In Silico and In Vitro Identification of Pan-Coronaviral Main Protease Inhibitors from a Large Natural Product Library
Abstract
:1. Introduction
2. Results
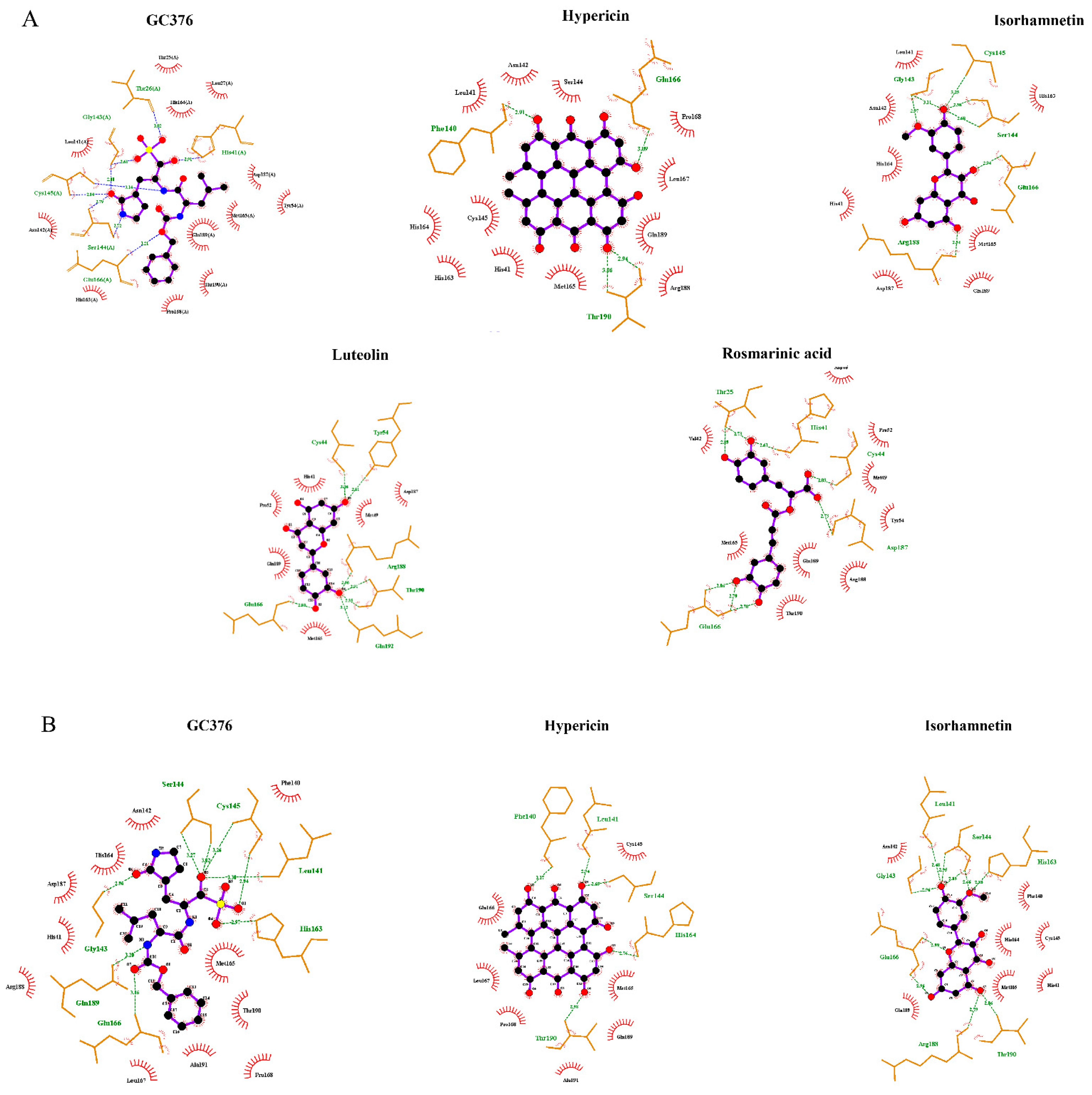
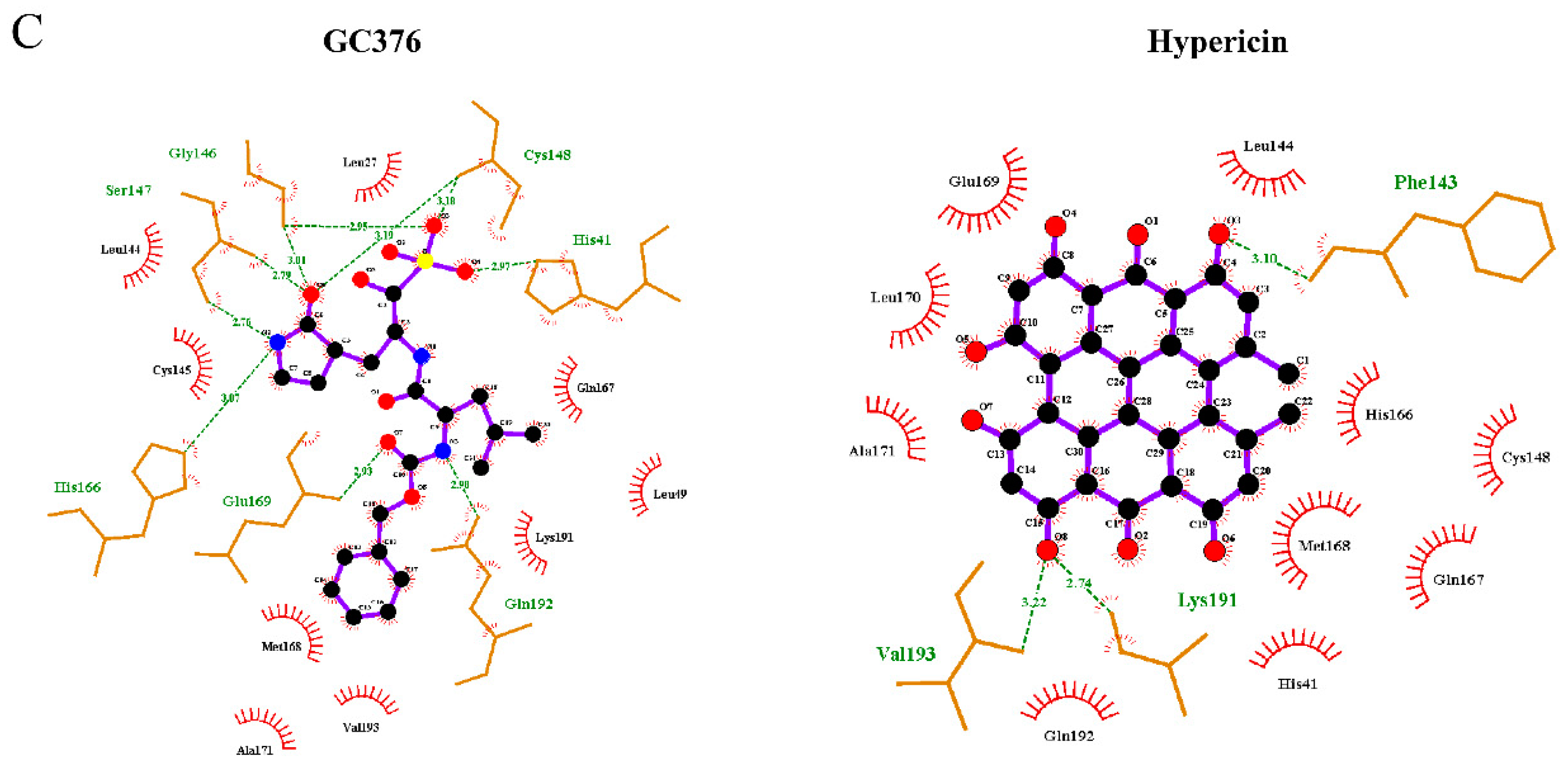
2.1. In Silico Studies
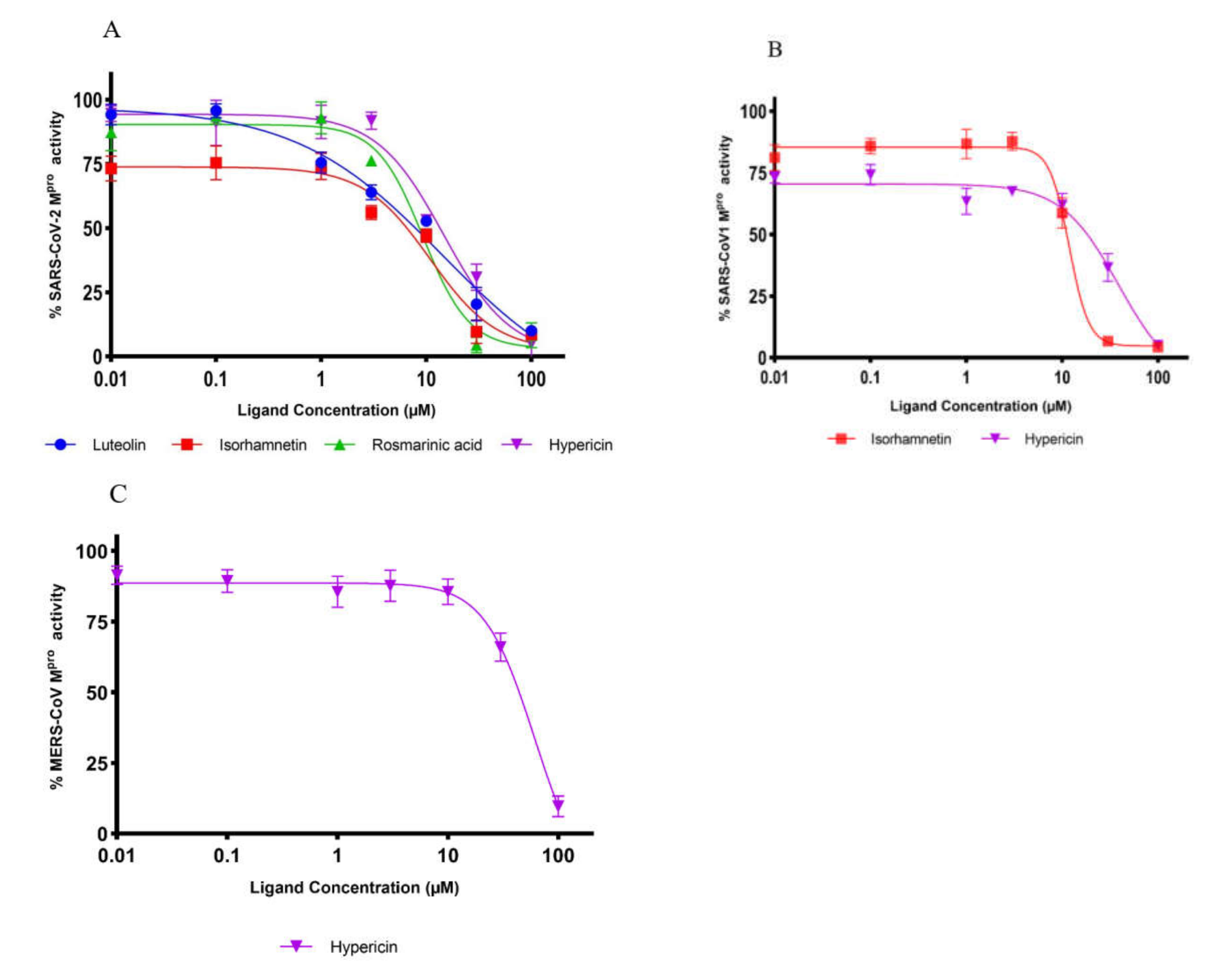
2.2. Inhibition of Mpro Enzyme Activity
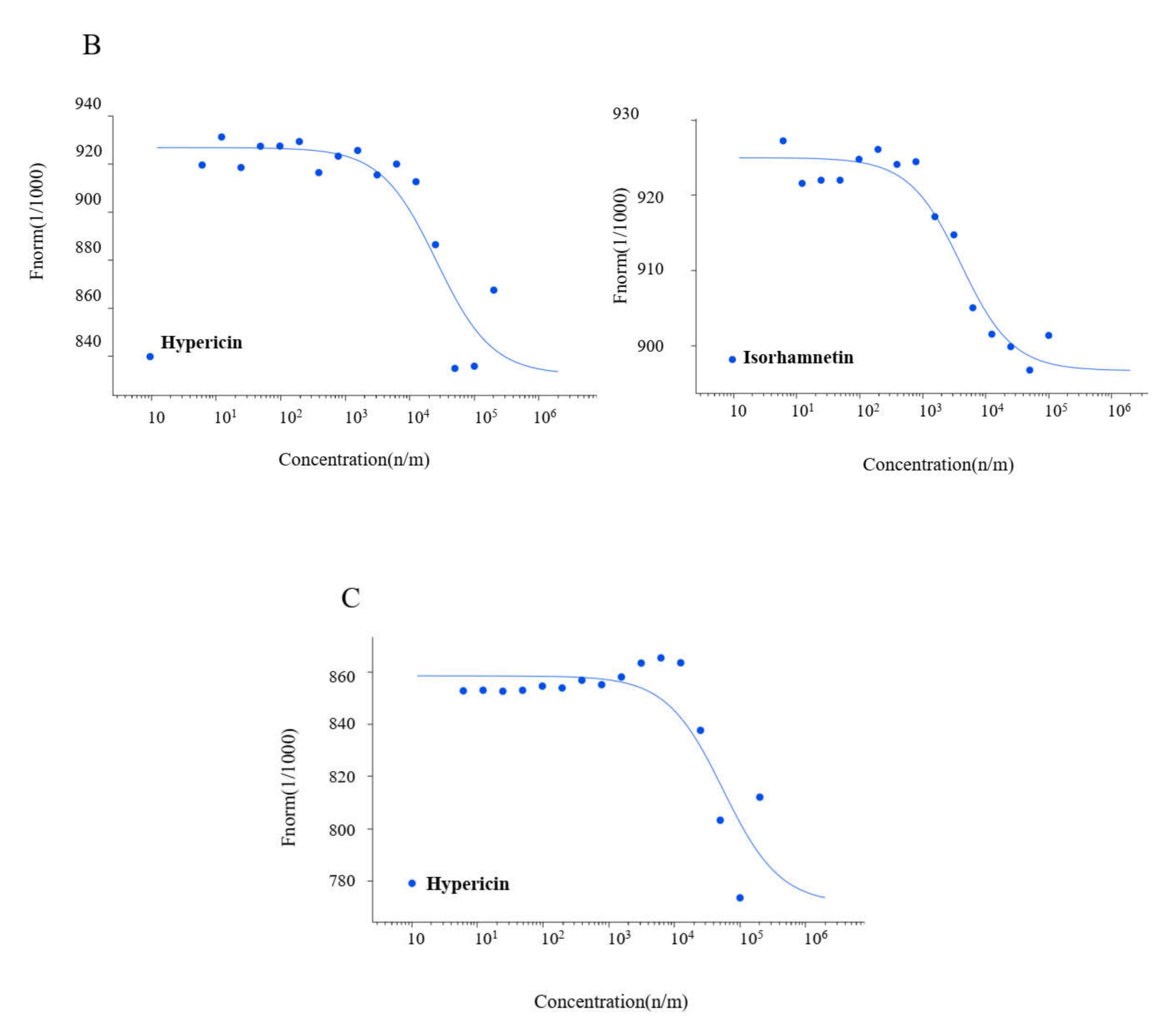
2.3. Microscale Thermophoresis
2.4. Binding of the Top Candidates
2.5. Cell Viability Assay
3. Discussion
4. Materials and Methods
4.1. Compounds
4.2. Virtual Screening
4.3. Sequence Alignment
4.4. Inhibition of Mpro Enzyme Activity
4.5. Molecular Docking
4.6. Microscale Thermophoresis
4.7. Cell Viability Assay
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- V’Kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Park, S.E. Epidemiology, virology, and clinical features of severe acute respiratory syndrome -coronavirus-2 (SARS-CoV-2; Coronavirus Disease-19). Korean J. Pediatr. 2020, 63, 119–124. [Google Scholar] [CrossRef] [Green Version]
- Totura, A.L.; Bavari, S. Broad-spectrum coronavirus antiviral drug discovery. Expert Opin. Drug Discov. 2019, 14, 397–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, S.; Yan, L.; Xu, W.; Agrawal, A.S.; Algaissi, A.; Tseng, C.-T.K.; Wang, Q.; Du, L.; Tan, W.; Wilson, I.A.; et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019, 5, eaav4580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menachery, V.D.; Yount, B.L., Jr.; Debbink, K.; Agnihothram, S.; Gralinski, L.E.; Plante, J.A.; Graham, R.L.; Scobey, T.; Ge, X.-Y.; Donaldson, E.F.; et al. A SARS-like Cluster of Circulating Bat Coronaviruses Shows Potential for Human Emergence. Nat. Med. 2015, 21, 1508–1513. [Google Scholar] [CrossRef]
- Xiao, K.; Zhai, J.; Feng, Y.; Zhou, N.; Zhang, X.; Zou, J.-J.; Li, N.; Guo, Y.; Li, X.; Shen, X.; et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature 2020, 583, 286–289. [Google Scholar] [CrossRef]
- Wacharapluesadee, S.; Tan, C.W.; Maneeorn, P.; Duengkae, P.; Zhu, F.; Joyjinda, Y.; Kaewpom, T.; Ni Chia, W.; Ampoot, W.; Lim, B.L.; et al. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat. Commun. 2021, 12, 972. [Google Scholar] [CrossRef]
- Grange, Z.L.; Goldstein, T.; Johnson, C.K.; Anthony, S.; Gilardi, K.; Daszak, P.; Olival, K.J.; O’Rourke, T.; Murray, S.; Olson, S.H.; et al. Ranking the risk of animal-to-human spillover for newly discovered viruses. Proc. Natl. Acad. Sci. USA 2021, 118, e2002324118. [Google Scholar] [CrossRef]
