A Clinical Update on SARS-CoV-2: Pathology and Development of Potential Inhibitors
Abstract
:1. Introduction
2. Pathology
2.1. Clinical Characteristics and Diagnosis of SARS-CoV-2 Infection
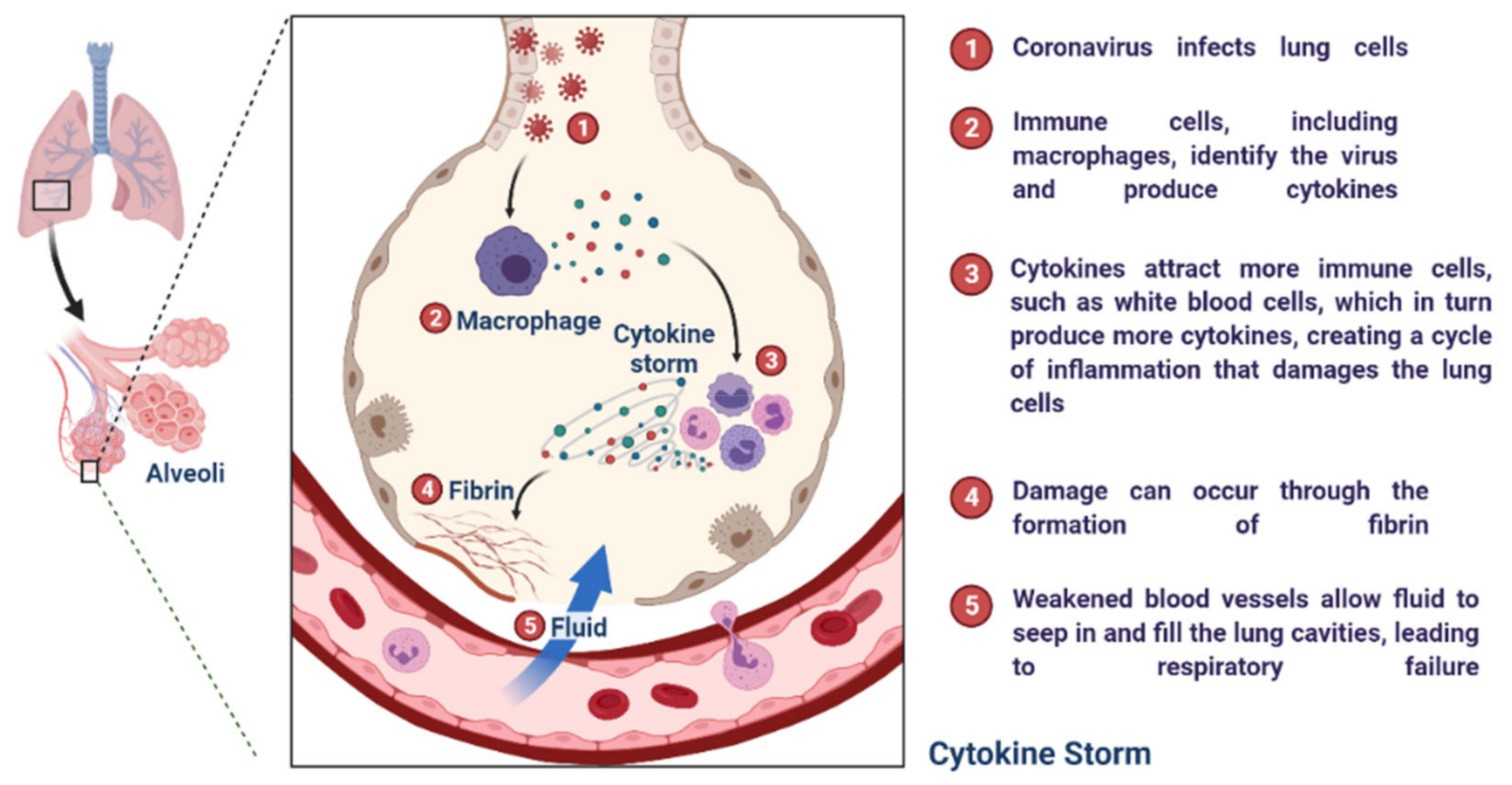
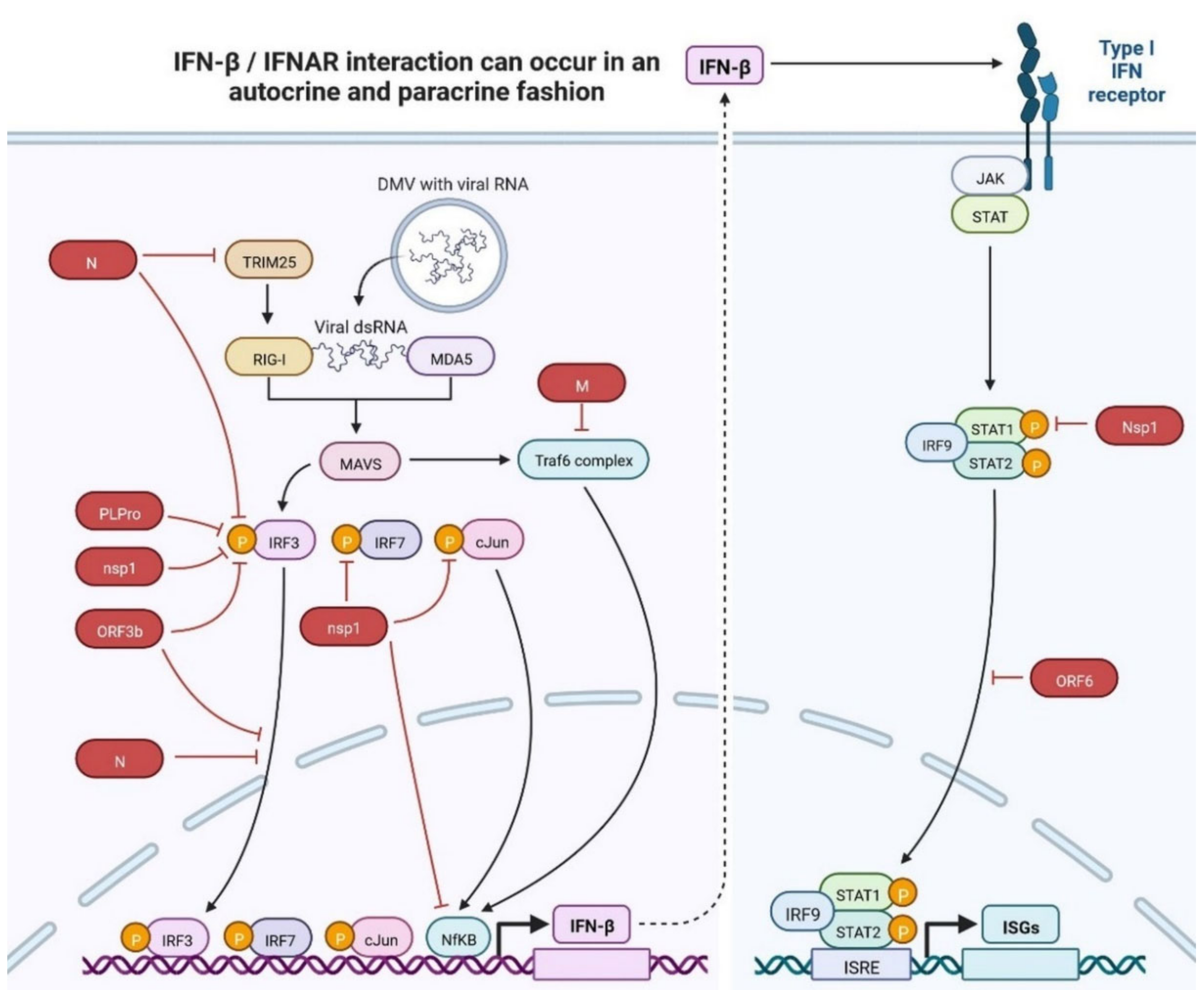
2.2. Immunopathology
3. New Variant Detection and Assessment
4. SARS-CoV-2 Drug and Treatment
4.1. Antivirals
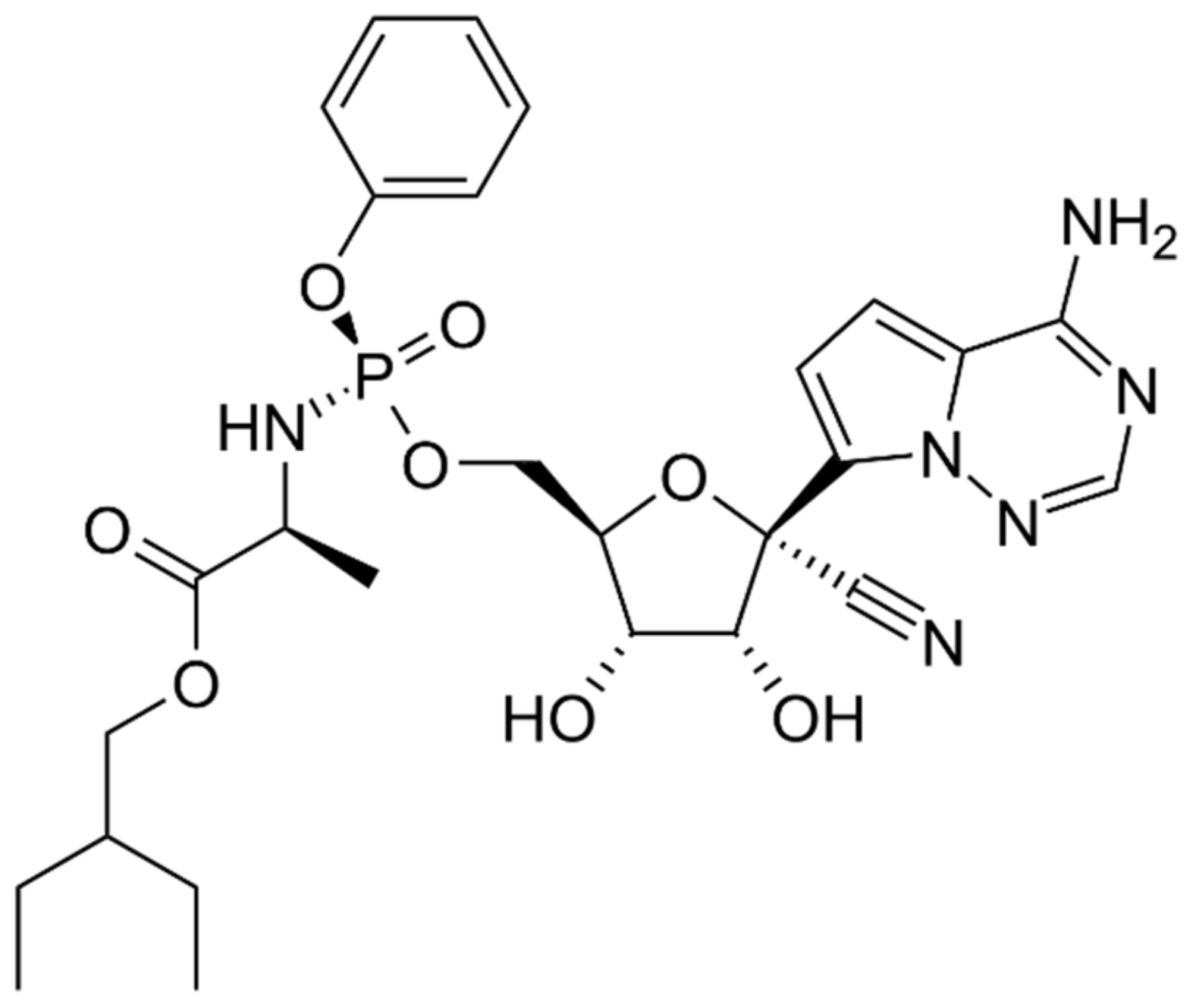
4.1.1. Molnupiravir (MK-4482)
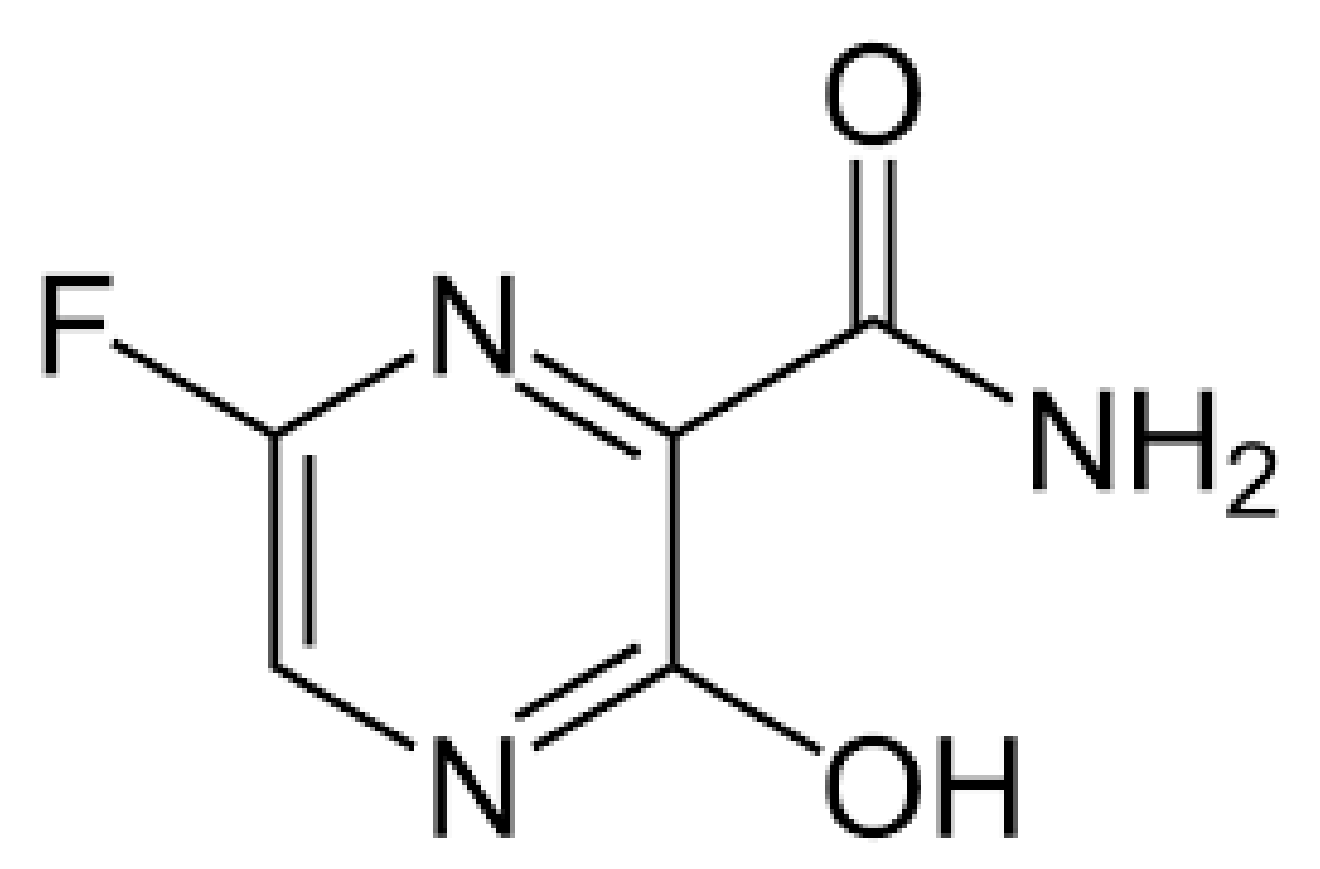
4.1.2. Favipiravir (Also Known as Avigan)
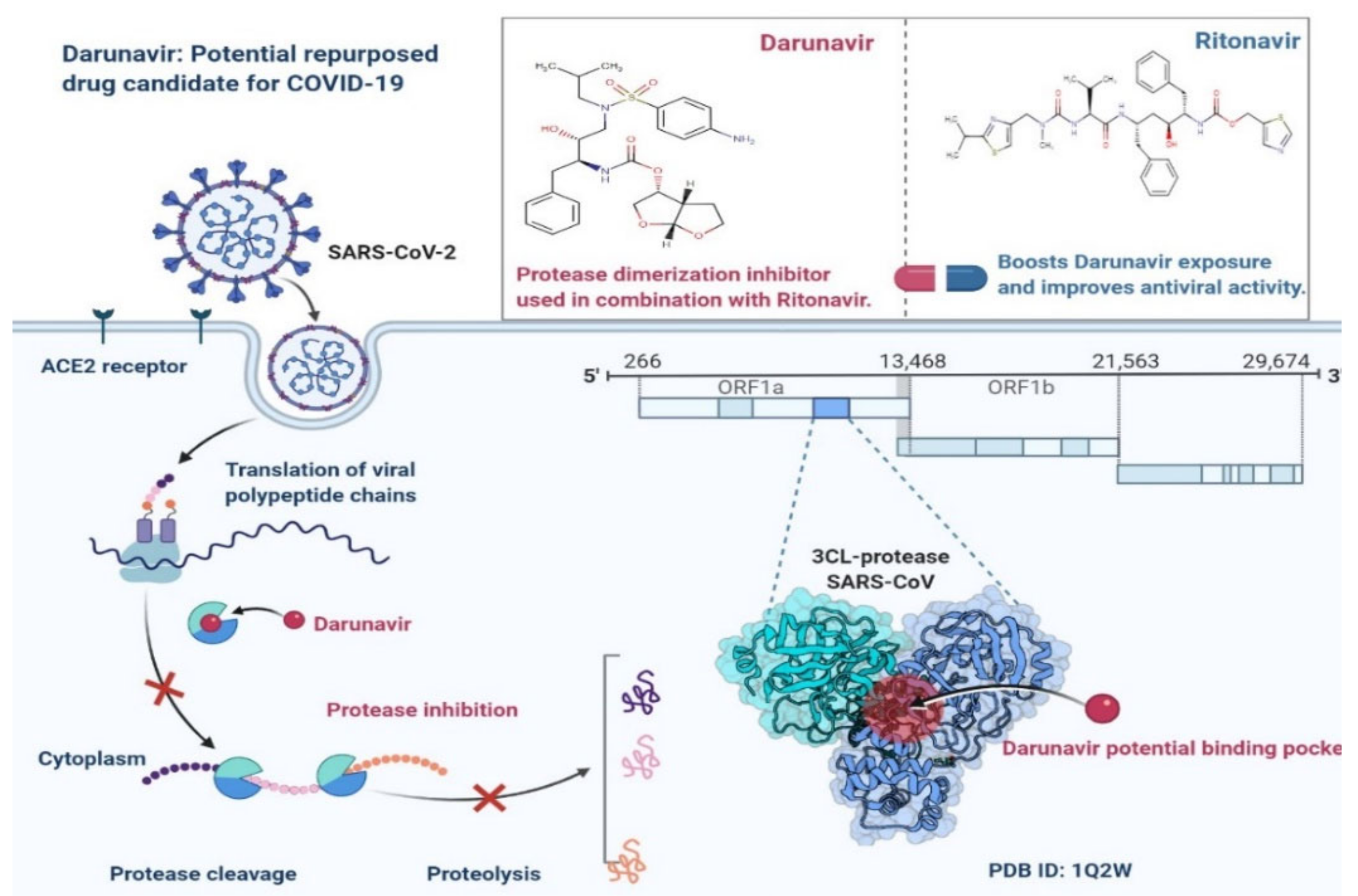
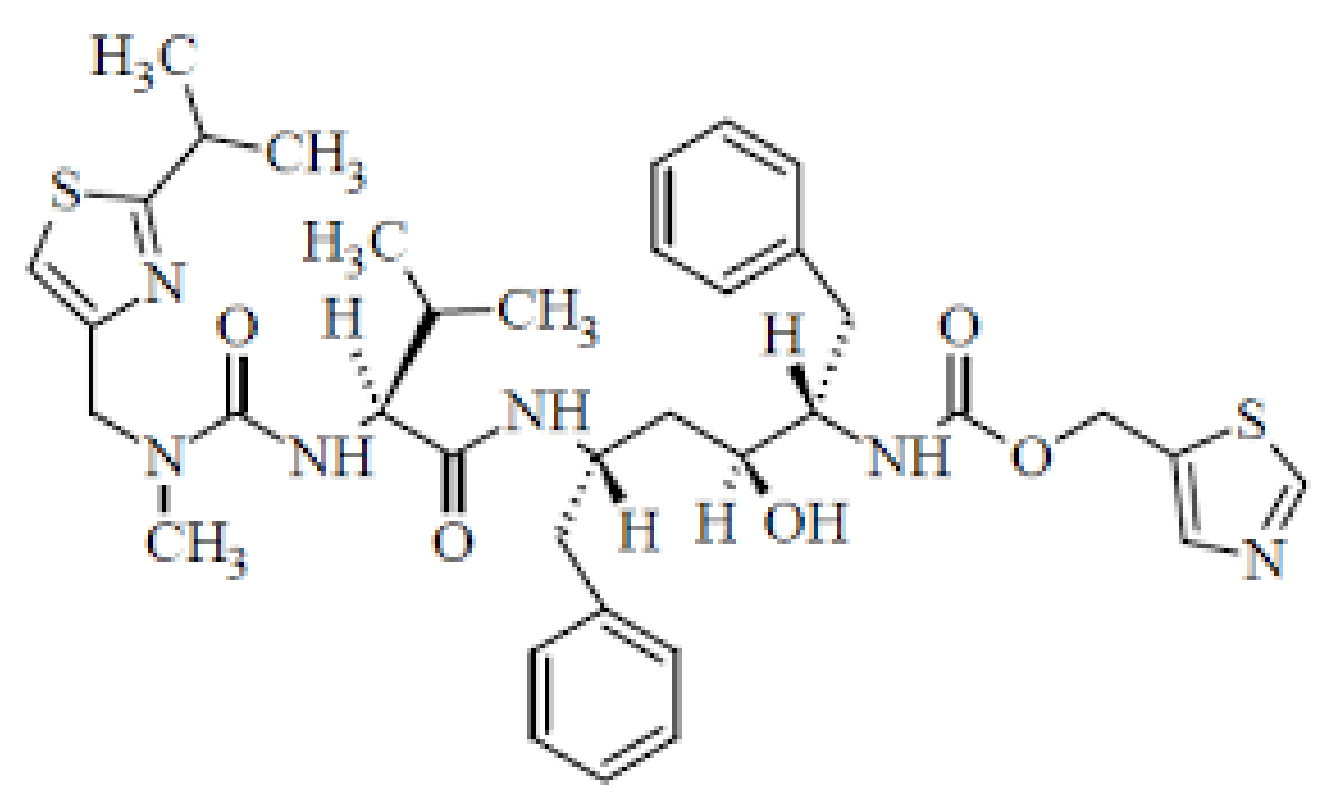
4.1.3. Ritonavir and Darunavir
4.1.4. PAXLOVID™ (PF-07321332)
4.1.5. Ivermectin
4.1.6. Remdesivir
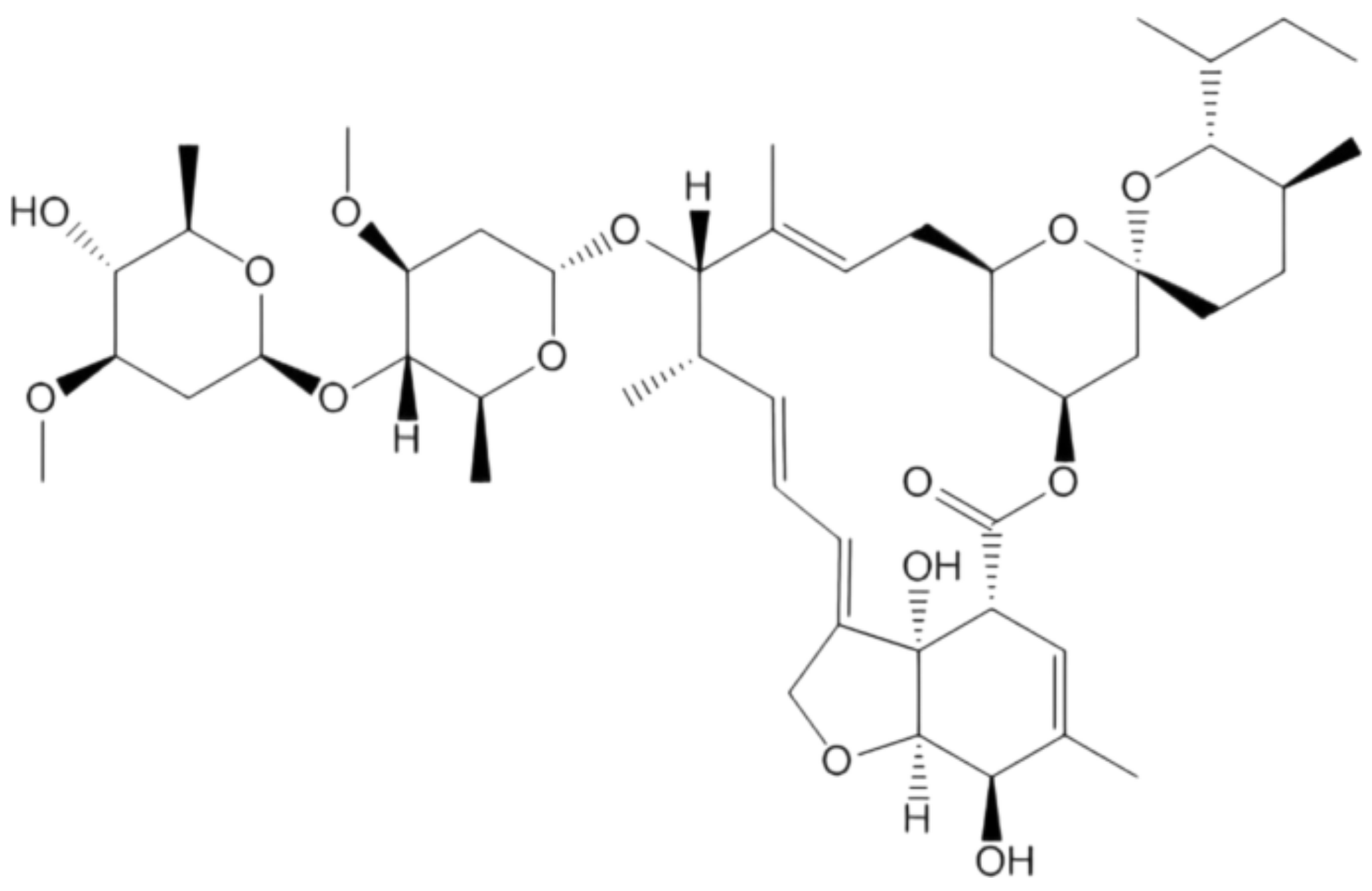
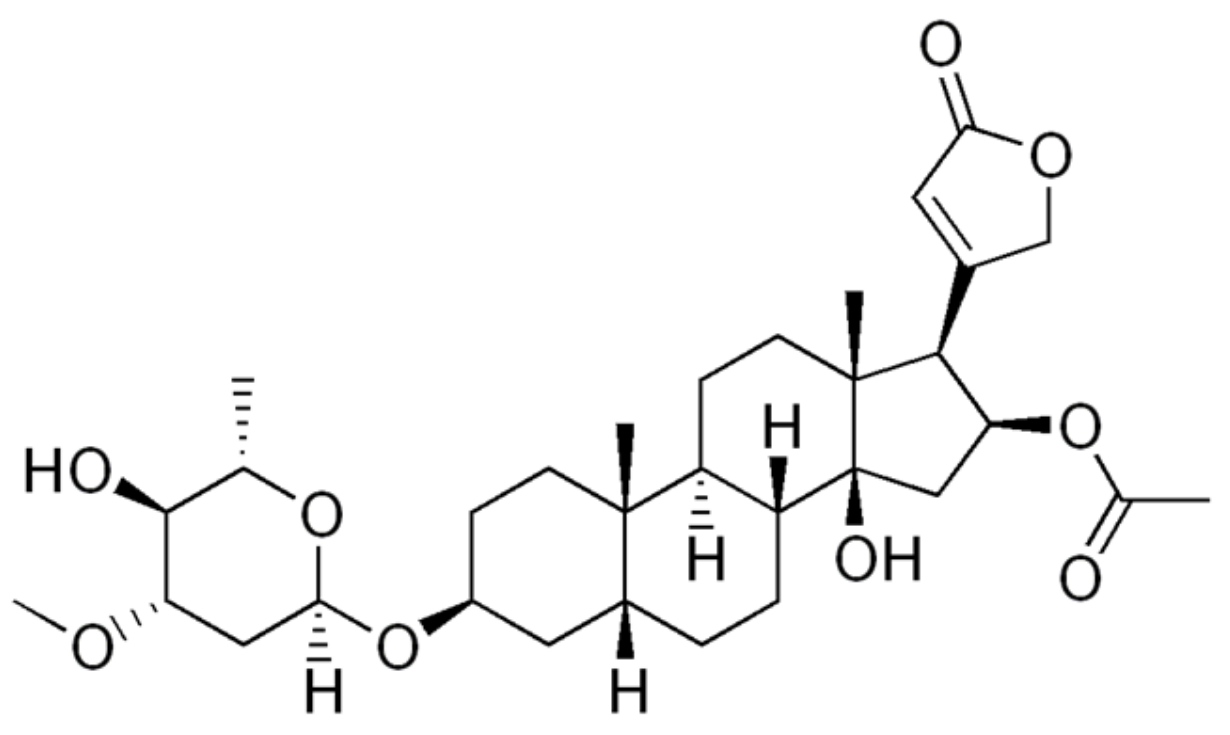
4.1.7. Oleandrin
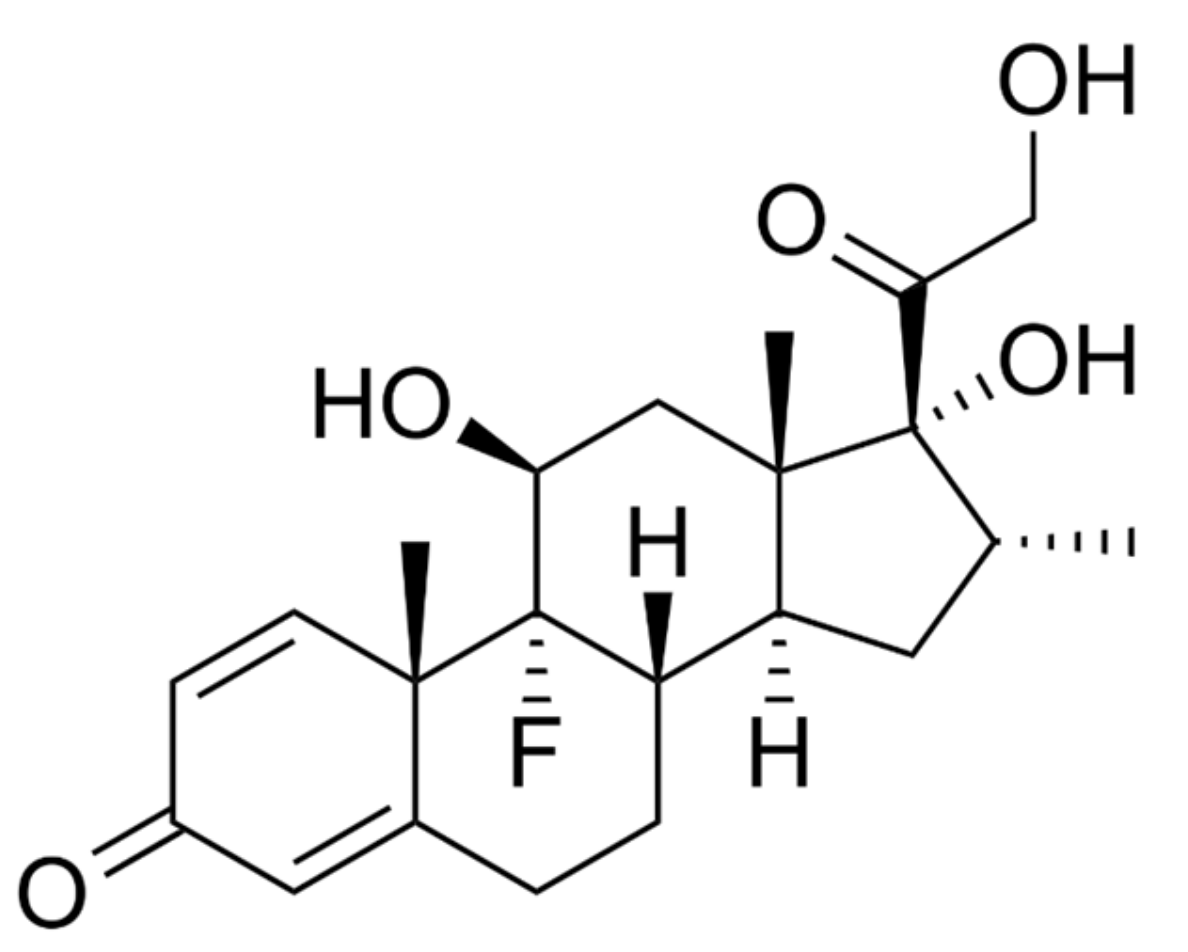
4.1.8. Dexamethasone
4.1.9. Fluvoxamine
4.2. Potential Immunotherapeutic Strategies for SARS-CoV-2
4.2.1. Baricitinib
4.2.2. Tocilizumab
