β-Elemene Reverses Gefitinib Resistance in NSCLC Cells by Inhibiting lncRNA H19-Mediated Autophagy
Abstract
1. Introduction
2. Results
2.1. Gefitinib Resistance Linked to Dysregulated Autophagy and Elevated lncRNA H19 Levels
2.2. Synergistic Effects of β-Elemene and Gefitinib on Apoptosis in Drug-Resistant Cells
2.3. Synergistic Antitumor Activity of β-Elemene and Gefitinib in Lung Cancer Models In Vivo
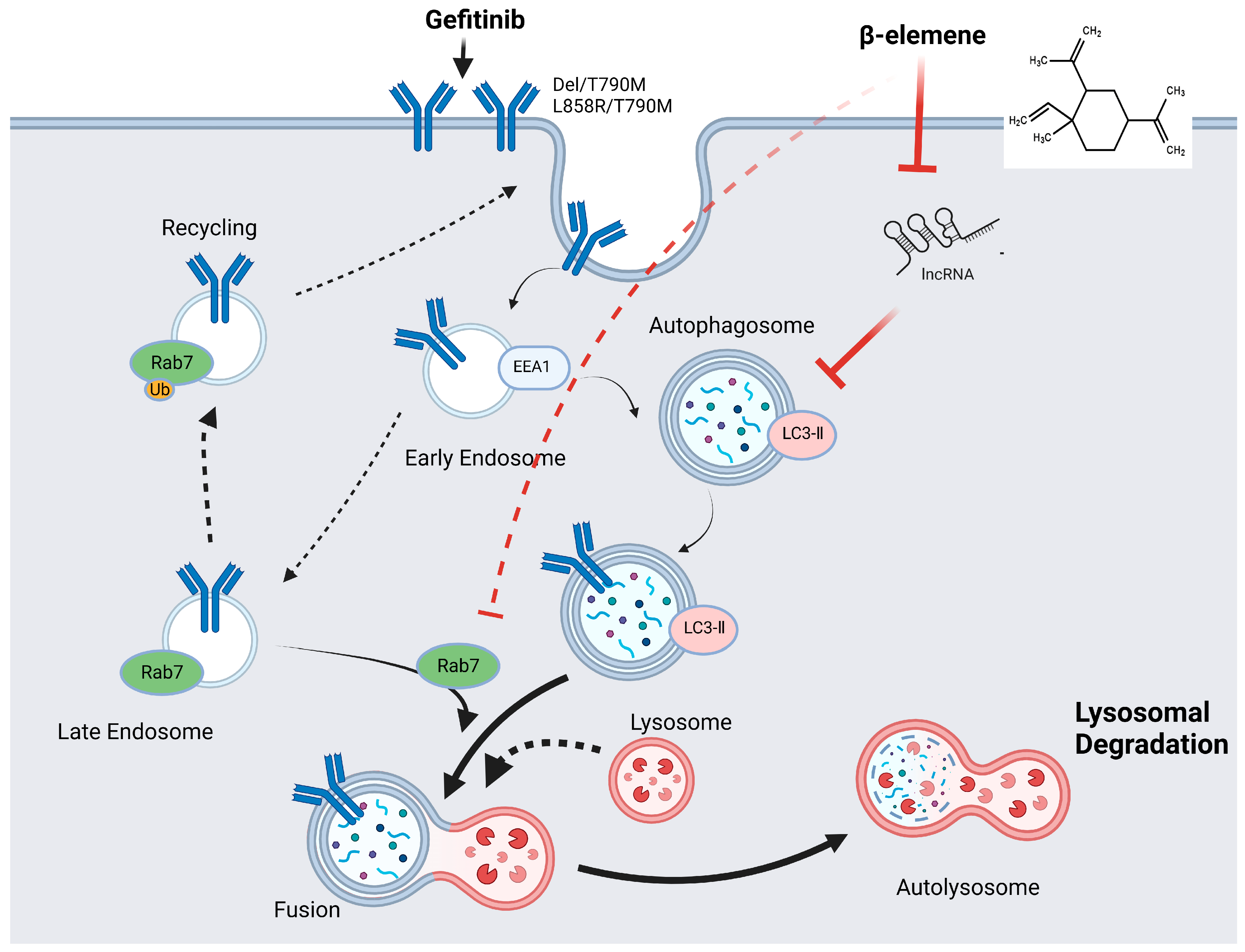
2.4. β-Elemene Counteracts Gefitinib-Induced Protective Autophagy in Lung Cancer Cells
2.5. Elucidating β-Elemene’s Modulation of lncRNA H19, Autophagy, and Gefitinib Resistance Dynamics
2.6. β-Elemene Inhibits EGFR Degradation Mediated by lncRNA H19-Associated Autophagy
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Reagentd and Antibodies
4.3. Cell Viability Assay
4.4. Cell Growth Evaluation
4.5. Apoptosis Analysis
4.6. Western Blot Analysis
4.7. Quantitative Real-Time RT-PCR
4.8. Measurement of Autophagic Flux
4.9. Hematoxylin-Eosin Staining
4.10. Immunofluorescence Assay
4.11. Lentivirus and Plasmid Transfection
4.12. Tumor Xenograft Model In Vivo
4.13. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer Statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Yatabe, Y.; Kerr, K.M.; Utomo, A.; Rajadurai, P.; Tran, V.K.; Du, X.; Chou, T.-Y.; Enriquez, M.L.D.; Lee, G.K.; Iqbal, J.; et al. Egfr Mutation Testing Practices within the Asia Pacific Region: Results of a Multicenter Diagnostic Survey. J. Thorac. Oncol. 2015, 10, 438–445. [Google Scholar] [CrossRef] [PubMed]
- Westover, D.; Zugazagoitia, J.; Cho, B.C.; Lovly, C.M.; Paz-Ares, L. Mechanisms of Acquired Resistance to First- and Second-Generation Egfr Tyrosine Kinase Inhibitors. Ann. Oncol. 2018, 29, i10–i19. [Google Scholar] [CrossRef] [PubMed]
- Noronha, V.; Patil, V.M.; Joshi, A.; Menon, N.; Chougule, A.; Mahajan, A.; Janu, A.; Purandare, N.; Kumar, R.; More, S.; et al. Gefitinib versus Gefitinib Plus Pemetrexed and Carboplatin Chemotherapy in Egfr-Mutated Lung Cancer. J. Clin. Oncol. 2020, 38, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-G.; Shih, J.-Y. Management of Acquired Resistance to Egfr Tki-Targeted Therapy in Advanced Non-Small Cell Lung Cancer. Mol. Cancer 2018, 17, 38. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef] [PubMed]
- Denton, D.; Kumar, S. Autophagy-Dependent Cell Death. Cell Death Differ. 2019, 26, 605–616. [Google Scholar] [CrossRef]
