Targeting Spike Glycoprotein S1 Mediated by NLRP3 Inflammasome Machinery and the Cytokine Releases in A549 Lung Epithelial Cells by Nanocurcumin
Abstract
:1. Introduction
2. Results
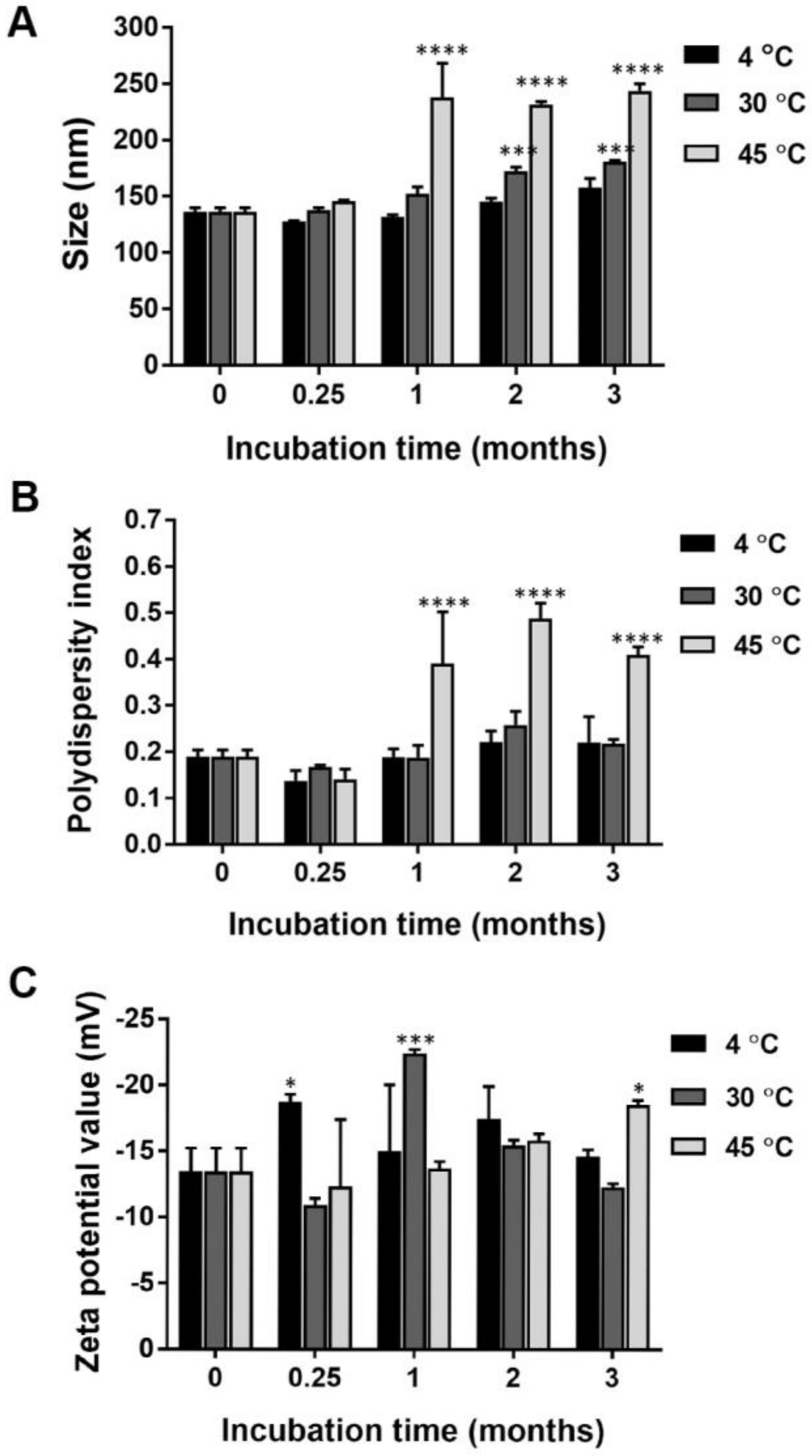
2.1. Particle Size, Size Distribution, and Surface Charge of Nanocurcumin
2.2. Encapsulation Efficiency of Nanocurcumin
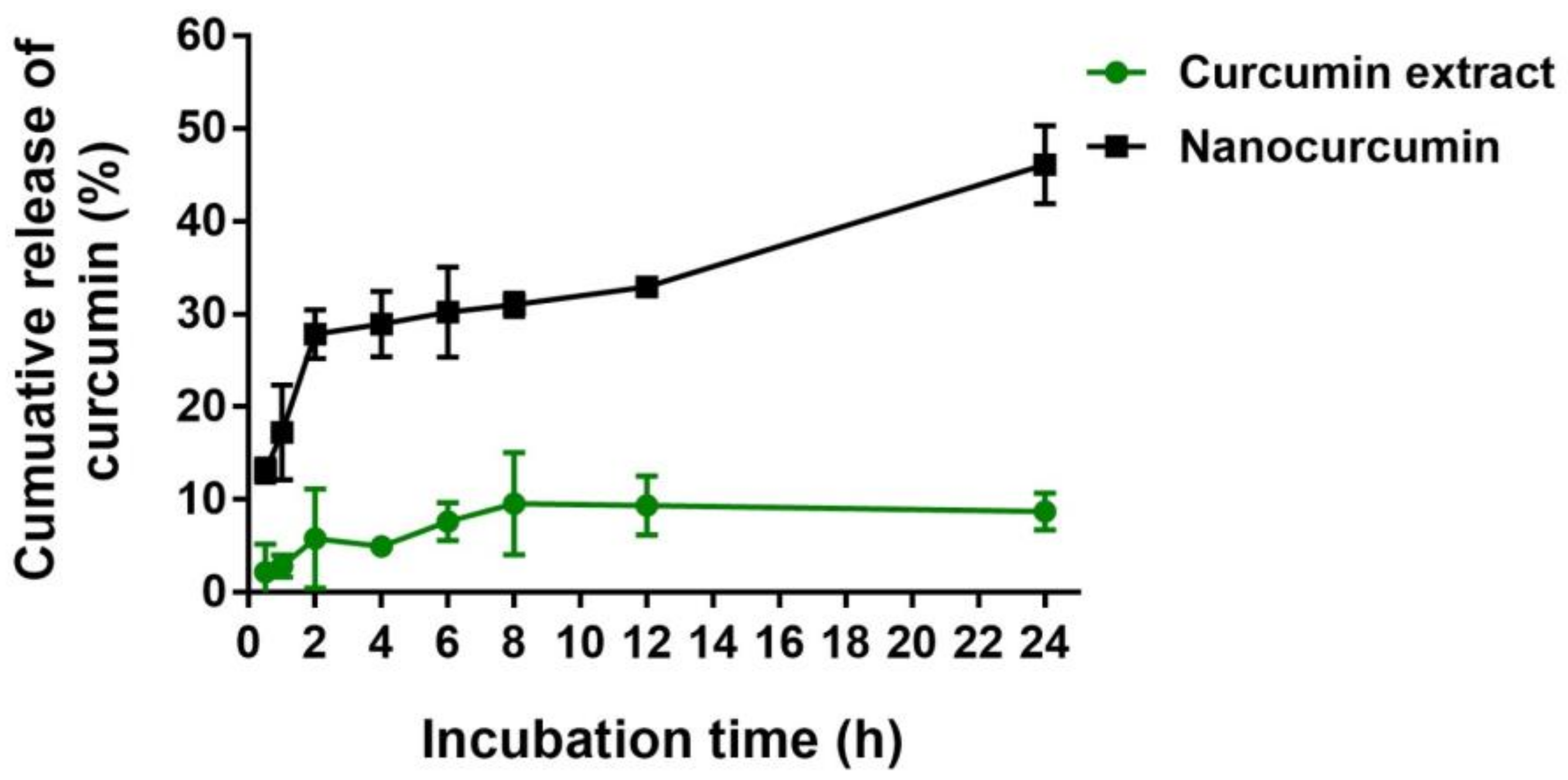
2.3. In Vitro Release of Curcumin from the Nanocurcumin
2.4. Chemical Stability of Nanocurcumin
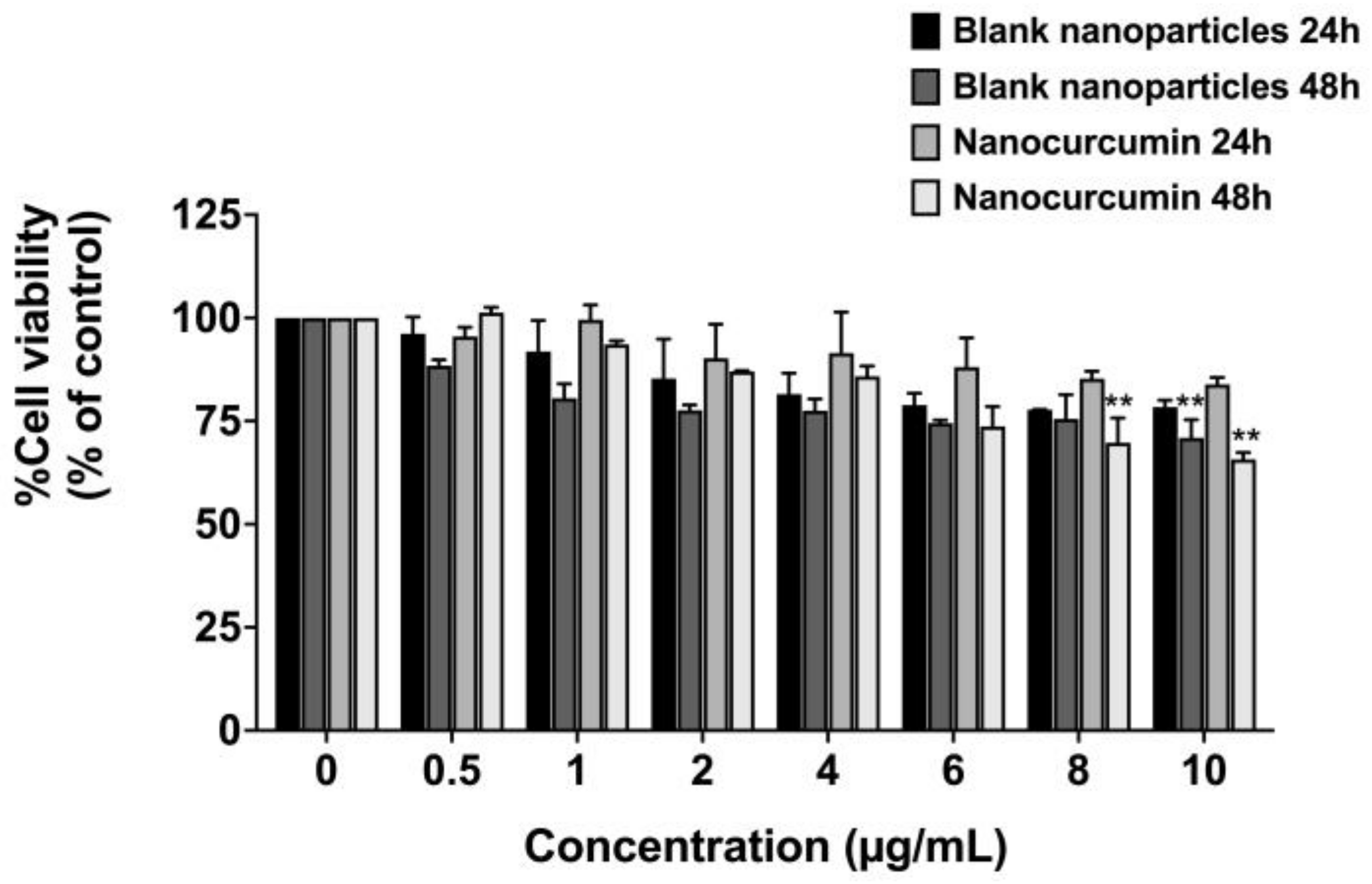
2.5. Effect of Nanocurcumin on Cell Viability of A549 Lung Epithelial Cells
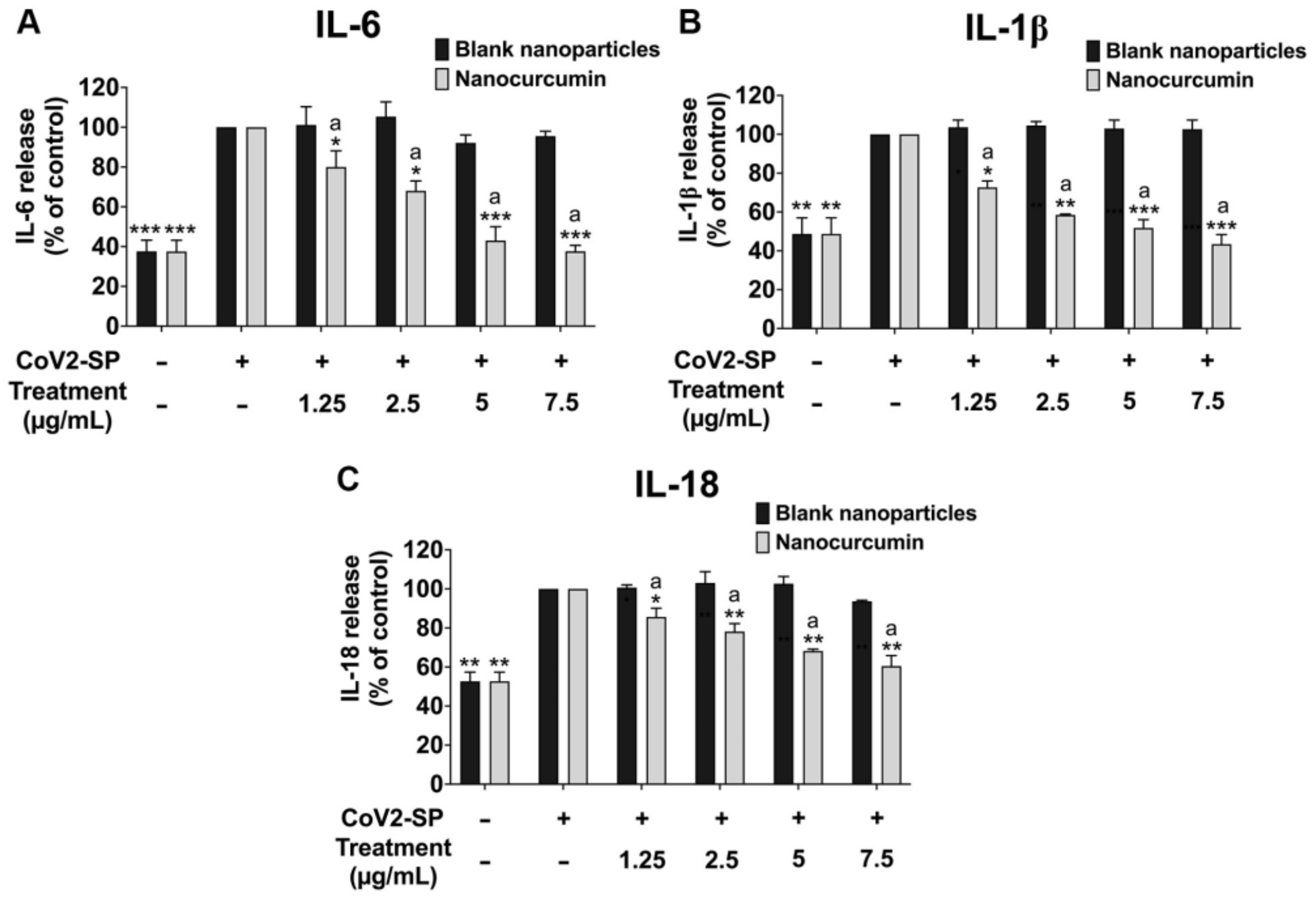
2.6. Effects of Nanocurcumin on the Inhibition of Pro-Inflammatory Cytokine Secretions and Inflammatory Gene Expressions in CoV2-SP-Induced A549 Cells
2.7. Inhibitory Effects of Nanocurcumin on NLRP3 Inflammasome Pathway in CoV2-SP-Induced A549 Cells
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Preparation and Characterization of Nanocurcumin
4.3. Physical and Chemical Stability of Nanocurcumin
4.4. In Vitro Release of Curcumin
4.5. Identification and Quantification of Curcumin Content in Nanocurcumin Using High-Performance Liquid Chromatography (HPLC)
4.6. Cell Viability Testing
4.7. Inhibition of Cytokine Releases by ELISA Test
4.8. Expression of IL-6, IL-1β, IL-18, and NLRP3 Genes by RT- qPCR Analysis
4.9. Western Blot Analysis
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Proal, A.D.; VanElzakker, M.B. Long COVID or post-acute sequelae of COVID-19 (PASC): An overview of biological factors that may contribute to persistent symptoms. Front. Microbiol. 2021, 12, 1494. [Google Scholar] [CrossRef]
- Davis, H.E.; McCorkell, L.; Vogel, J.M.; Topol, E.J. Long COVID: Major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 2023, 21, 133–146. [Google Scholar] [CrossRef]
- Bormann, M.; Alt, M.; Schipper, L.; van de Sand, L.; Le-Trilling, V.T.K.; Rink, L.; Heinen, N.; Madel, R.J.; Otte, M.; Wuensch, K. Turmeric root and its bioactive ingredient curcumin effectively neutralize SARS-CoV-2 in vitro. Viruses 2021, 13, 1914. [Google Scholar] [CrossRef] [PubMed]
