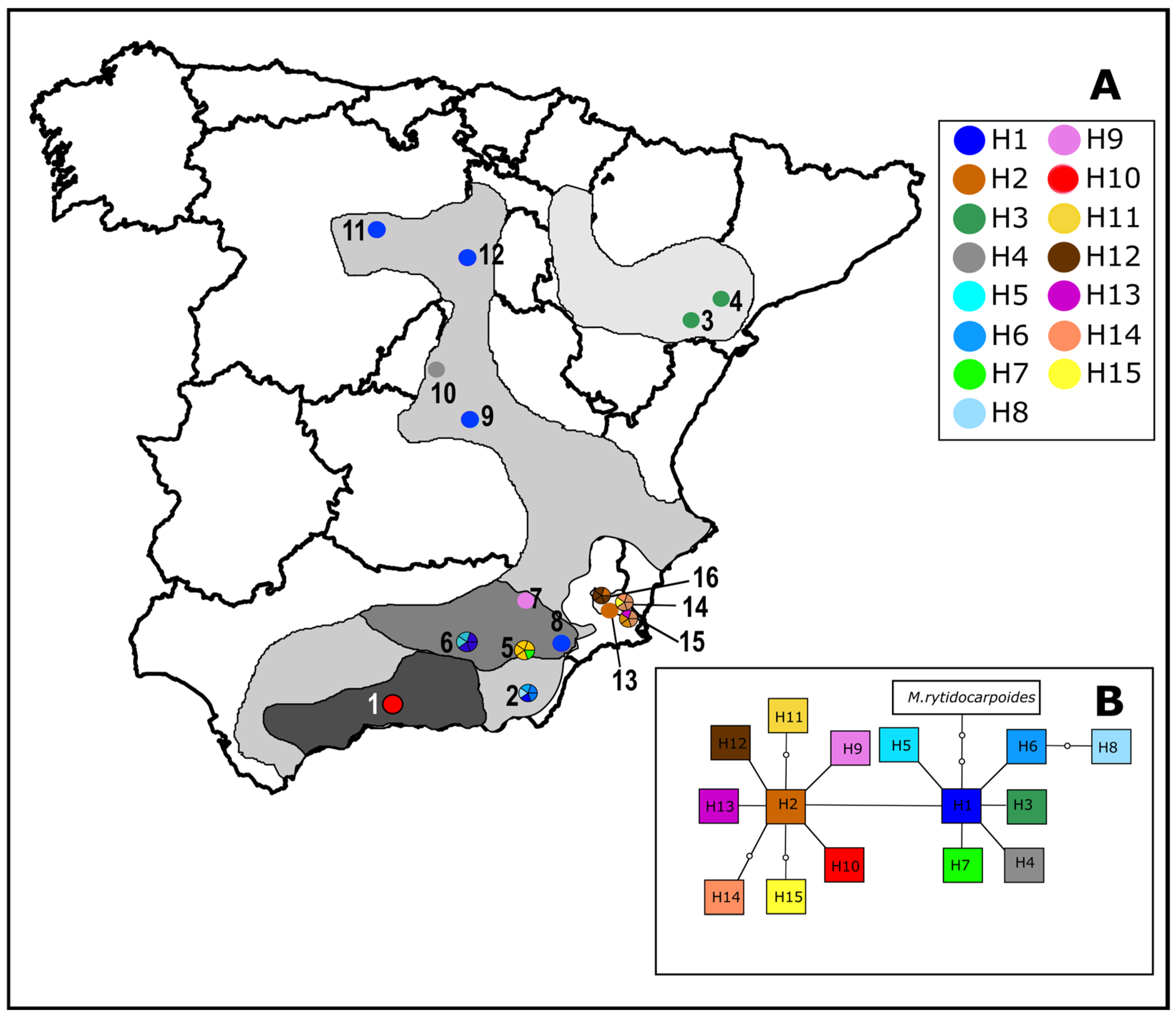
3.3. Plastid DNA Variation, Geographical Distribution and Phylogenetic Analysis
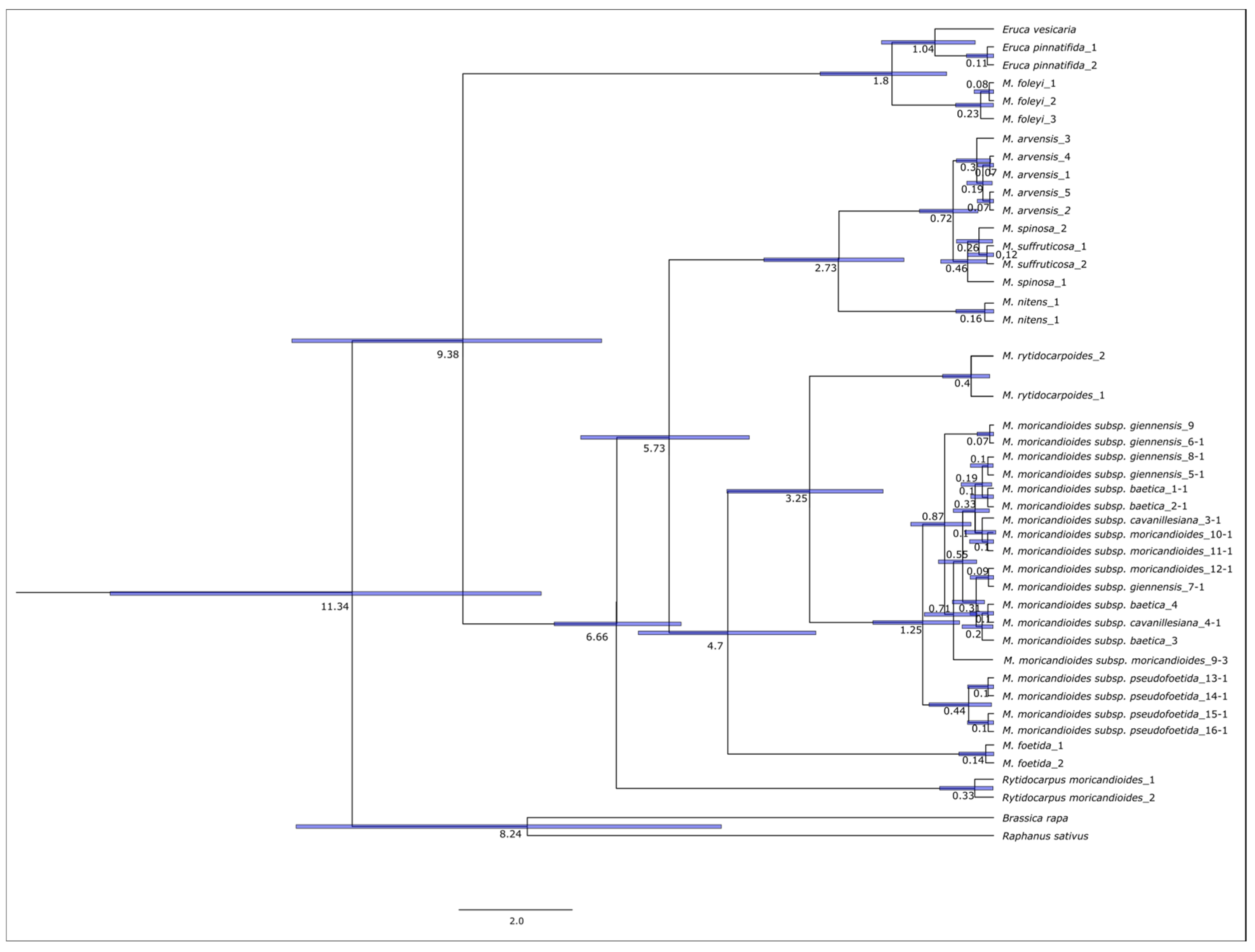
3.4. Estimation of Divergence Times
4. Discussion
4.1. Genetic Diversity and Rarity Patterns of Moricandia moricandioides
4.2. The Origin of the Present Phylogeographic Patterns of Moricandia moricandioides
4.3. Taxonomic Conclusions
4.4. Implications for Conservation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Schulz, O.E. Cruciferae. In Die Natürlichen Pflanzenfamilien; Engler, A., Harms, H., Eds.; Verlag von Wilhelm Engelmann: Leizpig, Germany, 1936; pp. 227–658. [Google Scholar]
- Perfectti, F.; Gómez, J.M.; González-Megías, A.; Abdelaziz, M.; Lorite, J. Molecular phylogeny and evolutionary history of Moricandia DC (Brassicaceae). PeerJ 2017, 5, e3964. [Google Scholar] [CrossRef] [PubMed]
- POWO. Plants of the World Online. Facilitated by the Royal Botanic Gardens, Kew. Available online: https://powo.science.kew.org/ (accessed on 23 October 2024).