4.2.3. Ruxolitinib
4.2.4. Sotrovimab
4.2.5. Casirivimab/Imdevimab
4.2.6. Tocilizumab
4.2.7. Bebtelovimab
4.2.8. REGEN-COV
4.2.9. Evusheld (Tixagevimab Co-Packaged with Cilgavimab)
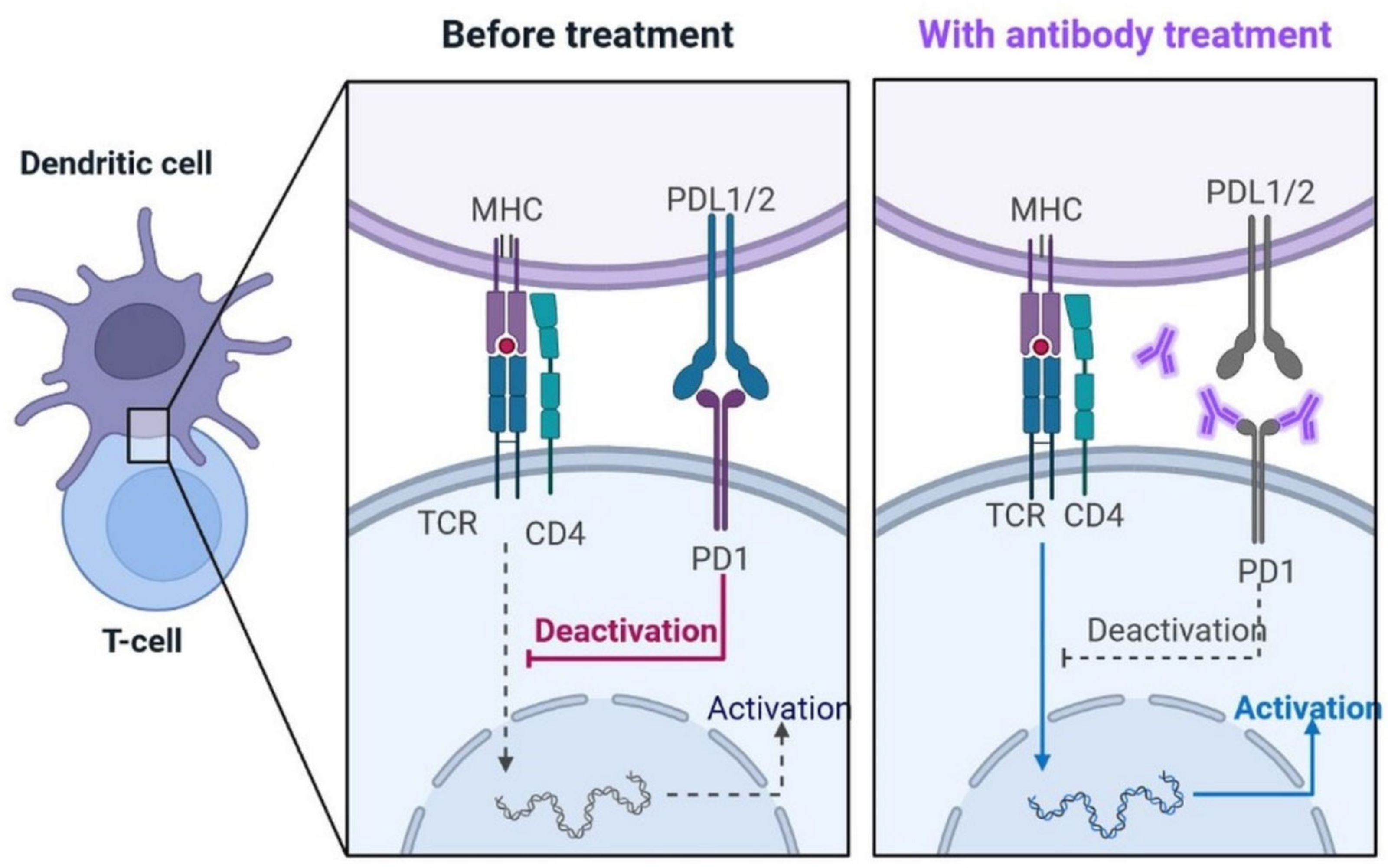
4.2.10. PD-1 Blocking Antibodies: A Potentially Repurposed Drug Candidate for COVID-19
4.3. Sedative Drug
Propofol-Lipuro 1%
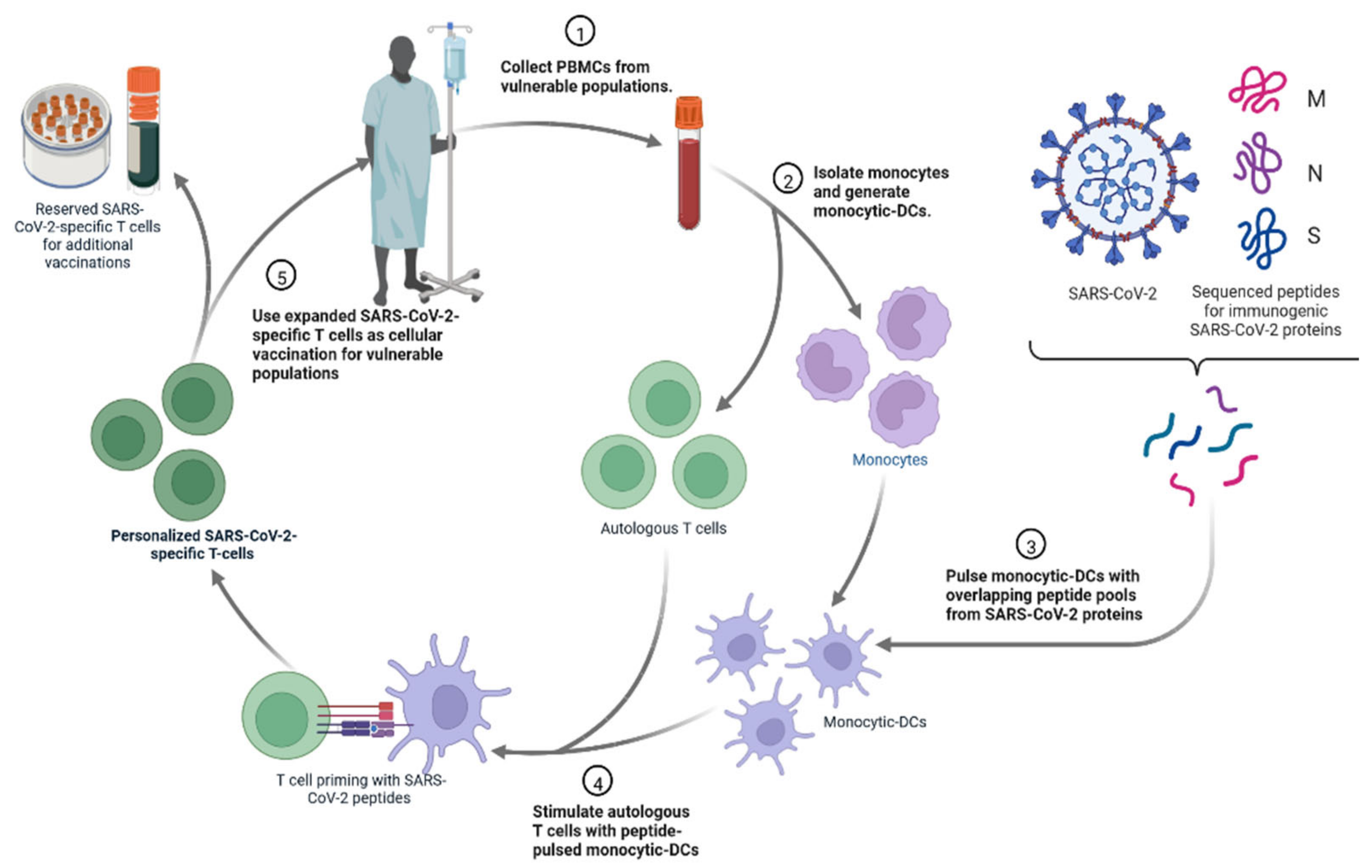
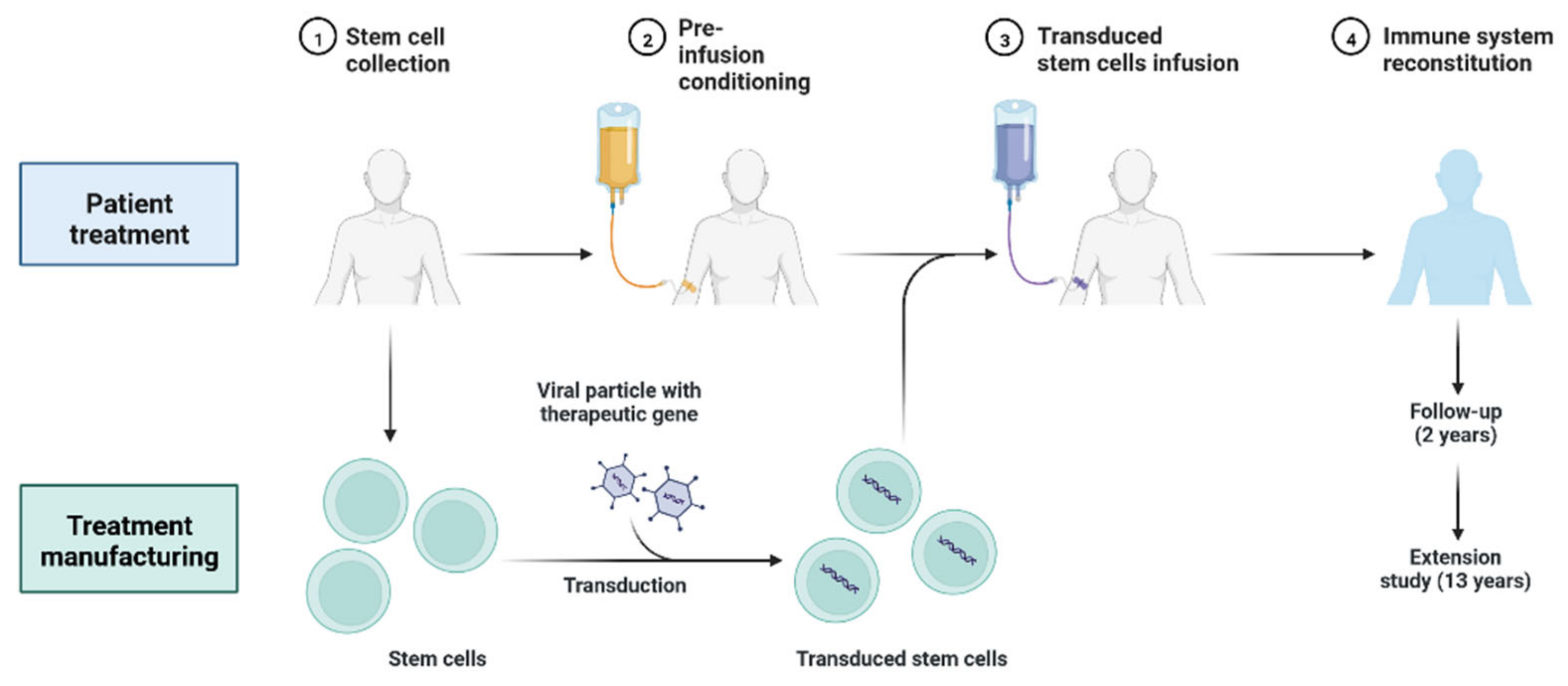
4.4. Personalized Cell Therapies to Combat COVID-19
4.5. Gene Therapy
4.6. Other Approaches for Drug Development
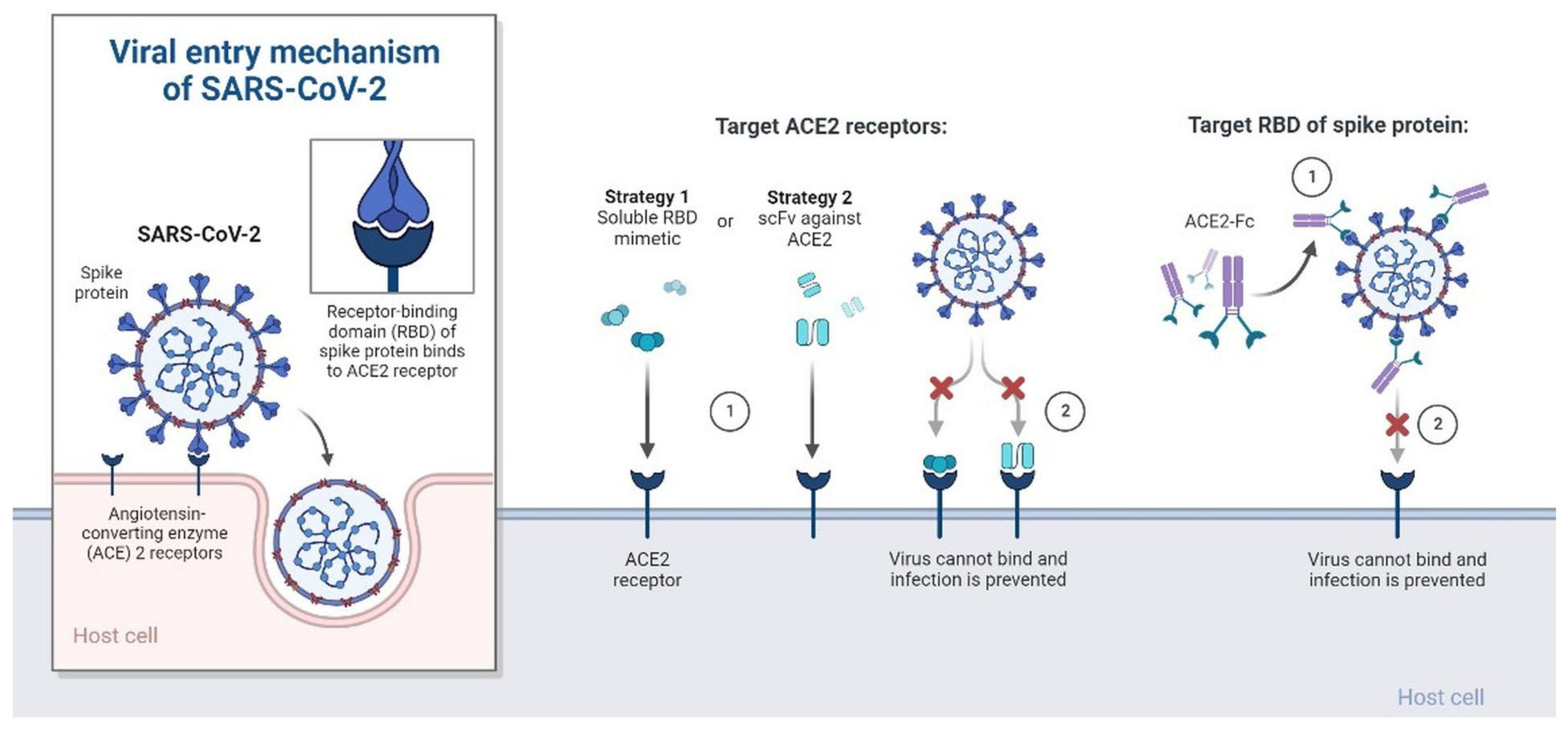
4.6.1. Recombinant ACE-2 Approaches
4.6.2. Renal Replacement Therapies (CRRT)
4.6.3. Cell-Free Biotherapeutic
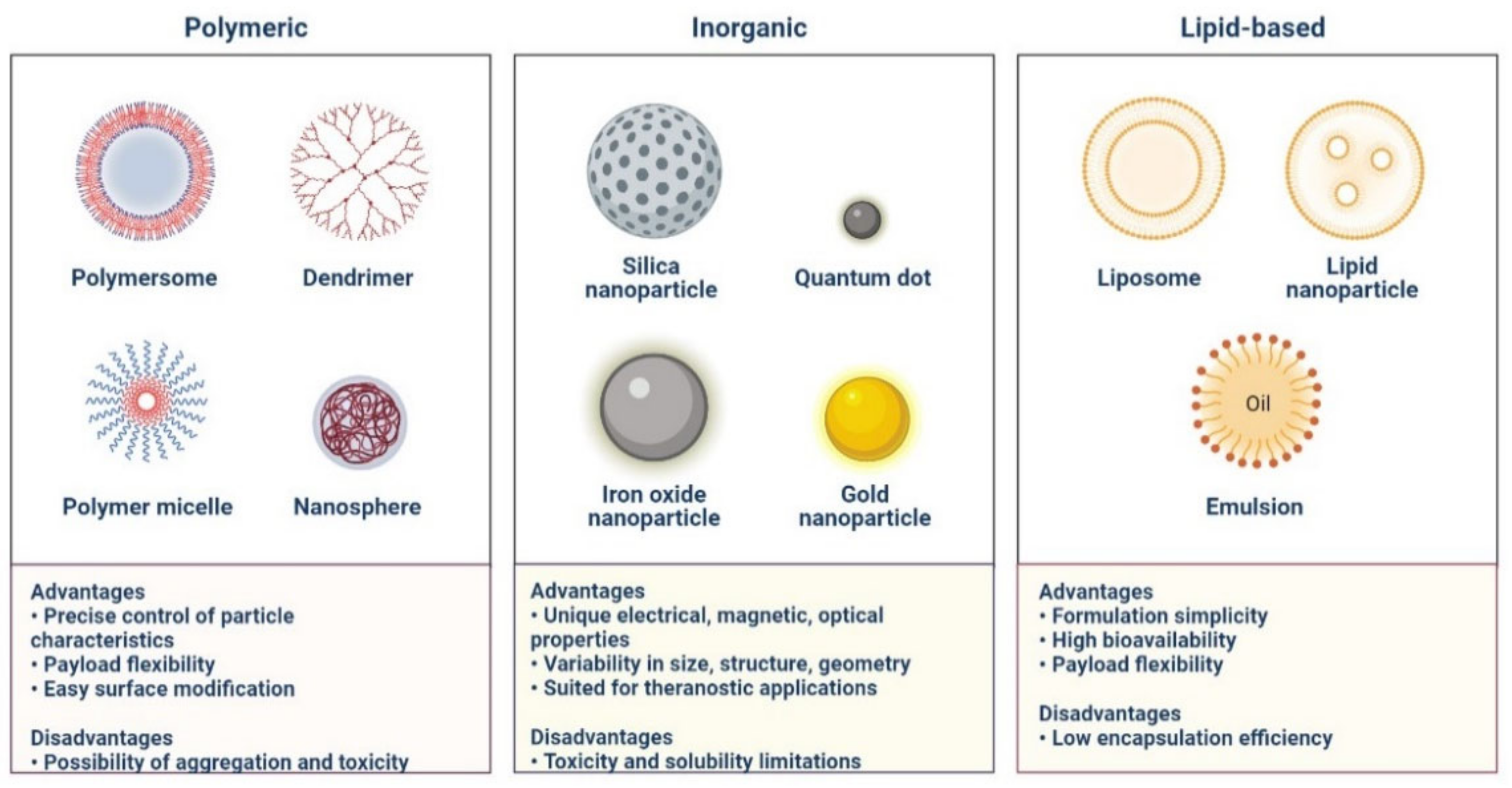
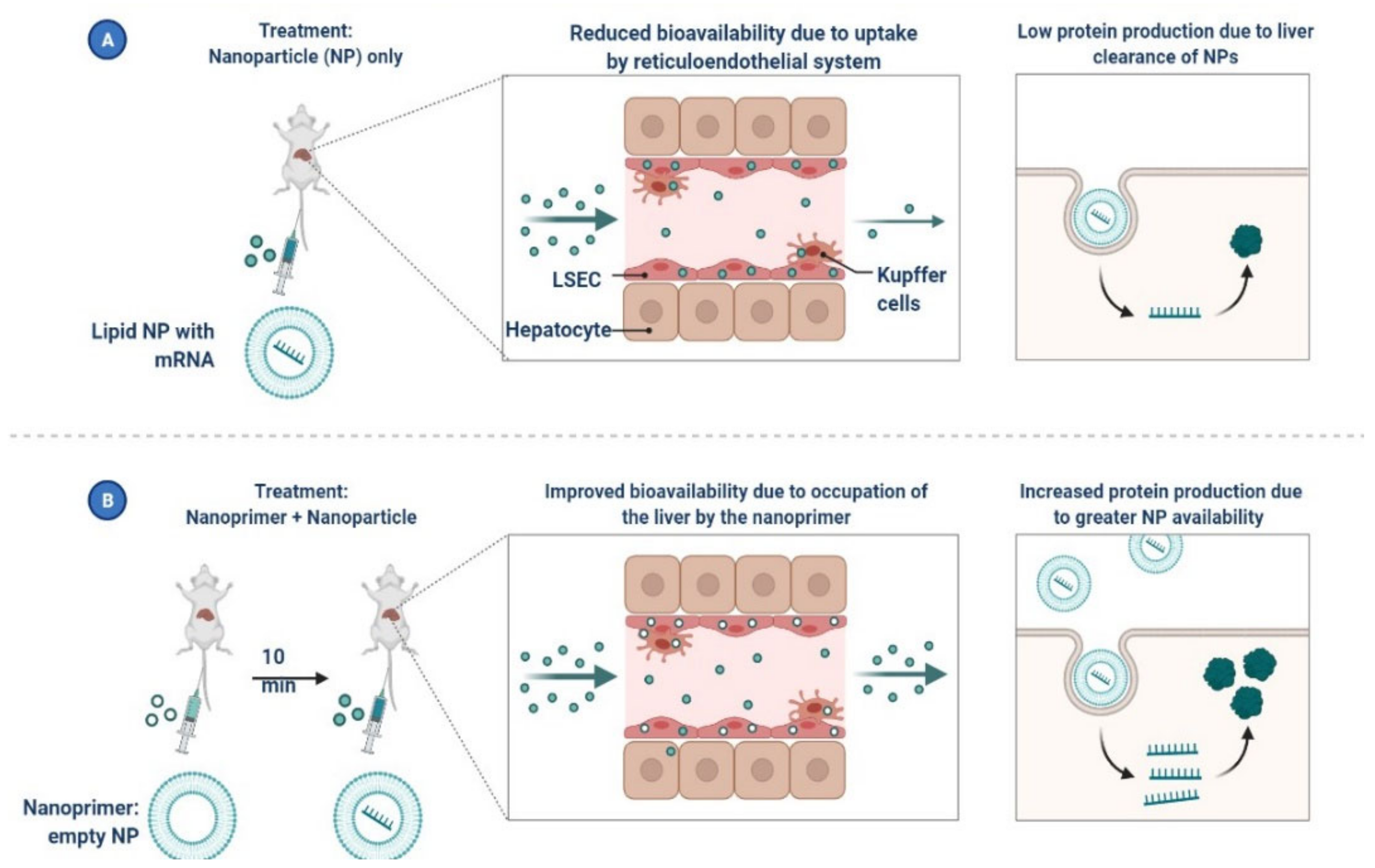
5. Nanomaterials for the Treatment of COVID-19
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abouelela, M.E.; Assaf, H.K.; Abdelhamid, R.A.; Elkhyat, E.S.; Sayed, A.M.; Oszako, T.; Belbahri, L.; El Zowalaty, A.E.; Abdelkader, M.S.A. Identification