- Johnson, C.K.; Hitchens, P.L.; Pandit, P.S.; Rushmore, J.; Evans, T.S.; Young, C.C.W.; Doyle, M.M. Global shifts in mammalian population trends reveal key predictors of virus spillover risk. Proc. R. Soc. B Biol. Sci. 2020, 287, 20192736. [Google Scholar] [CrossRef] [Green Version]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Harrison, A.G.; Lin, T.; Wang, P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol. 2020, 41, 1100–1115. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Kneller, D.W.; Phillips, G.; O’Neill, H.M.; Jedrzejczak, R.; Stols, L.; Langan, P.; Joachimiak, A.; Coates, L.; Kovalevsky, A. Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nat. Commun. 2020, 11, 3202. [Google Scholar] [CrossRef]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, C.; Sacco, M.D.; Hurst, B.; Townsend, J.A.; Hu, Y.; Szeto, T.; Zhang, X.; Tarbet, B.; Marty, M.; Chen, Y.; et al. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020, 30, 678–692. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, S.; Nitsche, C. The SARS-CoV-2 main protease as drug target. Bioorg. Med. Chem. Lett. 2020, 30, 127377. [Google Scholar] [CrossRef]
- Hilgenfeld, R. From SARS to MERS: Crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014, 281, 4085–4096. [Google Scholar] [CrossRef] [Green Version]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Drug Delivery Reviews Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Musarra-Pizzo, M.; Pennisi, R.; Ben-Amor, I.; Mandalari, G.; Sciortino, M. Antiviral Activity Exerted by Natural Products against Human Viruses. Viruses 2021, 13, 828. [Google Scholar] [CrossRef]
- Koehn, F.E.; Carter, G.T. The evolving role of natural products in drug discovery. Nat. Rev. Drug Discov. 2005, 4, 206–220. [Google Scholar] [CrossRef]
- Kadioglu, O.; Saeed, M.; Greten, H.J.; Efferth, T. Identification of novel compounds against three targets of SARS CoV-2 coronavirus by combined virtual screening and supervised machine learning. Comput. Biol. Med. 2021, 133, 104359. [Google Scholar] [CrossRef] [PubMed]
- Miskovsky, P. Hypericin—A New Antiviral and Antitumor Photosensitizer: Mechanism of Action and Interaction with Biological Macromolecules. Curr. Drug Targets 2002, 3, 55–84. [Google Scholar] [CrossRef]
- Fukuchi, K.; Okudaira, N.; Adachi, K.; Odai-Ide, R.; Watanabe, S.; Ohno, H.; Yamamoto, M.; Kanamoto, T.; Terakubo, S.; Nakashima, H.; et al. Antiviral and Antitumor Activity of Licorice Root Extracts. In Vivo 2016, 30, 777–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jennings, M.; Parks, R. Curcumin as an Antiviral Agent. Viruses 2020, 12, 1242. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Calvo, A.; de Oya, N.J.; Martin-Acebes, M.A.; Garcia-Moruno, E.; Saiz, J.-C. Antiviral Properties of the Natural Polyphenols Delphinidin and Epigallocatechin Gallate against the Flaviviruses West Nile Virus, Zika Virus, and Dengue Virus. Front. Microbiol. 2017, 8, 1314. [Google Scholar] [CrossRef]