- Hu, H.; Zhang, X.-W.; Li, L.; Hu, M.-N.; Hu, W.-Q.; Zhang, J.-Y.; Miao, X.-K.; Yang, W.-L.; Mou, L.-Y. Inhibition of Autophagy by Yc-1 Promotes Gefitinib Induced Apoptosis by Targeting Foxo1 in Gefitinib-Resistant Nsclc Cells. Eur. J. Pharmacol. 2021, 908, 174346. [Google Scholar] [CrossRef]
- Chen, P.; Huang, H.-P.; Wang, Y.; Jin, J.; Long, W.-G.; Chen, K.; Zhao, X.-H.; Chen, C.-G.; Li, J. Curcumin Overcome Primary Gefitinib Resistance in Non-Small-Cell Lung Cancer Cells through Inducing Autophagy-Related Cell Death. J. Exp. Clin. Cancer Res. 2019, 38, 254. [Google Scholar] [CrossRef]
- Liu, S.; Li, Q.; Li, G.; Zhang, Q.; Zhuo, L.; Han, X.; Zhang, M.; Chen, X.; Pan, T.; Yan, L.; et al. The Mechanism of M6a Methyltransferase Mettl3-Mediated Autophagy in Reversing Gefitinib Resistance in Nsclc Cells by β-Elemene. Cell Death Dis. 2020, 11, 969. [Google Scholar] [CrossRef]
- Tang, C.-Y.; Zhu, L.-X.; Yu, J.-D.; Chen, Z.; Gu, M.-C.; Mu, C.-F.; Liu, Q.; Xiong, Y. Effect of β-Elemene on the Kinetics of Intracellular Transport of D-Luciferin Potassium Salt (Abc Substrate) in Doxorubicin-Resistant Breast Cancer Cells and the Associated Molecular Mechanism. Eur. J. Pharm. Sci. 2018, 120, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Jamwal, R. Bioavailable Curcumin Formulations: A Review of Pharmacokinetic Studies in Healthy Volunteers. J. Integr. Med. 2018, 16, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, Z.; Sui, X.; Wu, Q.; Wang, J.; Xu, C. Elemene Injection as Adjunctive Treatment to Platinum-Based Chemotherapy in Patients with Stage Iii/Iv Non-Small Cell Lung Cancer: A Meta-Analysis Following the Prisma Guidelines. Phytomedicine 2019, 59, 152787. [Google Scholar] [CrossRef]
- Jiang, Z.; Jacob, J.A.; Loganathachetti, D.S.; Nainangu, P.; Chen, B. β-Elemene: Mechanistic Studies on Cancer Cell Interaction and Its Chemosensitization Effect. Front. Pharmacol. 2017, 8, 105. [Google Scholar] [CrossRef]
- Zhai, B.; Zhang, N.; Han, X.; Li, Q.; Zhang, M.; Chen, X.; Li, G.; Zhang, R.; Chen, P.; Wang, W.; et al. Molecular Targets of β-Elemene, a Herbal Extract Used in Traditional Chinese Medicine, and Its Potential Role in Cancer Therapy: A Review. Biomed. Pharmacother. 2019, 114, 108812. [Google Scholar] [CrossRef]
- Xu, C.; Jiang, Z.-B.; Shao, L.; Zhao, Z.-M.; Fan, X.-X.; Sui, X.; Yu, L.-L.; Wang, X.-R.; Zhang, R.-N.; Wang, W.-J.; et al. β-Elemene Enhances Erlotinib Sensitivity through Induction of Ferroptosis by Upregulating Lncrna H19 in Egfr-Mutant Non-Small Cell Lung Cancer. Pharmacol. Res. 2023, 191, 106739. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Pan, T.; Xiang, Y.; Zhang, M.; Feng, J.; Liu, S.; Duan, T.; Chen, P.; Zhai, B.; Chen, X.; et al. β-Elemene Reverses the Resistance of P53-Deficient Colorectal Cancer Cells to 5-Fluorouracil by Inducing Pro-Death Autophagy and Cyclin D3-Dependent Cycle Arrest. Front. Bioeng. Biotechnol. 2020, 8, 378. [Google Scholar] [CrossRef]