of Potential SARS-CoV-2 Main Protease and Spike Protein Inhibitors from the Genus Aloe: An In Silico Study for Drug Development. Molecules 2021, 26, 1767. [Google Scholar] [CrossRef] [PubMed]
- Overhoff, B.; Falls, Z.; Mangione, W.; Samudrala, R. A Deep-Learning Proteomic-Scale Approach for Drug Design. Pharmaceuticals 2021, 14, 1277. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Li, R.; Zhang, Z.; Wang, J.; Huang, Q.; Nie, D.; Fan, K.; Guo, W.; Zhao, Z.; Han, Z. A Review of Potential Therapeutic Strategies for COVID-19. Viruses 2022, 14, 2346. [Google Scholar] [CrossRef]
- Starshinova, A.; Malkova, A.; Zinchenko, U.; Kudlay, D.; Glushkova, A.; Dovgalyk, I.; Yablonskiy, P.; Shoenfeld, Y. Efficacy of Different Types of Therapy for COVID-19: A Comprehensive Review. Life 2021, 11, 753. [Google Scholar] [CrossRef]
- Gherghescu, I.; Delgado-Charro, M.B. The Biosimilar Landscape: An Overview of Regulatory Approvals by the EMA and FDA. Pharmaceutics 2021, 13, 48. [Google Scholar] [CrossRef] [PubMed]