- Cataneo, A.H.D.; Kuczera, D.; Koishi, A.C.; Zanluca, C.; Silveira, G.F.; De Arruda, T.B.; Suzukawa, A.A.; Bortot, L.O.; Dias-Baruffi, M.; Verri, W.A., Jr.; et al. The citrus flavonoid naringenin impairs the in vitro infection of human cells by Zika virus. Sci. Rep. 2019, 9, 16348. [Google Scholar] [CrossRef]
- Kai, H.; Obuchi, M.; Yoshida, H.; Watanabe, W.; Tsutsumi, S.; Park, Y.K.; Matsuno, K.; Yasukawa, K.; Kurokawa, M. In vitro and in vivo anti-influenza virus activities of flavonoids and related compounds as components of Brazilian propolis (AF-08). J. Funct. Foods 2014, 8, 214–223. [Google Scholar] [CrossRef]
- Cheng, S.-C.; Chang, G.-G.; Chou, C.-Y. Mutation of Glu-166 Blocks the Substrate-Induced Dimerization of SARS Coronavirus Main Protease. Biophys. J. 2010, 98, 1327–1336. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef] [Green Version]
- Tomar, S.; Johnston, M.L.; John, S.E.S.; Osswald, H.L.; Nyalapatla, P.R.; Paul, L.N.; Ghosh, A.K.; Denison, M.R.; Mesecar, A.D. Ligand-induced Dimerization of Middle East Respiratory Syndrome (MERS) Coronavirus nsp5 Protease (3CLpro): Implications for Nsp5 Regulation and the Development of Antivirals. J. Biol. Chem. 2015, 290, 19403–19422. [Google Scholar] [CrossRef] [Green Version]
- Ho, B.-L.; Cheng, S.-C.; Shi, L.; Wang, T.-Y.; Ho, K.-I.; Chou, C.-Y. Critical Assessment of the Important Residues Involved in the Dimerization and Catalysis of MERS Coronavirus Main Protease. PLoS ONE 2015, 10, e0144865. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Zia, K.; Ashraf, S.; Uddin, R.; Ul-Haq, Z. Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. J. Biomol. Struct. Dyn. 2021, 39, 2607–2616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, I.-L.; Mahindroo, N.; Liang, P.-H.; Peng, Y.-H.; Kuo, C.-J.; Tsai, K.-C.; Hsieh, H.-P.; Chao, Y.-S.; Wu, S.-Y. Structure-Based Drug Design and Structural Biology Study of Novel Nonpeptide Inhibitors of Severe Acute Respiratory Syndrome Coronavirus Main Protease. J. Med. Chem. 2006, 49, 5154–5161. [Google Scholar] [CrossRef] [PubMed]
- Birt, D.F.; Widrlechner, M.P.; Hammer, K.D.P.; Hillwig, M.L.; Wei, J.; Kraus, G.A.; Murphy, P.A.; McCoy, J.A.; Wurtele, E.S.; Neighbors, J.D.; et al. Hypericum in Infection: Identification of Anti-Viral and Anti-Inflammatory Constituents. Pharm. Biol. 2009, 47, 774–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Feng, R.; Muhammad, I.; Abbas, G.; Zhang, Y.; Ren, Y.; Huang, X.; Zhang, R.; Diao, L.; Wang, X.; et al. Protective effects of hypericin against infectious bronchitis virus induced apoptosis and reactive oxygen species in chicken embryo kidney cells. Poult. Sci. 2019, 98, 6367–6377. [Google Scholar] [CrossRef]