- Tahir, M.; Watts, R. Moricandia. In Wild Crop Relatives Genomic and Breeding Resources; Kole, C., Ed.; Springer: Berlin, Germany, 2011. [Google Scholar]
- De Bolòs, A. El género Moricandia en la Península Ibérica. An. Jard. Bot. Madr. 1946, 6, 451–461. [Google Scholar]
- Heywood, V. Moricandia DC. In Flora Europaea, Vol. I; Vaughan, J.G., MacLeod, A.J., Jones, B.M.G., Eds.; Academic Press: London, UK, 1964. [Google Scholar]
- Maire, R. Flore de L’Afrique du Nord, Vol. XIII; Editions Paul Lechevalier: Paris, France, 1967. [Google Scholar]
- Schulz, O.E. Cruciferae-Brassicaceae. Part II. Subtribes Cakilinae, Zillinae, Vellinae, Savignyinae and Moricandiinae. Pflanzer 1923, 82–85, 1–100. [Google Scholar]
- Gómez-Campo, C. Morphology and morphotaxonomy of the tribe Brassiceae. In Brassica Crops and Wild Allies: Biology and Breeding; Tsunoda, S., Hinata, K., Gómez-Campo, C., Eds.; Japan Scientific Soc. Press: Tokio, Japan, 1980; pp. 3–31. [Google Scholar]
- Warwick, S.I.; Black, L.D. Evaluation of the subtribes Moricandiinae, Savignyinae, Vellinae, and Zillinae (Brassicaceae, tribe Brassiceae) using chloroplast DNA restriction site variation. Canad. J. Bot. 1994, 72, 1692–1701. [Google Scholar] [CrossRef]
- Warwick, S.I.; Sauder, C.A. Phylogeny of tribe Brassiceae (Brassicaceae) based on chloroplast restriction site polymorphisms and nuclear ribosomal internal transcribed spacer and chloroplast trn L intron sequences. Canad. J. Bot. 2005, 83, 467–483. [Google Scholar] [CrossRef]
- Warwick, S.I.; Black, L.D. Phylogenetic implications of chloroplast DNA restriction site variation in subtribes Raphaninae and Cakilinae (Brassicaceae, tribe Brassiceae). Canad. J. Bot. 1997, 75, 960–973. [Google Scholar] [CrossRef]
- Bailey, C.D.; Koch, M.A.; Mayer, M.; Mummenhoff, K.; O’Kane, S.L.; Warwick, S.I.; Windham, M.D.; Al-Shehbaz, I.A. Toward a global phylogeny of the Brassicaceae. Mol. Biol. Evol. 2006, 23, 2142–2160. [Google Scholar] [CrossRef]
- Jiménez, J.F.; Sánchez-Gómez, P. Molecular taxonomy and genetic diversity of Moricandia moricandioides subsp. pseudofoetida compared to wild relatives. Plant Biosyst. 2012, 146, 99–105. [Google Scholar] [CrossRef]
- Schlüter, U.; Bräutigam, A.; Gowik, U.; Melzer, M.; Christin, P.-A.; Kurz, S.; Mettler-Altmann, T.; Weber, A.P.M. Photosynthesis in C3-C4 intermediate Moricandia species. J. Exp. Bot. 2016, 68, 191–206. [Google Scholar] [CrossRef]
- Couvreur, T.L.; Franzke, A.; Al-Shehbaz, I.A.; Bakker, F.T.; Koch, M.A.; Mummenhoff, K. Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae). Mol. Biol. Evol. 2010, 27, 55–71. [Google Scholar] [CrossRef]
- Sánchez-Gómez, P.; Vera, J.B.; Jiménez, J.F.; López, D.; Mota, J.F. Moricandia moricandioides subsp. cavanillesiana (Font Quer & A. Bolòs) Greuter & Burdet. In Diversidad Vegetal de las Yeseras Ibéricas. El Reto de los Archipiélagos Edáficos Para la Biología de la Conservación; Mota, J.F., Sánchez-Gómez, P., Guirado, J.S., Eds.; ADIF—Mediterráneo Asesores Consultores: Almería, Spain, 2011; pp. 245–247. [Google Scholar]