- Bauso, L.V.; Imbesi, C.; Irene, G.; Calì, G.; Bitto, A. New Approaches and Repurposed Antiviral Drugs for the Treatment of the SARS-CoV-2 Infection. Pharmaceuticals 2021, 14, 503. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, S.; Scaccabarozzi, D.; Signorini, L.; Perego, F.; Ilboudo, D.P.; Ferrante, P.; Delbue, S. The Use of Antimalarial Drugs against Viral Infection. Microorganisms 2020, 8, 85. [Google Scholar] [CrossRef] [Green Version]
- FDA Drug Competition Action Plan. Available online: https://www.fda.gov/drugs/guidance-compliance-regulatory-information/fda-drug-competition-action-plan (accessed on 30 October 2022).
- Queirós-Reis, L.; Gomes da Silva, P.; Gonçalves, J.; Brancale, A.; Bassetto, M.; Mesquita, J.R. SARS-CoV-2 Virus−Host Interaction: Currently Available Structures and Implications of Variant Emergence on Infectivity and Immune Response. Int. J. Mol. Sci. 2021, 22, 10836. [Google Scholar] [CrossRef]
- Ramesh, S.; Govindarajulu, M.; Parise, R.S.; Neel, L.; Shankar, T.; Patel, S.; Lowery, P.; Smith, F.; Dhanasekaran, M.; Moore, T. Emerging SARS-CoV-2 Variants: A Review of Its Mutations, Its Implications and Vaccine Efficacy. Vaccines 2021, 9, 1195. [Google Scholar] [CrossRef]
- Yang, L.; Li, J.; Guo, S.; Hou, C.; Liao, C.; Shi, L.; Ma, X.; Jiang, S.; Zheng, B.; Fang, Y.; et al. SARS-CoV-2 Variants, RBD Mutations, Binding Affinity, and Antibody Escape. Int. J. Mol. Sci. 2021, 22, 12114. [Google Scholar] [CrossRef]
- Majeed, A.; Lee, S. Applications of Machine Learning and High-Performance Computing in the Era of COVID-19. Appl. Syst. Innov. 2021, 4, 40. [Google Scholar] [CrossRef]
- Peyclit, L.; Yousfi, H.; Rolain, J.-M.; Bittar, F. Drug Repurposing in Medical Mycology: Identification of Compounds as Potential Antifungals to Overcome the Emergence of Multidrug-Resistant Fungi. Pharmaceuticals 2021, 14, 488. [Google Scholar] [CrossRef]