- Shih, C.-M.; Wu, C.-H.; Wu, W.-J.; Hsiao, Y.-M.; Ko, J.-L. Hypericin inhibits hepatitis C virus replication via deacetylation and down-regulation of heme oxygenase-1. Phytomedicine 2018, 46, 193–198. [Google Scholar] [CrossRef]
- Jacobson, J.M.; Feinman, L.; Liebes, L.; Ostrow, N.; Koslowski, V.; Tobia, A.; Cabana, B.E.; Lee, D.-H.; Spritzler, J.; Prince, A.M. Pharmacokinetics, Safety, and Antiviral Effects of Hypericin, a Derivative of St. John’s Wort Plant, in Patients with Chronic Hepatitis C Virus Infection. Antimicrob. Agents Chemother. 2001, 45, 517–524. [Google Scholar] [CrossRef] [Green Version]
- Lavie, G.; Valentine, F.; Levin, B.; Mazur, Y.; Gallo, G.; Lavie, D.; Weiner, D.; Meruelo, D. Studies of the mechanisms of action of the antiretroviral agents hypericin and pseudohypericin. Proc. Natl. Acad. Sci. USA 1989, 86, 5963–5967. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y. Raman spectroscopic study on structure of human immu-nodeficiency virus (HIV) and hypericin-induced photosen-sitive damage of HIV. Sci. China Ser. C Life Sci. 2005, 48, 117. [Google Scholar] [CrossRef]
- Gulick, R.M.; McAuliffe, V.; Holden-Wiltse, J.; Crumpacker, C.; Liebes, L.; Stein, D.S.; Meehan, P.; Hussey, S.; Forcht, J.; Valentine, F.T. Phase I Studies of Hypericin, the Active Compound in St. John’s Wort, as an Antiretroviral Agent in HIV-Infected Adults: AIDS Clinical Trials Group Protocols 150 and 258. Ann. Intern. Med. 1999, 130, 510–514. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, H.; Zou, M.; Oerlemans, R.; Shao, C.; Ren, Y.; Zhang, R.; Huang, X.; Li, G.; Cong, Y. Hypericin Inhibit Alpha-Coronavirus Replication by Targeting 3CL Protease. Viruses 2021, 13, 1825. [Google Scholar] [CrossRef] [PubMed]
- Pitsillou, E.; Liang, J.; Karagiannis, C.; Ververis, K.; Darmawan, K.K.; Ng, K.; Hung, A.; Karagiannis, T.C. Interaction of small molecules with the SARS-CoV-2 main protease in silico and in vitro validation of potential lead compounds using an enzyme-linked immunosorbent assay. Comput. Biol. Chem. 2020, 89, 107408. [Google Scholar] [CrossRef] [PubMed]
- Dellafiora, L.; Galaverna, G.; Cruciani, G.; Dall’Asta, C.; Bruni, R. On the Mechanism of Action of Anti-Inflammatory Activity of Hypericin: An In Silico Study Pointing to the Relevance of Janus Kinases Inhibition. Molecules 2018, 23, 3058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, K.; Gao, S.; Guo, J.; Ni, G.; Chen, Z.; Li, F.; Zhu, X.; Wen, Y.; Guo, Y. Hypericin-photodynamic therapy inhibits proliferation and induces apoptosis in human rheumatoid arthritis fibroblast-like synoviocytes cell line MH7A. Iran. J. Basic Med. Sci. 2018, 21, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Perinbam, K.; Mahendrakumar, M.; Seeni, S. Hypericin, an anthraquinone derivative of Hypericum hookerianum wight and Arn. (Hypericaceae) of Palni Hills, South India, exhibits anti-inflammatory property in lipopolysaccharide—stimulated raw 264.7 macrophages. Pharmacogn. Mag. 2018, 14, 378. [Google Scholar] [CrossRef]