- Wang, H.; Ma, Y. β-Elemene Alleviates Cisplatin Resistance in Oral Squamous Cell Carcinoma Cell Via Inhibiting Jak2/Stat3 Pathway in Vitro and in Vivo. Cancer Cell Int. 2022, 22, 244. [Google Scholar] [CrossRef]
- Zhang, S.; Guo, W. β-Elemene Enhances the Sensitivity of Osteosarcoma Cells to Doxorubicin Via Downregulation of Peroxiredoxin-1. OncoTargets Ther. 2021, 14, 3599–3609. [Google Scholar] [CrossRef]
- Deng, M.; Liu, B.; Song, H.; Yu, R.; Zou, D.; Chen, Y.; Ma, Y.; Lv, F.; Xu, L.; Zhang, Z.; et al. β-Elemene Inhibits the Metastasis of Multidrug-Resistant Gastric Cancer Cells through Mir-1323/Cbl-B/Egfr Pathway. Phytomedicine 2020, 69, 153184. [Google Scholar] [CrossRef]
- Li, Q.Q.; Lee, R.X.; Liang, H.; Wang, G.; Li, J.M.; Zhong, Y.; Reed, E. β-Elemene Enhances Susceptibility to Cisplatin in Resistant Ovarian Carcinoma Cells Via Downregulation of Ercc-1 and Xiap and Inactivation of Jnk. Int. J. Oncol. 2013, 43, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.-Z.; Li, X.-M.; Luo, L.-H.; Song, Z.-Q.; Gao, X.; Li, Z.-Q.; Su, J.-Y.; Liang, G.-B. β-Elemene Inhibits Stemness, Promotes Differentiation and Impairs Chemoresistance to Temozolomide in Glioblastoma Stem-Like Cells. Int. J. Oncol. 2014, 45, 699–709. [Google Scholar] [CrossRef]
- Prensner, J.R.; Chinnaiyan, A.M. The Emergence of Lncrnas in Cancer Biology. Cancer Discov. 2011, 1, 391–407. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Shoorei, H.; Bahroudi, Z.; Abak, A.; Taheri, M. The Role of H19 Lncrna in Conferring Chemoresistance in Cancer Cells. Biomed. Pharmacother. 2021, 138, 111447. [Google Scholar] [CrossRef]
- Raveh, E.; Matouk, I.J.; Gilon, M.; Hochberg, A. The H19 Long Non-Coding Rna in Cancer Initiation, Progression and Metastasis—A Proposed Unifying Theory. Mol. Cancer 2015, 14, 184. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Ma, Y.; Zhang, P.; Si, J.; Xiong, Y.; Yang, Y. Long Non-Coding Rna H19 Confers Resistance to Gefitinib Via Mir-148b-3p/Ddah1 Axis in Lung Adenocarcinoma. Anticancer Drugs 2020, 31, 44–54. [Google Scholar] [CrossRef]