- Salmerón-Sánchez, E.; Merlo, M.E.; Medina-Cazorla, J.M.; Pérez-García, F.J.; Martínez-Hernández, F.; Garrido-Becerra, J.A.; Mendoza-Fernández, A.J.; Valle, F.; Mota, J.F. Variability, genetic structure and phylogeography of the dolomitophilous species Convolvulus boissieri (Convolvulaceae) in the Baetic ranges, inferred from AFLPs, plastid DNA and ITS sequences. Bot. J. Linn. Soc. 2014, 176, 506–523. [Google Scholar] [CrossRef]
- Martínez-Nieto, M.I.; Segarra-Moragues, J.G.; Merlo, E.; Martínez-Hernández, F.; Mota, J.F. Genetic diversity, genetic structure and phylogeography of the Iberian endemic Gypsophila struthium (Caryophyllaceae) as revealed by AFLP and plastid DNA sequences: Connecting habitat fragmentation and diversification. Bot. J. Linn. Soc. 2013, 173, 654–675. [Google Scholar] [CrossRef]
- Salmerón-Sánchez, E.; Martínez-Nieto, M.I.; Martínez-Hernández, F.; Garrido-Becerra, J.A.; Mendoza-Fernández, A.J.; de Carrasco, C.G.; Ramos-Miras, J.J.; Lozano, R.; Merlo, M.E. Ecology, genetic diversity and phylogeography of the Iberian endemic plant Jurinea pinnata (Lag.) DC.(Compositae) on two special edaphic substrates: Dolomite and gypsum. Plant Soil 2014, 374, 233–250. [Google Scholar] [CrossRef]
- Salmerón-Sánchez, E.; Martínez-Ortega, M.M.; Mota, J.F.; Peñas, J. A complex history of edaphic habitat islands on the Iberian Peninsula: Phylogeography of the halo-gypsophyte Jacobaea auricula (Asteraceae). Bot. J. Linn. Soc. 2017, 185, 376–392. [Google Scholar] [CrossRef]
- Blanco-Sánchez, M.; Moore, M.J.; Ramos-Muñoz, M.; Pías, B.; García-Fernández, A.; Prieto, M.; Plaza, L.; Isabel, I.; Escudero, A.; Matesanz, S. Phylogeography of a gypsum endemic plant across its entire distribution range in the western Mediterranean. Am. J. Bot. 2021, 108, 443–460. [Google Scholar] [CrossRef]
- Nieto Feliner, G. Patterns and processes in plant phylogeography in the Mediterranean Basin. A review. Perspect. Plant Ecol. Evol. Syst. 2014, 16, 265–278. [Google Scholar] [CrossRef]
- Vitales, D.; García-Fernández, A.; Garnatje, T.; Vallès, J.; Cowan, R.S.; Fay, M.F.; Pellicer, J. Conservation genetics of the rare Iberian endemic Cheirolophus uliginosus (Asteraceae). Bot. J. Linn. Soc. 2015, 179, 151–171. [Google Scholar] [CrossRef]
- Médail, F.; Diadema, K. Glacial refugia influence plant diversity patterns in the Mediterranean Basin. J. Biogeogr. 2009, 36, 1333–1345. [Google Scholar] [CrossRef]
- Zietkiewicz, E.; Rafalski, A.; Labuda, D. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 1994, 20, 176–183. [Google Scholar] [CrossRef]
- Williams, J.G.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990, 18, 6531–6535. [Google Scholar] [CrossRef]
- Rafalski, J.A.; Vogel, J.M.; Morgante, M.; Powell, W.; Andre, C.; Tingey, S.V. Generating and using DNA markers in plants. In Nonmammalian Genomic Analysis: A Practical Guide; Brain, B., Lai, E., Eds.; Academic Press: San Diego, CA, USA, 1998; pp. 75–134. [Google Scholar] [CrossRef]
- Wolfe, A.D.; Xiang, Q.Y.; Kephart, S.R. Assessing hybridization in natural populations of Penstemon (Scrophulariaceae) using hypervariable intersimple sequence repeat (ISSR) bands. Mol. Ecol. 1998, 7, 1107–1125. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, A.D.; Randle, C.P. Relationships within and among species of the holoparasitic genus Hyobanche (Orobanchaceae) inferred from ISSR banding patterns and nucleotide sequences. Syst. Bot. 2001, 26, 120–130. [Google Scholar]