- Zhuang, D.; Ibrahim, A.K. Deep Learning for Drug Discovery: A Study of Identifying High Efficacy Drug Compounds Using a Cascade Transfer Learning Approach. Appl. Sci. 2021, 11, 7772. [Google Scholar] [CrossRef]
- Agrawal, L.; Poullikkas, T.; Eisenhower, S.; Monsanto, C.; Bakku, R.K.; Chen, M.-H.; Kalra, R.S. Viroinformatics-Based Analysis of SARS-CoV-2 Core Proteins for Potential Therapeutic Targets. Antibodies 2021, 10, 3. [Google Scholar] [CrossRef]
- Miyazaki, M.; Yanagida, R.; Nakashima, A.; Matsuo, K.; Moriwaki, N.; Uchiyama, M.; Yamada, Y.; Hirata, H.; Kushima, H.; Kinoshita, Y.; et al. Evaluation of Remdesivir for Mildly to Moderately Ill Patients with COVID-19: A Single-Arm, Single-Center, Retrospective Study. Medicina 2022, 58, 1007. [Google Scholar] [CrossRef] [PubMed]
- Magazine, N.; Zhang, T.; Wu, Y.; McGee, M.C.; Veggiani, G.; Huang, W. Mutations and Evolution of the SARS-CoV-2 Spike Protein. Viruses 2022, 14, 640. [Google Scholar] [CrossRef] [PubMed]