- Levsh, O.; Pluskal, T.; Carballo, V.; Mitchell, A.J.; Weng, J.-K. Independent evolution of rosmarinic acid biosynthesis in two sister families under the Lamiids clade of flowering plants. J. Biol. Chem. 2019, 294, 15193–15205. [Google Scholar] [CrossRef]
- Tsukamoto, Y.; Ikeda, S.; Uwai, K.; Taguchi, R.; Chayama, K.; Sakaguchi, T.; Narita, R.; Yao, W.-L.; Takeuchi, F.; Otakaki, Y.; et al. Rosmarinic acid is a novel inhibitor for Hepatitis B virus replication targeting viral epsilon RNA-polymerase interaction. PLoS ONE 2018, 13, e0197664. [Google Scholar] [CrossRef] [Green Version]
- Mahalapbutr, P.; Sangkhawasi, M.; Kammarabutr, J.; Chamni, S.; Rungrotmongkol, T. Rosmarinic Acid as a Potent Influenza Neuraminidase Inhibitor: In Vitro and In Silico Study. Curr. Top. Med. Chem. 2020, 20, 2046–2055. [Google Scholar] [CrossRef]
- Hsieh, C.-F.; Jheng, J.-R.; Lin, G.-H.; Chen, Y.-L.; Ho, J.-Y.; Liu, C.-J.; Hsu, K.-Y.; Chen, Y.-S.; Chan, Y.F.; Yu, H.-M.; et al. Rosmarinic acid exhibits broad anti-enterovirus A71 activity by inhibiting the interaction between the five-fold axis of capsid VP1 and cognate sulfated receptors. Emerg. Microbes Infect. 2020, 9, 1194–1205. [Google Scholar] [CrossRef]
- Elebeedy, D.; Elkhatib, W.F.; Kandeil, A.; Ghanem, A.; Kutkat, O.; Alnajjar, R.; Saleh, M.A.; El Maksoud, A.I.A.; Badawy, I.; Al-Karmalawy, A.A. Anti-SARS-CoV-2 activities of tanshinone IIA, carnosic acid, rosmarinic acid, salvianolic acid, baicalein, and glycyrrhetinic acid between computational and in vitro insights. RSC Adv. 2021, 11, 29267–29286. [Google Scholar] [CrossRef]
- Jiang, K.; Ma, X.; Guo, S.; Zhang, T.; Zhao, G.; Wu, H.; Wang, X.; Deng, G. Anti-inflammatory Effects of Rosmarinic Acid in Lipopolysaccharide-Induced Mastitis in Mice. Inflammation 2017, 41, 437–448. [Google Scholar] [CrossRef]
- Jin, B.-R.; Chung, K.-S.; Cheon, S.-Y.; Lee, M.; Hwang, S.; Hwang, S.N.; Rhee, K.-J.; An, H.-J. Rosmarinic acid suppresses colonic inflammation in dextran sulphate sodium (DSS)-induced mice via dual inhibition of NF-κB and STAT3 activation. Sci. Rep. 2017, 7, srep46252. [Google Scholar] [CrossRef]
- Luo, C.; Zou, L.; Sun, H.; Peng, J.; Gao, C.; Bao, L.; Ji, R.; Jin, Y.; Sun, S. A Review of the Anti-Inflammatory Effects of Rosmarinic Acid on Inflammatory Diseases. Front. Pharmacol. 2020, 11, 153. [Google Scholar] [CrossRef]
- Gong, G.; Guan, Y.-Y.; Zhang, Z.-L.; Rahman, K.; Wang, S.-J.; Zhou, S.; Luan, X.; Zhang, H. Isorhamnetin: A review of pharmacological effects. Biomed. Pharmacother. 2020, 128, 110301. [Google Scholar] [CrossRef]