- Grimalt, M.; García-Martínez, S.; Carbonell, P.; Hernández, F.; Legua, P.; Almansa, M.S.; Amorós, A. Relationships between chemical composition, antioxidant activity and genetic analysis with ISSR markers in flower buds of caper plants (Capparis spinosa L.) of two subspecies spinosa and rupestris of Spanish cultivars. Genet. Resour. Crop Evol. 2022, 69, 1451–1469. [Google Scholar] [CrossRef]
- Bodo Slotta, T.A.; Porter, D.M. Genetic variation within and between Iliamna corei and I. remota (Malvaceae): Implications for species delimitation. Bot. J. Linn. Soc. 2006, 151, 345–354. [Google Scholar] [CrossRef]
- Galván, M.Z.; Lanteri, A.A.; Menendez Sevillano, M.D.C.; Balatti, P.A. Molecular characterisation of wild populations and landraces of common bean from northwestern Argentina. Plant Biosyst. 2010, 144, 365–372. [Google Scholar] [CrossRef]
- Ramírez-Rodriguez, R.; Jiménez, J.F.; Amich, F.; Sánchez-Gómez, P. Plastid phylogeography of Delphinium fissum subsp. sordidum and the series Fissa (Ranunculaceae) in the Iberian Peninsula: Implications for conservation. Bot. Lett. 2019, 166, 345–355. [Google Scholar] [CrossRef]
- Salmerón-Sánchez, E.; Fuertes-Aguilar, J.; Španiel, S.; Pérez-García, F.J.; Merlo, E.; Garrido-Becerra, J.A.; Mota, J.F. Plant evolution in alkaline magnesium-rich soils: A phylogenetic study of the Mediterranean genus Hormathophylla (Cruciferae: Alysseae) based on nuclear and plastid sequences. PLoS ONE 2018, 13, e0208307. [Google Scholar] [CrossRef]
- Richardson, J.E.; Pennington, R.T.; Pennington, T.D.; Hollingsworth, P.M. Rapid diversification of a species-rich genus of neotropical rain forest trees. Science 2001, 293, 2242–2245. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–321. [Google Scholar] [CrossRef]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef]
- Grosberg, R.K.; Levitan, D.R.; Cameron, B.B. Characterization of genetic structure and genealogies using RAPD-PCR markers: A random primer for the novice and nervous. In Molecular Zoology. Advances, Strategies, and Protocols; Ferraris, J.D., Palumbi, S.R., Eds.; Wiley-Liss: New York, NY, USA, 1996; pp. 67–100. [Google Scholar]
- Lynch, M.M.; Milligan, B.G. Analysis of population genetic structure with RAPD markers. Mol. Ecol. 1994, 3, 91–99. [Google Scholar] [CrossRef]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Schönswetter, P.; Tribsch, A. Vicariance and dispersal in the alpine perennial Bupleurum stellatum L. (Apiaceae). Taxon 2005, 54, 725–732. [Google Scholar] [CrossRef]
- Crisp, M.D.; Laffan, S.; Linder, H.P.; Monro, A. Endemism in the Australian flora. J. Biogeogr. 2001, 28, 183–198. [Google Scholar] [CrossRef]