- Rattis, B.A.; Ramos, S.G.; Celes, M. Curcumin as a Potential Treatment for COVID-19. Front. Pharmacol. 2021, 12, 1068. [Google Scholar] [CrossRef] [PubMed]
- Vahedian-Azimi, A.; Abbasifard, M.; Rahimi-Bashar, F.; Guest, P.C.; Majeed, M.; Mohammadi, A.; Banach, M.; Jamialahmadi, T.; Sahebkar, A. Effectiveness of curcumin on outcomes of hospitalized COVID-19 patients: A systematic review of clinical trials. Nutrients 2022, 14, 256. [Google Scholar] [CrossRef]
- Tahmasebi, S.; El-Esawi, M.A.; Mahmoud, Z.H.; Timoshin, A.; Valizadeh, H.; Roshangar, L.; Varshoch, M.; Vaez, A.; Aslani, S.; Navashenaq, J.G. Immunomodulatory effects of nanocurcumin on Th17 cell responses in mild and severe COVID-19 patients. J. Cell. Physiol. 2021, 236, 5325–5338. [Google Scholar] [CrossRef]
- Pawar, K.S.; Mastud, R.N.; Pawar, S.K.; Pawar, S.S.; Bhoite, R.R.; Bhoite, R.R.; Kulkarni, M.V.; Deshpande, A.R. Oral curcumin with piperine as adjuvant therapy for the treatment of COVID-19: A randomized clinical trial. Front. Pharmacol. 2021, 12, 1056. [Google Scholar] [CrossRef] [PubMed]
- Dourado, D.; Freire, D.T.; Pereira, D.T.; Amaral-Machado, L.; Alencar, É.N.; de Barros, A.L.B.; Egito, E.S.T. Will curcumin nanosystems be the next promising antiviral alternatives in COVID-19 treatment trials? Biomed. Pharmacother. 2021, 139, 111578. [Google Scholar] [CrossRef]
- Saber-Moghaddam, N.; Salari, S.; Hejazi, S.; Amini, M.; Taherzadeh, Z.; Eslami, S.; Rezayat, S.M.; Jaafari, M.R.; Elyasi, S. Oral nano-curcumin formulation efficacy in management of mild to moderate hospitalized coronavirus disease-19 patients: An open label nonrandomized clinical trial. Phytother. Res. 2021, 35, 2616–2623. [Google Scholar] [CrossRef]
- Valizadeh, H.; Abdolmohammadi-Vahid, S.; Danshina, S.; Gencer, M.Z.; Ammari, A.; Sadeghi, A.; Roshangar, L.; Aslani, S.; Esmaeilzadeh, A.; Ghaebi, M. Nano-curcumin therapy, a promising method in modulating inflammatory cytokines in COVID-19 patients. Int. Immunopharmacol. 2020, 89, 107088. [Google Scholar] [CrossRef]