- Haghi, G.; Safaei, A.; Ghomi, J.S. Identification and determination of flavonoids in leaf, dried aqueous and dried hydroalcoholic extract of Artemisia absinthium by HPLC. Iran. J. Basic Med. Sci. 2004, 3, 89–90. [Google Scholar]
- Dayem, A.A.; Choi, H.Y.; Kim, Y.B.; Cho, S.-G. Antiviral Effect of Methylated Flavonol Isorhamnetin against Influenza. PLoS ONE 2015, 10, e0121610. [Google Scholar] [CrossRef] [Green Version]
- Zhan, Y.; Ta, W.; Tang, W.; Hua, R.; Wang, J.; Wang, C.; Lu, W. Potential antiviral activity of isorhamnetin against SARS-CoV-2 spike pseudotyped virus in vitro. Drug Dev. Res. 2021, 82, 1124–1130. [Google Scholar] [CrossRef]
- Dou, W.; Zhang, J.; Li, H.; Kortagere, S.; Sun, K.; Ding, L.; Ren, G.; Wang, Z.; Mani, S. Plant flavonol isorhamnetin attenuates chemically induced inflammatory bowel disease via a PXR-dependent pathway. J. Nutr. Biochem. 2014, 25, 923–933. [Google Scholar] [CrossRef] [Green Version]
- Imran, M.; Rauf, A.; Abu-Izneid, T.; Nadeem, M.; Shariati, M.A.; Khan, I.A.; Imran, A.; Orhan, I.E.; Rizwan, M.; Atif, M.; et al. Luteolin, a flavonoid, as an anticancer agent: A review. Biomed. Pharmacother. 2019, 112, 108612. [Google Scholar] [CrossRef]
- Lin, Y.; Shi, R.; Wang, X.; Shen, H.-M. Luteolin, a Flavonoid with Potential for Cancer Prevention and Therapy. Curr. Cancer Drug Targets 2008, 8, 634–646. [Google Scholar] [CrossRef]
- Bai, L.; Nong, Y.; Shi, Y.; Liu, M.; Yan, L.; Shang, J.; Huang, F.; Lin, Y.; Tang, H. Luteolin Inhibits Hepatitis B Virus Replication through Extracellular Signal-Regulated Kinase-Mediated Down-Regulation of Hepatocyte Nuclear Factor 4α Expression. Mol. Pharm. 2015, 13, 568–577. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Qian, S.; Qian, P.; Li, X. Antiviral activity of luteolin against Japanese encephalitis virus. Virus Res. 2016, 220, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Ma, L.; Wang, H.; Wu, S.; Huang, H.; Gu, Z.; Jiang, J.; Li, Y. Luteolin decreases the yield of influenza A virus in vitro by interfering with the coat protein I complex expression. J. Nat. Med. 2019, 73, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Mehla, R.; Bivalkar-Mehla, S.; Chauhan, A. A Flavonoid, Luteolin, Cripples HIV-1 by Abrogation of Tat Function. PLoS ONE 2011, 6, e27915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueda, H.; Yamazaki, C.; Yamazaki, M. Luteolin as an Anti-Inflammatory and Anti-Allergic Constituent of Perilla. Biol. Pharm. Bull. 2002, 25, 1197–1202. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-Y.; Peng, W.-H.; Tsai, K.-D.; Hsu, S.-L. Luteolin suppresses inflammation-associated gene expression by blocking NF-κB and AP-1 activation pathway in mouse alveolar macrophages. Life Sci. 2007, 81, 1602–1614. [Google Scholar] [CrossRef]