- Ehrich, D. AFLPdat: A collection of R functions for convenient handling of AFLP data. Mol. Ecol. Notes 2006, 6, 603–604. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Rosenberg, N.A.; Donnelly, P. Association mapping in structured populations. Am. J. Hum. Genet. 2000, 67, 170–181. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Wen, W. Documentation for Structure Software: Version 2; University of Chicago: Chicago, IL, USA, 2004. [Google Scholar]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, N.A. DISTRUCT: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Templeton, A.R.; Crandall, K.A.; Sing, C.F. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 1992, 132, 619–633. [Google Scholar] [CrossRef]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef]
- Pattengale, N.D.; Alipour, M.; Bininda-Emonds, O.R.; Moret, B.M.; Stamatakis, A. How many bootstrap replicates are necessary? In Annual International Conference on Research in Computational Molecular Biology; Batzoglou, S., Ed.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 184–200. [Google Scholar] [CrossRef]
- Alfaro, M.E.; Zoller, S.; Lutzoni, F. Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol. Biol. Evol. 2003, 20, 255–266. [Google Scholar] [CrossRef]
- Drummond, A.J.; Ho, S.Y.W.; Phillips, M.J.; Rambaut, A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006, 4, e88. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. Figtree ver 1.4.4; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, UK, 2018. [Google Scholar]
- Cuenot, Y.; Gómez, J.M.; González-Megias, A.; Pannell, J.R.; Torices, R. Characterization of microsatellite markers for Moricandia moricandioides (Brassicaceae) and related species. Appl. Plant Sci. 2018, 6, e01172. [Google Scholar] [CrossRef]
- Nybom, H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol. 2004, 13, 1143–1155. [Google Scholar] [CrossRef]
- Valtueña, F.J.; Fernández-Mazuecos, M.; Rodríguez-Riaño, T.; López, J.; Ortega-Olivencia, A. Repeated jumps from Northwest Africa to the European continent: The case of peripheral populations of an annual plant. J. Syst. Evol. 2020, 58, 487–503. [Google Scholar] [CrossRef]
- Díaz-Pérez, A.; Sequeira, M.; Santos-Guerra, A.; Catalán, P. Multiple colonizations, in situ speciation, and volcanism-associated stepping-stone dispersals shaped the phylogeography of the Macaronesian red fescues (Festuca L., Gramineae). Syst. Biol. 2008, 57, 732–749. [Google Scholar] [CrossRef]
- Pérez-Collazos, E.; Sanchez-Gómez, P.; Jiménez, J.F.; Catalán, P. The phylogeographical history of the Iberian steppe plant Ferula loscosii (Apiaceae): A test of the abundant-centre hypothesis. Mol. Ecol. 2009, 18, 848–861. [Google Scholar] [CrossRef]
- Alarcón, M.; Vargas, P.; Sáez, L.; Molero, J.; Aldasoro, J.J. Genetic diversity of mountain plants: Two migration episodes of Mediterranean Erodium (Geraniaceae). Mol. Phylogenet. Evol. 2012, 63, 866–876. [Google Scholar] [CrossRef]
- Garnatje, T.; Pérez-Collazos, E.; Pellicer, J.; Catalán, P. Balearic insular isolation and large continental spread framed the phylogeography of the western Mediterranean Cheirolophus intybaceus sl (Asteraceae). Plant Biol. 2013, 15, 166–175. [Google Scholar] [CrossRef]
- Thompson, J.D. Plant Evolution in the Mediterranean; Oxford University Press: Oxford, UK, 2005. [Google Scholar] [CrossRef]
- Valente, L.M.; Savolainen, V.; Vargas, P. Unparalleled rates of species diversification in Europe. Proc. Biol. Sci. 2010, 277, 1489–1496. [Google Scholar] [CrossRef] [PubMed]