- Dei Cas, M.; Ghidoni, R. Dietary curcumin: Correlation between bioavailability and health potential. Nutrients 2019, 11, 2147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, V.K.; Prateeksha; Singh, S.P.; Singh, B.N.; Rao, C.V.; Barik, S.K. Nanocurcumin Potently Inhibits SARS-CoV-2 Spike Protein-Induced Cytokine Storm by Deactivation of MAPK/NF-κB Signaling in Epithelial Cells. ACS Appl. Bio Mater. 2022, 5, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Hamming, I.; Timens, W.; Bulthuis, M.; Lely, A.; Navis, G.v.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. J. Pathol. Soc. Great Br. Irel. 2004, 203, 631–637. [Google Scholar] [CrossRef]
- Harcourt, J.; Tamin, A.; Lu, X.; Kamili, S.; Sakthivel, S.K.; Murray, J.; Queen, K.; Tao, Y.; Paden, C.R.; Zhang, J. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg. Infect. Dis. 2020, 26, 1266–1273. [Google Scholar] [CrossRef] [PubMed]
- Gamage, A.M.; Tan, K.S.; Chan, W.O.; Liu, J.; Tan, C.W.; Ong, Y.K.; Thong, M.; Andiappan, A.K.; Anderson, D.E.; Wang, D.Y. Infection of human Nasal Epithelial Cells with SARS-CoV-2 and a 382-nt deletion isolate lacking ORF8 reveals similar viral kinetics and host transcriptional profiles. PLoS Pathog. 2020, 16, e1009130. [Google Scholar] [CrossRef] [PubMed]
- Lukassen, S.; Chua, R.L.; Trefzer, T.; Kahn, N.C.; Schneider, M.A.; Muley, T.; Winter, H.; Meister, M.; Veith, C.; Boots, A.W. SARS-CoV-2 receptor ACE 2 and TMPRSS 2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020, 39, e105114. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Ryu, G.; Shin, H.-W. SARS-CoV-2 infection of airway epithelial cells. Immune Netw. 2021, 21, e3. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [Green Version]
- Cahaya, S.T.H.D. Stability of curcumin in turmeric oleoresin-β-cyclodextrin inclusion complex during storage. Malays. J. Anal. Sci. 2009, 13, 1–5. [Google Scholar]
- Gholami, L.; Momtazi-Borojeni, A.A.; Malaekeh-Nikouei, B.; Nikfar, B.; Amanolahi, F.; Mohammadi, A.; Oskuee, R.K. Selective Cellular Uptake and Cytotoxicity of Curcumin-encapsulated SPC and HSPC Liposome Nanoparticles on Human Bladder Cancer Cells. Curr. Pharm. Des. 2023, 29, 1046–1058. [Google Scholar] [PubMed]