- Franza, L.; Carusi, V.; Nucera, E.; Pandolfi, F. Luteolin, inflammation and cancer: Special emphasis on gut microbiota. BioFactors 2021, 47, 181–189. [Google Scholar] [CrossRef]
- Theoharides, T.C. Luteolin as a therapeutic option for multiple sclerosis. J. Neuroinflamm. 2009, 6, 29. [Google Scholar] [CrossRef] [Green Version]
- Xia, N.; Chen, G.; Liu, M.; Ye, X.; Pan, Y.; Ge, J.; Mao, Y.; Wang, H.; Wang, J.; Xie, S. Anti-inflammatory effects of luteolin on experimental autoimmune thyroiditis in mice. Exp. Ther. Med. 2016, 12, 4049–4054. [Google Scholar] [CrossRef] [Green Version]
- Zeino, M.; Saeed, M.E.M.; Kadioglu, O.; Efferth, T. The ability of molecular docking to unravel the controversy and challenges related to P-glycoprotein—a well-known, yet poorly understood drug transporter. Investig. New Drugs 2014, 32, 618–625. [Google Scholar] [CrossRef]
- Abdelfatah, S.; Berg, A.; Böckers, M.; Efferth, T. A selective inhibitor of the Polo-box domain of Polo-like kinase 1 identified by virtual screening. J. Adv. Res. 2019, 16, 145–156. [Google Scholar] [CrossRef]
- Abdelfatah, S.; Fleischer, E.; Klinger, A.; Wong, V.K.W.; Efferth, T. Identification of inhibitors of the polo-box domain of polo-like kinase 1 from natural and semisynthetic compounds. Investig. New Drugs 2019, 38, 1–9. [Google Scholar] [CrossRef]
- Efferth, T. Complex Interactions between Phytochemicals. The Multi-Target Therapeutic Concept of Phytotherapy. Curr. Drug Targets 2011, 12, 122–132. [Google Scholar] [CrossRef]
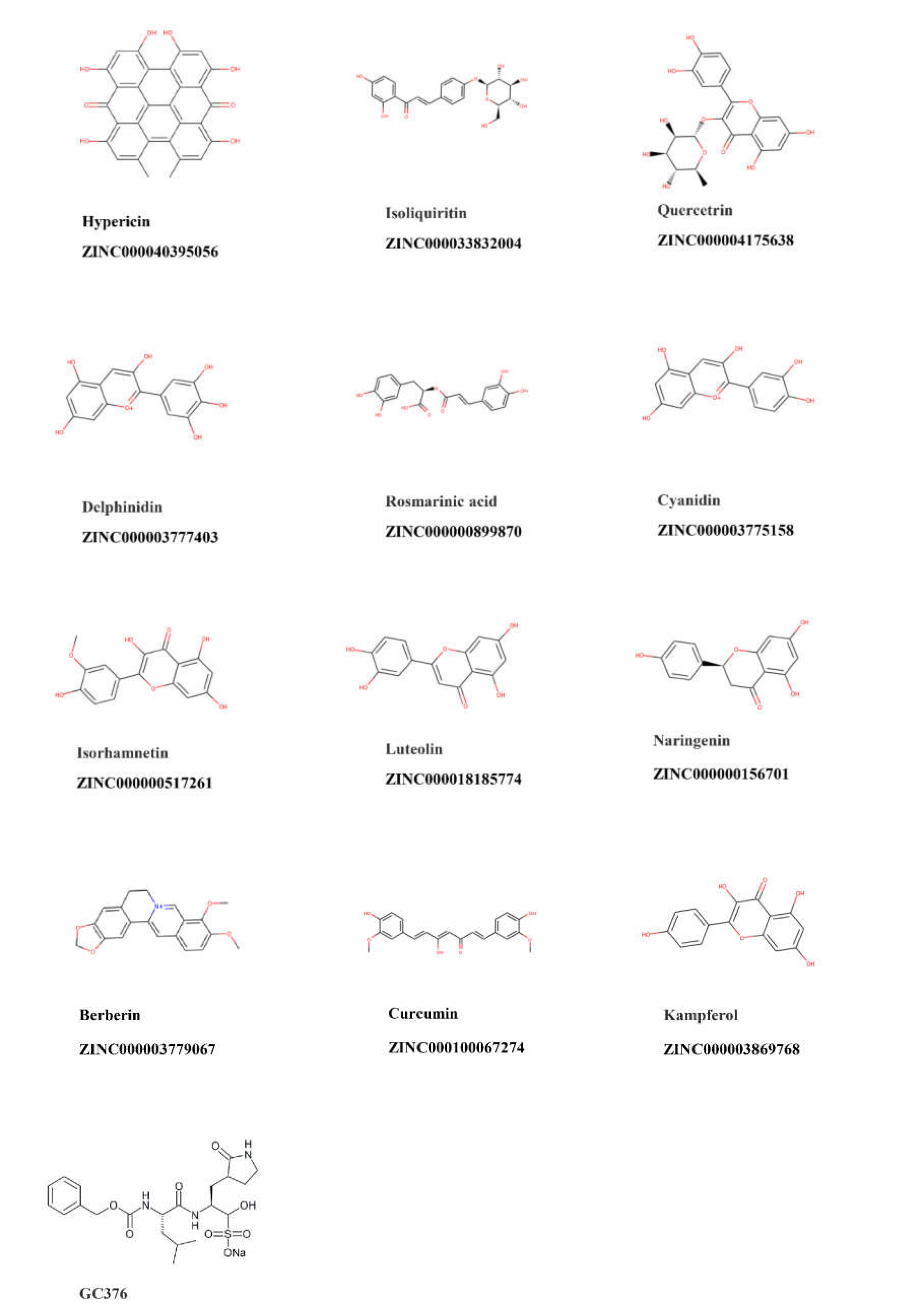







| Compound | PyRx Binding Affinity (kcal/mol) | Lowest Binding Energy (kcal/mol) | Predicted Ki (nM) |
|---|---|---|---|
| Hypericin | −8.70 | −12.44 ± <0.01 | 0.762 ± 1.34 |
| Curcumin | −7.90 | −12.48 ± 0.04 | 0.679 ± 11.7 |
| Isoliquiritin | −7.60 | −11.69 ± 0.02 | 2.64 ± 0.11 |
| Quercetin | −9.20 | −10.72 ± 0.03 | 13.90 ± 0.80 |
| Rosmarinic acid | −7.80 | −9.98 ± 0.08 | 42.62 ± 3.25 |
| Delphinidin | −8.50 | −9.23 ± <0.01 | 170.92 ± 0.06 |
| Cyanidin | −8.20 | −9.13 ± <0.01 | 203.87 ± 0.04 |
| Isorhamnetin | −8.20 | −9.06 ± <0.01 | 237.99 ± 11.59 |
| Luteolin | −8.10 | −9.01 ± <0.01 | 247.73 ± 1.21 |
| Kaempferol | −8.00 | −8.77 ± <0.01 | 375.12 ± 0.05 |
| Berberine | −8.10 | −8.07 ± <0.01 | 1210 ± 0.01 |
| Naringenin | −7.80 | −8.00 ± 0.04 | 1370 ± 0.1 |
| GC376 (positive control) | −8.00 | −12.58 ± 0.29 | 0.70 ± 0.42 |
| Human Coronavirus | % Identity with SARS-CoV-2 |
|---|---|
| SARS-CoV-1 | 96.08 |
| MERS-CoV | 50.83 |
| HcoV-NL63 | 49.17 |
| HcoV-OC43 | 48.51 |
| HcoV-HKU1 | 44.04 |
| HcoV-229E | 41.06 |
| Compound | IC50 Value (µM) (mean ± SD) | ||
|---|---|---|---|
| SARS-CoV-2 Mpro | SARS-CoV-1 Mpro | MERS-CoV Mpro | |