- Zhang, R.; Pan, T.; Xiang, Y.; Zhang, M.; Xie, H.; Liang, Z.; Chen, B.; Xu, C.; Wang, J.; Huang, X.; et al. Curcumenol Triggered Ferroptosis in Lung Cancer Cells Via Lncrna H19/Mir-19b-3p/Fth1 Axis. Bioact. Mater. 2022, 13, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xie, S.; Yang, J.; Xiong, H.; Jia, Y.; Zhou, Y.; Chen, Y.; Ying, X.; Chen, C.; Ye, C.; et al. The Long Noncoding Rna H19 Promotes Tamoxifen Resistance in Breast Cancer Via Autophagy. J. Hematol. Oncol. 2019, 12, 81. [Google Scholar] [CrossRef]
- Uribe, M.L.; Marrocco, I.; Yarden, Y. Egfr in Cancer: Signaling Mechanisms, Drugs, and Acquired Resistance. Cancers 2021, 13, 2748. [Google Scholar] [CrossRef]
- Vöing, K.; Michgehl, U.; Mertens, N.D.; Picciotto, C.; Maywald, M.-L.; Goretzko, J.; Waimann, S.; Gilhaus, K.; Rogg, M.; Schell, C.; et al. Disruption of the Rab7-Dependent Final Common Pathway of Endosomal and Autophagic Processing Results in a Severe Podocytopathy. J. Am. Soc. Nephrol. 2023, 34, 1191–1206. [Google Scholar] [CrossRef]
- Wu, P.-S.; Lin, M.-H.; Hsiao, J.-C.; Lin, P.-Y.; Pan, S.-H.; Chen, Y.-J. Egfr-T790m Mutation-Derived Interactome Rerouted Egfr Translocation Contributing to Gefitinib Resistance in Non-Small Cell Lung Cancer. Mol. Cell Proteom. 2023, 22, 100624. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: Globocan Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Paez, J.G.; Jänne, P.A.; Lee, J.C.; Tracy, S.; Greulich, H.; Gabriel, S.; Herman, P.; Kaye, F.J.; Lindeman, N.; Boggon, T.J.; et al. Egfr Mutations in Lung Cancer: Correlation with Clinical Response to Gefitinib Therapy. Science 2004, 304, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-L.; Zhou, C.; Hu, C.-P.; Feng, J.; Lu, S.; Huang, Y.; Li, W.; Hou, M.; Shi, J.H.; Lee, K.Y.; et al. Afatinib Versus Cisplatin Plus Gemcitabine for First-Line Treatment of Asian Patients with Advanced Non-Small-Cell Lung Cancer Harbouring Egfr Mutations (Lux-Lung 6): An Open-Label, Randomised Phase 3 Trial. Lancet Oncol. 2014, 15, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Jackman, D.; Pao, W.; Riely, G.J.; Engelman, J.A.; Kris, M.G.; Jänne, P.A.; Lynch, T.; Johnson, B.E.; Miller, V.A. Clinical Definition of Acquired Resistance to Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2010, 28, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Levy, J.M.M.; Towers, C.G.; Thorburn, A. Targeting Autophagy in Cancer. Nat. Rev. Cancer 2017, 17, 528–542. [Google Scholar] [CrossRef] [PubMed]
- Rybstein, M.D.; Bravo-San Pedro, J.M.; Kroemer, G.; Galluzzi, L. The Autophagic Network and Cancer. Nat. Cell Biol. 2018, 20, 243–251. [Google Scholar] [CrossRef]
- Galluzzi, L.; Pietrocola, F.; Levine, B.; Kroemer, G. Metabolic Control of Autophagy. Cell 2014, 159, 1263–1276. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Kanzawa, T.; Sawaya, R.; Kondo, S. The Role of Autophagy in Cancer Development and Response to Therapy. Nat. Rev. Cancer 2005, 5, 726–734. [Google Scholar] [CrossRef]