- Fernández-De-Las-Peñas, C.; Notarte, K.I.; Peligro, P.J.; Velasco, J.V.; Ocampo, M.J.; Henry, B.M.; Arendt-Nielsen, L.; Torres-Macho, J.; Plaza-Manzano, G. Long-COVID Symptoms in Individuals Infected with Different SARS-CoV-2 Variants of Concern: A Systematic Review of the Literature. Viruses 2022, 14, 2629. [Google Scholar] [CrossRef] [PubMed]
- Ramadori, G.P. SARS-CoV-2-Infection (COVID-19): Clinical Course, Viral Acute Respiratory Distress Syndrome (ARDS) and Cause(s) of Death. Med Sci. 2022, 10, 58. [Google Scholar] [CrossRef]
- Pustake, M.; Tambolkar, I.; Giri, P.; Gandhi, C. SARS, MERS and COVID-19: An overview and comparison of clinical, laboratory and radiological features. J. Fam. Med. Prim. Care 2022, 11, 10–17. [Google Scholar] [CrossRef]
- Jiang, W.; Ji, W.; Zhang, Y.; Xie, Y.; Chen, S.; Jin, Y.; Duan, G. An Update on Detection Technologies for SARS-CoV-2 Variants of Concern. Viruses 2022, 14, 2324. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Coronavirus COVID-19 Dashboard. Available online: https://covid19.who.int (accessed on 8 December 2021).