| Hypericin | 23.30 ± 1.21 | 19.43 ± 3.11 | 49.65 ± 5.41 |
| Rosmarinic acid | 9.43 ± 0.46 | n.a. | n.a. |
| Isorhamnetin | 8.42 ± 1.15 | 13.13 ± 1.78 | n.a. |
| Luteolin | 11.81 ± 1.27 | n.a. | n.a. |
| Compound | Kd Value (µM) | ||
|---|---|---|---|
| SARS-CoV-2 Mpro | SARS-CoV-1 Mpro | MERS-CoV Mpro | |
| Hypericin | 7.73 ± 6.50 | 25.49 ± 13.61 | 54.91 ± 13.80 |
| Rosmarinic acid | 15.47 ± 4.77 | n.a. | n.a. |
| Isorhamnetin | 4.37 ± 3.90 | 3.60 ± 2.60 | n.a. |
| Luteolin | 13.41 ± 2.70 | n.a. | n.a. |
| Compound | Lowest Binding Energy (kcal/mol) | ||||||
|---|---|---|---|---|---|---|---|
| SARS-CoV-2 | SARS-CoV-1 | MERS-CoV | HCoV-HKU1 | HCoV-NL63 | HCoV-OC43 | HCoV-229E | |
| Hypericin | −12.44 ± <0.01 | −11.53 ± 0.005 | −11.98 ± 1.77 | −9.11 ± < 0.01 | −9.77 ± 0.31 | −12.99 ± <0.01 | −10.65 ± <0.01 |
| Rosmarinic acid | −9.90 ± 0.08 | −9.80 ± 0.03 | −10.12 ± 0.21 | −10.48 ± 0.31 | −10.18 ± 0.11 | −10.06 ± 0.06 | −10.61 ± 0.05 |
| Isorhamnetin | −9.06 ± <0.01 | −8.83 ± <0.01 | −8.59 ± <0.01 | −8.50 ± <0.01 | −8.57 ± <0.01 | −8.33 ± <0.01 | −8.19 ± 0.01 |
| Luteolin | −9.01 ± <0.01 | −7.66 ± <0.01 | −7.67 ± 0.06 | −7.65 ± <0.01 | −9.25 ± <0.01 | −8.21 ± <0.01 | −8.02 ± <0.01 |
| GC376 (positive control) | −12.58 ± 0.29 | −12.17 ± 0.27 | −13.65 ± 0.44 | −12.78 ± 0.5 | −11.04 ± 0.09 | −12.28 ± 0.05 | −11.52 ± 0.09 |
| Compound | CC50 Value (µM) |
|---|---|
| (mean ± SD) | |
| Hypericin | 55.46 ± 2.2 |
| Isorhamnetin | 36.80 ± 3.4 |
| Rosmarinic acid | n.a. |
| Luteolin | n.a. |
| Compound | Therapeutic Index | ||
|---|---|---|---|
| SARS-CoV-2 Mpro | SARS-CoV-1 Mpro | MERS-CoV Mpro | |
| Hypericin | 2.38 | 2.85 | 1.11 |
| Isorhamnetin | 4.37 | 2.80 | n.a. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahhamzehei, N.; Abdelfatah, S.; Efferth, T. In Silico and In Vitro Identification of Pan-Coronaviral Main Protease Inhibitors from a Large Natural Product Library. Pharmaceuticals 2022, 15, 308. https://doi.org/10.3390/ph15030308
Shahhamzehei N, Abdelfatah S, Efferth T. In Silico and In Vitro Identification of Pan-Coronaviral Main Protease Inhibitors from a Large Natural Product Library. Pharmaceuticals. 2022; 15(3):308. https://doi.org/10.3390/ph15030308
Chicago/Turabian StyleShahhamzehei, Nasim, Sara Abdelfatah, and Thomas Efferth. 2022. "In Silico and In Vitro Identification of Pan-Coronaviral Main Protease Inhibitors from a Large Natural Product Library" Pharmaceuticals 15, no. 3: 308. https://doi.org/10.3390/ph15030308
APA StyleShahhamzehei, N., Abdelfatah, S., & Efferth, T. (2022). In Silico and In Vitro Identification of Pan-Coronaviral Main Protease Inhibitors from a Large Natural Product Library. Pharmaceuticals, 15(3), 308. https://doi.org/10.3390/ph15030308