- Chiangnoon, R.; Samee, W.; Uttayarat, P.; Jittachai, W.; Ruksiriwanich, W.; Sommano, S.R.; Athikomkulchai, S.; Chittasupho, C. Phytochemical Analysis, Antioxidant, and Wound Healing Activity of Pluchea indica L.(Less) Branch Extract Nanoparticles. Molecules 2022, 27, 635. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Jutková, A.; Šrámková, P.; Lenkavská, L.; Huntošová, V.; Chorvát, D.; Miškovský, P.; Jancura, D.; Kronek, J. Unravelling the excellent chemical stability and bioavailability of solvent responsive curcumin-loaded 2-ethyl-2-oxazoline-grad-2-(4-dodecyloxyphenyl)-2-oxazoline copolymer nanoparticles for drug delivery. Biomacromolecules 2018, 19, 2459–2471. [Google Scholar] [CrossRef]
- Rodrigues, T.S.; de Sá, K.S.; Ishimoto, A.Y.; Becerra, A.; Oliveira, S.; Almeida, L.; Gonçalves, A.V.; Perucello, D.B.; Andrade, W.A.; Castro, R. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J. Exp. Med. 2021, 218, e20201707. [Google Scholar] [CrossRef]
- Dos Santos Bezerra, R.; Valença, I.N.; de Cassia Ruy, P.; Ximenez, J.P.; da Silva Junior, W.A.; Covas, D.T.; Kashima, S.; Slavov, S.N. The novel coronavirus SARS-CoV-2: From a zoonotic infection to coronavirus disease 2019. J. Med. Virol. 2020, 92, 2607–2615. [Google Scholar] [CrossRef]
- Desforges, M.; Le Coupanec, A.; Dubeau, P.; Bourgouin, A.; Lajoie, L.; Dubé, M.; Talbot, P.J. Human coronaviruses and other respiratory viruses: Underestimated opportunistic pathogens of the central nervous system? Viruses 2019, 12, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Churilov, L.P.; Normatov, M.G.; Utekhin, V.J. Molecular Mimicry between SARS-CoV-2 and Human Endocrinocytes: A Prerequisite of Post-COVID-19 Endocrine Autoimmunity? Pathophysiology 2022, 29, 486–494. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef] [Green Version]
- Schroeder, J.T.; Bieneman, A.P. The S1 subunit of the SARS-CoV-2 spike protein activates human monocytes to produce cytokines linked to COVID-19: Relevance to galectin-3. Front. Immunol. 2022, 13, 1238. [Google Scholar] [CrossRef]