- Guzmán, B.; Vargas, P. Historical biogeography and character evolution of Cistaceae (Malvales) based on analysis of plastid rbcL and trnL-trnF sequences. Org. Divers. Evol. 2009, 9, 83–99. [Google Scholar] [CrossRef]
- Fauquette, S.; Suc, J.P.; Guiot, J.; Diniz, F.; Feddi, N.; Zheng, Z.; Bessais, E.; Drivaliari, A. Climate and biomes in the West Mediterranean area during the Pliocene. Palaeogeogr. Palaeoclimatol. Palaeoecol. 1999, 152, 15–36. [Google Scholar] [CrossRef]
- Hewitt, G. The genetic legacy of the Quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef]
- Jiménez-Moreno, G.; Fauquette, S.; Suc, J.P. Miocene to Pliocene vegetation reconstruction and climate estimates in the Iberian Peninsula from pollen data. Rev. Palaeobot. Palynol. 2010, 162, 403–415. [Google Scholar] [CrossRef]
- Gutiérrez Larena, B.; Fuertes Aguilar, J.; Nieto Feliner, G. Glacial-induced altitudinal migrations in Armeria (Plumbaginaceae) inferred from patterns of chloroplast DNA haplotype sharing. Mol. Ecol. 2002, 11, 1965–1974. [Google Scholar] [CrossRef] [PubMed]
- Nieto Feliner, G.; Gutiérrez Larena, B.; Fuertes Aguilar, J. Fine scale geographical structure, intra-individual polymorphism and recombination in nuclear ribosomal internal transcribed spacers in Armeria (Plumbaginaceae). Ann Bot. 2004, 93, 189–200. [Google Scholar] [CrossRef]
- Fuertes-Aguilar, J.; Gutiérrez Larena, B.; Nieto Feliner, G. Genetic and morphological diversity in Armeria (Plumbaginaceae) is shaped by glacial cycles in Mediterranean refugia. An. Jard. Bot. Madr. 2011, 68, 175–197. [Google Scholar] [CrossRef]
- Rajakaruna, N. Lessons on evolution from the study of edaphic specialization. Bot. Rev. 2018, 84, 39–78. [Google Scholar] [CrossRef]
- Cowling, R.M.; Rundel, P.W.; Lamont, B.B.; Arroyo, M.K.; Arianoutsou, M. Plant diversity in mediterranean-climate regions. Tree 1996, 11, 362–366. [Google Scholar] [CrossRef]
- Givnish, T.J. Ecology of plant speciation. Taxon 2010, 59, 1326–1366. [Google Scholar] [CrossRef]
- Sobrino-Vesperinas, E. Interfertility in the genus Moricandia DC. Lagascalia 1997, 19, 839–844. [Google Scholar]
- Sobrino-Vesperinas, E. Moricandia DC. In Flora Iberica. Vol. 4; Castroviejo, S., Aedo, C., Laínz, M., Muñoz Garmendia, F., Nieto Feliner, G., Paiva, J., Benedí, C., Eds.; Real Jardín Botánico, CSIC: Madrid, Spain, 1993; pp. 338–344. [Google Scholar]

| Geogr. Coord | ISSR Analysis | cpDNA Analysis | nrITS Analysis | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pop. | Pop. Code | SUBSP. | Long. | Lat. | ISSR Samples | PL | PLP | He | cpDNA Samples | Hap | Hd | Nd | Hgroup | Haplotype | ITS Samples | Ribotype |
| Antequera, Málaga | 1 | baetica | 37.01 | −4.3 | 20 | 42 | 60 | 0.18199 | 5 | 1 | 0 | 0 | 2 | 10 (5) | 1 | I |
| Canjáyar, Almeria | 2 | baetica | 36.59 | −2.41 | 20 | 37 | 52.9 | 0.16115 | 5 | 3 | 0.8 | 0.00177 | 1 | 1 (2), 6 (2), 8 (2) | 1 | I |
| Σ | 0.17157 | 10 | 4 | 0.733 | 0.0025 | 1.2 | 1, 6, 8, 10 | |||||||||
| Ascó, Tarragona | 3 | cavanillesiana | 41.1 | 0.33 | 20 | 36 | 51.4 | 0.14307 | 5 | 1 | 0 | 0 | 1 | 3 | 1 | |
| Granja Escarp, Lleida | 4 | cavanillesiana | 41.09 | 0.15 | 20 | 31 | 44.3 | 0.1026 | 5 | 1 | 0 | 0 | 1 | 3 | 1 | I |