- Selvakumaran, M.; Amaravadi, R.K.; Vasilevskaya, I.A.; O’Dwyer, P.J. Autophagy Inhibition Sensitizes Colon Cancer Cells to Antiangiogenic and Cytotoxic Therapy. Clin. Cancer Res. 2013, 19, 2995–3007. [Google Scholar] [CrossRef]
- Ma, C.; Zhang, X.; Mo, X.; Yu, Y.; Xiao, Z.; Wu, J.; Ding, L.; Lei, C.; Zhu, Y.; Zhang, H. Xie-Bai-San Increases Nsclc Cells Sensitivity to Gefitinib by Inhibiting Beclin-1 Mediated Autophagosome Formation. Phytomedicine 2024, 125, 155351. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Esmaeili, M.; Taheri, M. H19 Lncrna: Roles in Tumorigenesis. Biomed. Pharmacother. 2020, 123, 109774. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, M.; Moosavi, M.S.; Abed, H.M.; Dehghani, M.; Aalipour, M.; Heydari, E.A.; Behroozaghdam, M.; Entezari, M.; Salimimoghadam, S.; Gunduz, E.S.; et al. Long Non-Coding Rna (Lncrna) H19 in Human Cancer: From Proliferation and Metastasis to Therapy. Pharmacol. Res. 2022, 184, 106418. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Qi, M.; Fei, X.; Wang, X.; Wang, K. Lncrna H19: A Novel Oncogene in Multiple Cancers. Int. J. Biol. Sci. 2021, 17, 3188–3208. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Zhang, Y.; Liu, W.; Huang, Y.; Shen, X.; Jing, R.; Pu, J.; Wang, X.; Ju, S.; Cong, H.; et al. Lncrna H19 Overexpression Induces Bortezomib Resistance in Multiple Myeloma by Targeting Mcl-1 Via Mir-29b-3p. Cell Death Dis. 2019, 10, 106. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Han, D.; Yuan, Z.; Hu, H.; Zhao, Z.; Yang, R.; Jin, Y.; Zou, C.; Chen, Y.; Wang, G.; et al. Long Non-Coding Rna H19 Confers 5-Fu Resistance in Colorectal Cancer by Promoting Sirt1-Mediated Autophagy. Cell Death Dis. 2018, 9, 1149. [Google Scholar] [CrossRef] [PubMed]
- Luan, W.; Zhou, Z.; Ni, X.; Xia, Y.; Wang, J.; Yan, Y.; Xu, B. Long Non-Coding Rna H19 Promotes Glucose Metabolism and Cell Growth in Malignant Melanoma Via Mir-106a-5p/E2f3 Axis. J. Cancer Res. Clin. Oncol. 2018, 144, 531–542. [Google Scholar] [CrossRef]
- Peperstraete, E.; Lecerf, C.; Collette, J.; Vennin, C.; Raby, L.; Völkel, P.; Angrand, P.-O.; Winter, M.; Bertucci, F.; Finetti, P.; et al. Enhancement of Breast Cancer Cell Aggressiveness by Lncrna H19 and Its Mir-675 Derivative: Insight into Shared and Different Actions. Cancers 2020, 12, 1730. [Google Scholar] [CrossRef]