- Semmarath, W.; Mapoung, S.; Umsumarng, S.; Arjsri, P.; Srisawad, K.; Thippraphan, P.; Yodkeeree, S.; Dejkriengkraikul, P. Cyanidin-3-O-glucoside and peonidin-3-O-glucoside-rich fraction of black rice germ and bran suppresses inflammatory responses from SARS-CoV-2 spike glycoprotein S1-induction in vitro in A549 lung cells and THP-1 macrophages via inhibition of the NLRP3 inflammasome pathway. Nutrients 2022, 14, 2738. [Google Scholar]
- Arjsri, P.; Srisawad, K.; Mapoung, S.; Semmarath, W.; Thippraphan, P.; Umsumarng, S.; Yodkeeree, S.; Dejkriengkraikul, P. Hesperetin from Root Extract of Clerodendrum petasites S. Moore Inhibits SARS-CoV-2 Spike Protein S1 Subunit-Induced NLRP3 Inflammasome in A549 Lung Cells via Modulation of the Akt/MAPK/AP-1 Pathway. Int. J. Mol. Sci. 2022, 23, 10346. [Google Scholar] [CrossRef] [PubMed]
- Dissook, S.; Umsumarng, S.; Mapoung, S.; Semmarath, W.; Arjsri, P.; Srisawad, K.; Dejkriengkraikul, P. Luteolin-rich fraction from Perilla frutescens seed meal inhibits spike glycoprotein S1 of SARS-CoV-2-induced NLRP3 inflammasome lung cell inflammation via regulation of JAK1/STAT3 pathway: A potential anti-inflammatory compound against inflammation-induced long-COVID. Front. Med. 2022, 9, 1072056. [Google Scholar]
- Suresh, M.V.; Francis, S.; Aktay, S.; Kralovich, G.; Raghavendran, K. Therapeutic potential of curcumin in ARDS and COVID-19. Clin. Exp. Pharmacol. Physiol. 2023, 50, 267–276. [Google Scholar] [CrossRef]
- Kushwaha, A.D.; Mishra, K.; Singh, M.; Ganju, L.; Saraswat, D. Nanocurcumin formulation: A possible therapeutic agent for post COVID inflammatory syndrome. Immunopharmacol. Immunotoxicol. 2022, 44, 141–146. [Google Scholar] [CrossRef]
- Ehi-Eromosele, C. The effect of polyethylene glycol (PEG) coating on the magneto-structural properties and colloidal stability of CO0. 8Mg0. 2Fe2O4 nanoparticles for potential biomedical applications. Dig. J. Nanomater. Biostruct. 2016, 11, 7–14. [Google Scholar]