| Σ | 0.122835 | 10 | 1 | 0 | 0 | 1 | 3 | |||||||||
| Galera, Granada | 5 | giennensis | 37.36 | −2.4 | 20 | 46 | 65.7 | 0.22134 | 5 | 2 | 0.4 | 0.00152 | 1.2 | 7 (1), 11 (4) | 1 | I |
| Huelma, Jaén | 6 | giennensis | 37.39 | −3.27 | 20 | 51 | 72.9 | 0.2603 | 5 | 2 | 0.6 | 0.00076 | 1 | 1 (3), 5 (2) | 1 | IV |
| Moratalla, Murcia | 7 | giennensis | 38.1 | −2.12 | 20 | 44 | 62.9 | 0.23398 | 5 | 1 | 0 | 0 | 2 | 9 (5) | 1 | I |
| Lorca, Murcia | 8 | giennensis | 37.52 | −1.56 | 20 | 44 | 62.9 | 0.23799 | 5 | 1 | 0 | 0 | 1 | 1 (5) | 1 | I |
| Σ | 0.2384025 | 20 | 5 | 0.763 | 0.00249 | 1.2 | 1, 5, 7, 9, 11 | |||||||||
| Valdemoro, Cuenca | 9 | moricandioides | 40.15 | −2.37 | 20 | 36 | 51.4 | 0.12841 | 5 | 1 | 0 | 0 | 1 | 1 (5) | 1 | III |
| Humosa, Madrid | 10 | moricandioides | 37.01 | −4.3 | 20 | 41 | 58.6 | 0.18451 | 5 | 1 | 0 | 0 | 1 | 4 (5) | 1 | I |
| Near Palencia | 11 | moricandioides | 41.56 | −4.25 | 20 | 39 | 55.7 | 0.15399 | 5 | 1 | 0 | 0 | 1 | 1 (5) | 1 | I |
| South of Almazan, Soria | 12 | moricandioides | 41.17 | −2.23 | 10 | 33 | 47.1 | 0.13725 | 5 | 1 | 0 | 0 | 1 | 1 (5) | 1 | I |
| Σ | 0.15104 | 20 | 2 | 0.395 | 0.0005 | 1 | 1, 4 | |||||||||
| Emb. Rodeos, Murcia | 13 | pseudofoetida | 38.2 | −1.17 | 20 | 46 | 65.7 | 0.22377 | 5 | 1 | 0 | 0 | 2 | 2 (5) | 1 | II |
| Fortuna, Murcia | 14 | pseudofoetida | 38.12 | −1.04 | 20 | 56 | 80 | 0.2767 | 5 | 3 | 0.7 | 0.00101 | 2 | 2 (3), 15 (1), 16 (1) | 1 | II |
| El Garruchal, Murcia | 15 | pseudofoetida | 37.56 | −1.03 | 20 | 48 | 68.6 | 0.22371 | 5 | 3 | 0.8 | 0.00243 | 2 | 2 (2), 13 (2), 14 (1) | 1 | II |
| Ricote, Murcia | 16 | pseudofoetida | 38.08 | −1.22 | 14 | 49 | 70 | 0.24874 | 5 | 2 | 0.4 | 0.00051 | 2 | 2 (1), 12 (4) | 1 | II |
| Σ | 0.24323 | 20 | 6 | 0.674 | 0.00141 | 2 | 2, 12, 13, 14, 15, 16 | I | ||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiménez, J.F.; Salmerón-Sánchez, E.; Mota, J.F.; Sánchez-Gómez, P. The Genetic Diversity and Phylogeography of the Iberian Endemic Steppe Plant Moricandia moricandioides (Boiss.) Heywood, Inferred from ISSR, Plastid DNA, and ITS Sequences. Diversity 2025, 17, 310. https://doi.org/10.3390/d17050310
Jiménez JF, Salmerón-Sánchez E, Mota JF, Sánchez-Gómez P. The Genetic Diversity and Phylogeography of the Iberian Endemic Steppe Plant Moricandia moricandioides (Boiss.) Heywood, Inferred from ISSR, Plastid DNA, and ITS Sequences. Diversity. 2025; 17(5):310. https://doi.org/10.3390/d17050310
Chicago/Turabian StyleJiménez, Juan F., Esteban Salmerón-Sánchez, Juan F. Mota, and Pedro Sánchez-Gómez. 2025. "The Genetic Diversity and Phylogeography of the Iberian Endemic Steppe Plant Moricandia moricandioides (Boiss.) Heywood, Inferred from ISSR, Plastid DNA, and ITS Sequences" Diversity 17, no. 5: 310. https://doi.org/10.3390/d17050310
APA StyleJiménez, J. F., Salmerón-Sánchez, E., Mota, J. F., & Sánchez-Gómez, P. (2025). The Genetic Diversity and Phylogeography of the Iberian Endemic Steppe Plant Moricandia moricandioides (Boiss.) Heywood, Inferred from ISSR, Plastid DNA, and ITS Sequences. Diversity, 17(5), 310. https://doi.org/10.3390/d17050310