- Jiang, M.-C.; Ni, J.-J.; Cui, W.-Y.; Wang, B.-Y.; Zhuo, W. Emerging Roles of Lncrna in Cancer and Therapeutic Opportunities. Am. J. Cancer Res. 2019, 9, 1354–1366. [Google Scholar]
- Shinde, S.R.; Maddika, S. Pten Modulates Egfr Late Endocytic Trafficking and Degradation by Dephosphorylating Rab7. Nat. Commun. 2016, 7, 10689. [Google Scholar] [CrossRef]
- Eminaga, S.; Teekakirikul, P.; Seidman, C.E.; Seidman, J.G. Detection of Cell Proliferation Markers by Immunofluorescence Staining and Microscopy Imaging in Paraffin-Embedded Tissue Sections. Curr. Protoc. Mol. Biol. 2016, 115, 14.25.1–14.25.14. [Google Scholar] [CrossRef] [PubMed]









| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| GAPDH | 5′-GCACCGTCAAGGCTGAGAAC-3′ | 5′-TGGTGAAGAACGCCAGTGGA-3′ |
| LncRNA H19 | 5′-TCCCAGAACCCACAACATGAA-3′ | 5′-TTCACCTTCCAGAGCCGATTC-3′ |
| LC3B | 5′-GAGAAGCAGCTTCCTGTTCTGG-3′ | 5′-GTGTCCGTTCACCAACAGGAAG-3′ |
| Beclin-1 | 5′-CTGGACACTCAGCTCAACGTCA-3′ | CTCTAGTGCCAGCTCCTTTAGC-3′ |
| ATG5 | 5′-GCAGATGGACAGTTGCACACAC-3′ | 5′-GAGGTGTTTCCAACATTGGCTCA-3′ |
| ATG7 | 5′-CGTTGCCCACAGCATCATCTTC-3′ | 5′-CACTGAGGTTCACCATCCTTGG-3′ |
| ATG12 | 5′-GGGAAGGACTTACGGATGTCTC-3′ | 5′-AGGAGTGTCTCCCACAGCCTTT-3′ |
| AKT | 5′-TGGACTACCTGCACTCGGAGAA-3′ | 5′-GTGCCGCAAAAGGTCTTCATGG-3′ |
| mTOR | 5′-AGCATCGGATGCTTAGGAGTGG-3′ | 5′-CAGCCAGTCATCTTTGGAGACC-3′ |
| TSC1 | 5′-CTGGACAGACTGATACAGCAGG-3′ | 5′-TGCGGATCTCATCTGAAGGAGG-3′ |
| PTEN | 5′-TGAGTTCCCTCAGCCGTTACCT-3′ | 5′-GAGGTTTCCTCTGGTCCTGGTA-3′ |
| ULK1 | 5′-GCAAGGACTCTTCCTGTGACAC-3′ | 5′-CCACTGCACATCAGGCTGTCTG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, R.; Zheng, Y.; Zhu, Q.; Gu, X.; Xiang, B.; Gu, X.; Xie, T.; Sui, X. β-Elemene Reverses Gefitinib Resistance in NSCLC Cells by Inhibiting lncRNA H19-Mediated Autophagy. Pharmaceuticals 2024, 17, 626. https://doi.org/10.3390/ph17050626
Zhang R, Zheng Y, Zhu Q, Gu X, Xiang B, Gu X, Xie T, Sui X. β-Elemene Reverses Gefitinib Resistance in NSCLC Cells by Inhibiting lncRNA H19-Mediated Autophagy. Pharmaceuticals. 2024; 17(5):626. https://doi.org/10.3390/ph17050626
Chicago/Turabian StyleZhang, Ruonan, Yintao Zheng, Qianru Zhu, Xiaoqing Gu, Bo Xiang, Xidong Gu, Tian Xie, and Xinbing Sui. 2024. "β-Elemene Reverses Gefitinib Resistance in NSCLC Cells by Inhibiting lncRNA H19-Mediated Autophagy" Pharmaceuticals 17, no. 5: 626. https://doi.org/10.3390/ph17050626
APA StyleZhang, R., Zheng, Y., Zhu, Q., Gu, X., Xiang, B., Gu, X., Xie, T., & Sui, X. (2024). β-Elemene Reverses Gefitinib Resistance in NSCLC Cells by Inhibiting lncRNA H19-Mediated Autophagy. Pharmaceuticals, 17(5), 626. https://doi.org/10.3390/ph17050626