- Hasan, M.; Elkhoury, K.; Belhaj, N.; Kahn, C.; Tamayol, A.; Barberi-Heyob, M.; Arab-Tehrany, E.; Linder, M. Growth-inhibitory effect of chitosan-coated liposomes encapsulating curcumin on MCF-7 breast cancer cells. Mar. Drugs 2020, 18, 217. [Google Scholar] [CrossRef]
- Gao, X.; Yi, X.; Liu, Z.; Dong, X.; Xia, G.; Zhang, X.; Shen, X. Comparative study on curcumin loaded in golden pompano (trachinotus blochii) head phospholipid and soybean lecithin liposomes: Preparation, characteristics and anti-inflammatory properties. Molecules 2021, 26, 2328. [Google Scholar] [CrossRef]
- Bonaccorso, A.; Pellitteri, R.; Ruozi, B.; Puglia, C.; Santonocito, D.; Pignatello, R.; Musumeci, T. Curcumin loaded polymeric vs. lipid nanoparticles: Antioxidant effect on normal and hypoxic olfactory ensheathing cells. Nanomaterials 2021, 11, 159. [Google Scholar] [CrossRef]
- Mazumder, S.; Dewangan, A.K.; Pavurala, N. Enhanced dissolution of poorly soluble antiviral drugs from nanoparticles of cellulose acetate based solid dispersion matrices. Asian J. Pharm. Sci 2017, 12, 532–541. [Google Scholar] [CrossRef]
- Müller, R.H.; Peters, K. Nanosuspensions for the formulation of poorly soluble drugs: I. Preparation by a size-reduction technique. Int. J. Pharm. 1998, 160, 229–237. [Google Scholar] [CrossRef]
- Jia, L. Nanoparticle formulation increases oral bioavailability of poorly soluble drugs: Approaches, experimental evidences and theory. Curr. Nanosci. 2005, 1, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Shafiei, M.S.; Longoria, C.; Schoggins, J.W.; Savani, R.C.; Zaki, H. SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-κB pathway. eLife 2021, 10, e68563. [Google Scholar] [CrossRef] [PubMed]
- Barilli, A.; Visigalli, R.; Ferrari, F.; Bianchi, M.G.; Dall’Asta, V.; Rotoli, B.M. Immune-mediated inflammatory responses of alveolar epithelial cells: Implications for covid-19 lung pathology. Biomedicines 2022, 10, 618. [Google Scholar] [CrossRef]
- Kelley, N.; Jeltema, D.; Duan, Y.; He, Y. The NLRP3 inflammasome: An overview of mechanisms of activation and regulation. Int. J. Mol. Sci. 2019, 20, 3328. [Google Scholar] [CrossRef] [Green Version]
- Jo, E.-K.; Kim, J.K.; Shin, D.-M.; Sasakawa, C. Molecular mechanisms regulating NLRP3 inflammasome activation. Cell. Mol. Immunol. 2016, 13, 148–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dos Santos, G.; Kutuzov, M.A.; Ridge, K.M. The inflammasome in lung diseases. Am. J. Physiol. Lung Cell Mol. Physiol. 2012, 303, 627–633. [Google Scholar] [CrossRef] [Green Version]
- Aboudounya, M.M.; Heads, R.J. COVID-19 and toll-like receptor 4 (TLR4): SARS-CoV-2 may bind and activate TLR4 to increase ACE2 expression, facilitating entry and causing hyperinflammation. Mediat. Inflamm. 2021, 2021, 8874339. [Google Scholar] [CrossRef]
- Sohn, K.M.; Lee, S.-G.; Kim, H.J.; Cheon, S.; Jeong, H.; Lee, J.; Kim, I.S.; Silwal, P.; Kim, Y.J.; Paik, S. COVID-19 patients upregulate toll-like receptor 4-mediated inflammatory signaling that mimics bacterial sepsis. J. Korean Med. Sci. 2020, 35, e343. [Google Scholar] [CrossRef]
- Manik, M.; Singh, R.K. Role of toll-like receptors in modulation of cytokine storm signaling in SARS-CoV-2-induced COVID-19. J. Med. Virol. 2022, 94, 869–877. [Google Scholar] [CrossRef]
- Hemmat, N.; Asadzadeh, Z.; Ahangar, N.K.; Alemohammad, H.; Najafzadeh, B.; Derakhshani, A.; Baghbanzadeh, A.; Baghi, H.B.; Javadrashid, D.; Najafi, S. The roles of signaling pathways in SARS-CoV-2 infection; lessons learned from SARS-CoV and MERS-CoV. Arch. Virol. 2021, 166, 675–696. [Google Scholar] [CrossRef]
- Boonrueang, N.; Kiattisin, K.; Ampasavate, C. Effects of Antioxidants and Sun-Screening Agent on the Photostability of Curcumin. In Proceedings of the 10th International Conference on Nutrition and Physical Activity in Ageing, Obesity, and Cancer, Chonburi, Thailand, 18–20 December 2019; pp. 32–42. [Google Scholar]
- Pradhan, N.; Parbin, S.; Kausar, C.; Kar, S.; Mawatwal, S.; Das, L.; Deb, M.; Sengupta, D.; Dhiman, R.; Patra, S.K. Paederia foetida induces anticancer activity by modulating chromatin modification enzymes and altering pro-inflammatory cytokine gene expression in human prostate cancer cells. Food Chem. Toxicol. 2019, 130, 161–173. [Google Scholar] [CrossRef]
- Plotnikova, M.A.; Klotchenko, S.A.; Vasin, A.V. Development of a multiplex quantitative PCR assay for the analysis of human cytokine gene expression in influenza A virus-infected cells. J. Immunol. Methods 2016, 430, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kong, H.; Zeng, X.; Liu, W.; Wang, Z.; Yan, X.; Wang, H.; Xie, W. Activation of NLRP3 inflammasome enhances the proliferation and migration of A549 lung cancer cells. Oncol. Rep. 2016, 35, 2053–2064. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Time (min) | A | B | C |
|---|---|---|---|
| 0 | 8.25 | 83.50 | 8.25 |
| 7 | 8.25 | 83.50 | 8.25 |
| 12 | 55.00 | 40.00 | 5.00 |
| 27 | 55.00 | 40.00 | 5.00 |
| 28 | 8.25 | 83.50 | 8.25 |
| 30 | 8.25 | 83.50 | 8.25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chittasupho, C.; Srisawad, K.; Arjsri, P.; Phongpradist, R.; Tingya, W.; Ampasavate, C.; Dejkriengkraikul, P. Targeting Spike Glycoprotein S1 Mediated by NLRP3 Inflammasome Machinery and the Cytokine Releases in A549 Lung Epithelial Cells by Nanocurcumin. Pharmaceuticals 2023, 16, 862. https://doi.org/10.3390/ph16060862
Chittasupho C, Srisawad K, Arjsri P, Phongpradist R, Tingya W, Ampasavate C, Dejkriengkraikul P. Targeting Spike Glycoprotein S1 Mediated by NLRP3 Inflammasome Machinery and the Cytokine Releases in A549 Lung Epithelial Cells by Nanocurcumin. Pharmaceuticals. 2023; 16(6):862. https://doi.org/10.3390/ph16060862
Chicago/Turabian StyleChittasupho, Chuda, Kamonwan Srisawad, Punnida Arjsri, Rungsinee Phongpradist, Wipawan Tingya, Chadarat Ampasavate, and Pornngarm Dejkriengkraikul. 2023. "Targeting Spike Glycoprotein S1 Mediated by NLRP3 Inflammasome Machinery and the Cytokine Releases in A549 Lung Epithelial Cells by Nanocurcumin" Pharmaceuticals 16, no. 6: 862. https://doi.org/10.3390/ph16060862
APA StyleChittasupho, C., Srisawad, K., Arjsri, P., Phongpradist, R., Tingya, W., Ampasavate, C., & Dejkriengkraikul, P. (2023). Targeting Spike Glycoprotein S1 Mediated by NLRP3 Inflammasome Machinery and the Cytokine Releases in A549 Lung Epithelial Cells by Nanocurcumin. Pharmaceuticals, 16(6), 862. https://doi.org/10.3390/ph16060862