- Kaivola, J.; Nyman, T.A.; Matikainen, S. Inflammasomes and SARS-CoV-2 Infection. Viruses 2021, 13, 2513. [Google Scholar] [CrossRef] [PubMed]
- Di Sante, G.; Buonsenso, D.; De Rose, C.; Tredicine, M.; Palucci, I.; De Maio, F.; Camponeschi, C.; Bonadia, N.; Biasucci, D.; Pata, D.; et al. Immunopathology of SARS-CoV-2 Infection: A Focus on T Regulatory and B Cell Responses in Children Compared with Adults. Children 2022, 9, 681. [Google Scholar] [CrossRef] [PubMed]
- Rabaan, A.A.; Al-Ahmed, S.H.; Muhammad, J.; Khan, A.; Sule, A.A.; Tirupathi, R.; Mutair, A.A.; Alhumaid, S.; Al-Omari, A.; Dhawan, M.; et al. Role of Inflammatory Cytokines in COVID-19 Patients: A Review on Molecular Mechanisms, Immune Functions, Immunopathology and Immunomodulatory Drugs to Counter Cytokine Storm. Vaccines 2021, 9, 436. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Chi, S.; Dmytruk, K.; Dmytruk, O.; Tan, S. Coronaviral Infection and Interferon Response: The Virus-Host Arms Race and COVID-19. Viruses 2022, 14, 1349. [Google Scholar] [CrossRef] [PubMed]
- Howard, F.H.N.; Kwan, A.; Winder, N.; Mughal, A.; Collado-Rojas, C.; Muthana, M. Understanding Immune Responses to Viruses—Do Underlying Th1/Th2 Cell Biases Predict Outcome? Viruses 2022, 14, 1493. [Google Scholar] [CrossRef]
- Reed, S.G.; Ager, A. Immune Responses to IAV Infection and the Roles of L-Selectin and ADAM17 in Lymphocyte Homing. Pathogens 2022, 11, 150. [Google Scholar] [CrossRef]
- Rubio-Casillas, A.; Redwan, E.M.; Uversky, V.N. SARS-CoV-2: A Master of Immune Evasion. Biomedicines 2022, 10, 1339. [Google Scholar] [CrossRef]
- Lim, H.X.; Masomian, M.; Khalid, K.; Kumar, A.U.; MacAry, P.A.; Poh, C.L. Identification of B-Cell Epitopes for Eliciting Neutralizing Antibodies against the SARS-CoV-2 Spike Protein through Bioinformatics and Monoclonal Antibody Targeting. Int. J. Mol. Sci. 2022, 23, 4341. [Google Scholar] [CrossRef]
- Morales-Núñez, J.J.; Muñoz-Valle, J.F.; Torres-Hernández, P.C.; Hernández-Bello, J. Overview of Neutralizing Antibodies and Their Potential in COVID-19. Vaccines 2021, 9, 1376. [Google Scholar] [CrossRef]
- Kherabi, Y.; Launay, O.; Luong Nguyen, L.B. COVID-19 Vaccines against Omicron Variant: Real-World Data on Effectiveness. Viruses 2022, 14, 2086. [Google Scholar] [CrossRef]
- Redwan, E.M.; Elrashdy, F.; Aljabali, A.A.A.; Baetas-da-Cruz, W.; Barh, D.; Brufsky, A.M.; Hassan, S.S.; Lundstrom, K.; Serrano-Aroca, Á.; Takayama, K.; et al. Would New SARS-CoV-2 Variants Change the War against COVID-19? Epidemiologia 2022, 3, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Berno, G.; Fabeni, L.; Matusali, G.; Gruber, C.E.M.; Rueca, M.; Giombini, E.; Garbuglia, A.R. SARS-CoV-2 Variants Identification: Overview of Molecular Existing Methods. Pathogens 2022, 11, 1058. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Mangalaganesh, S.; Wilson, L.O.W.; Kuiper, M.J.; Drew, T.W.; Vasan, S.S. Tracking Co-Occurrence of N501Y, P681R, and Other Key Mutations in SARS-CoV-2 Spike for Surveillance. Zoonotic Dis. 2022, 2, 147–162. [Google Scholar] [CrossRef]
- Emmelot, M.E.; Vos, M.; Boer, M.C.; Rots, N.Y.; de Wit, J.; van Els, C.A.C.M.; Kaaijk, P. Omicron BA.1 Mutations in SARS-CoV-2 Spike Lead to Reduced T-Cell Response in Vaccinated and Convalescent Individuals. Viruses 2022, 14, 1570. [Google Scholar] [CrossRef] [PubMed]
- Basile, K.; Rockett, R.J.; McPhie, K.; Fennell, M.; Johnson-Mackinnon, J.; Agius, J.E.; Fong, W.; Rahman, H.; Ko, D.; Donavan, L.; et al. Improved Neutralisation of the SARS-CoV-2 Omicron Variant following a Booster Dose of Pfizer-BioNTech (BNT162b2) COVID-19 Vaccine. Viruses 2022, 14, 2023. [Google Scholar] [CrossRef] [PubMed]
- Kandeel, M.; Mohamed, M.E.M.; Abd El-Lateef, H.M.A.; Venugopala, K.N.; El-Beltagi, H.S. Omicron variant genome evolution and phylogenetics. J. Med Virol. 2022, 94, 1627–1632. [Google Scholar] [CrossRef]
- Bellamkonda, N.; Lambe, U.P.; Sawant, S.; Nandi, S.S.; Chakraborty, C.; Shukla, D. Immune Response to SARS-CoV-2 Vaccines. Biomedicines 2022, 10, 1464. [Google Scholar] [CrossRef]
- Ng, T.I.; Correia, I.; Seagal, J.; DeGoey, D.A.; Schrimpf, M.R.; Hardee, D.J.; Noey, E.L.; Kati, W.M. Antiviral Drug Discovery for the Treatment of COVID-19 Infections. Viruses 2022, 14, 961. [Google Scholar] [CrossRef]
- Heustess, A.M.; Allard, M.A.; Thompson, D.K.; Fasinu, P.S. Clinical Management of COVID-19: A Review of Pharmacological Treatment Options. Pharmaceuticals 2021, 14, 520. [Google Scholar] [CrossRef]
- Focosi, D.; Franchini, M.; Pirofski, L.-A.; Burnouf, T.; Fairweather, D.; Joyner, M.J.; Casadevall, A. COVID-19 Convalescent Plasma Is More than Neutralizing Antibodies: A Narrative Review of Potential Beneficial and Detrimental Co-Factors. Viruses 2021, 13, 1594. [Google Scholar] [CrossRef]
- Kim, K.; Bae, K.S.; Kim, H.S.; Lee, W.-Y. Effectiveness of Mesenchymal Stem Cell Therapy for COVID-19-Induced ARDS Patients: A Case Report. Medicina 2022, 58, 1698. [Google Scholar] [CrossRef] [PubMed]
- Almagro, J.C.; Mellado-Sánchez, G.; Pedraza-Escalona, M.; Pérez-Tapia, S.M. Evolution of Anti-SARS-CoV-2 Therapeutic Antibodies. Int. J. Mol. Sci. 2022, 23, 9763. [Google Scholar] [CrossRef] [PubMed]
- Castro e Silva, A.; Bernardes, A.T.; Barbosa, E.A.G.; das Chagas, I.A.S.; Dáttilo, W.; Reis, A.B.; Ribeiro, S.P. Successive Pandemic Waves with Different Virulent Strains and the Effects of Vaccination for SARS-CoV-2. Vaccines 2022, 10, 343. [Google Scholar] [CrossRef] [PubMed]
- Voulgaridi, I.; Sarrou, S.; Dadouli, A.; Peristeri, A.-M.; Nasika, A.; Onoufriadis, I.; Kyritsi, M.A.; Anagnostopoulos, L.; Theodoridou, A.; Avakian, I.; et al. Intensity of Humoral Immune Responses, Adverse Reactions, and Post-Vaccination Morbidity after Adenovirus Vector-Based and mRNA Anti-COVID-19 Vaccines. Vaccines 2022, 10, 1268. [Google Scholar] [CrossRef]
- Hsu, J.-Y.; Mao, Y.-C.; Liu, P.-Y.; Lai, K.-L. Pharmacology and Adverse Events of Emergency-Use Authorized Medication in Moderate to Severe COVID-19. Pharmaceuticals 2021, 14, 955. [Google Scholar] [CrossRef]
- Esposito, R.; Mirra, D.; Sportiello, L.; Spaziano, G.; D’Agostino, B. Overview of Antiviral Drug Therapy for COVID-19: Where Do We Stand? Biomedicines 2022, 10, 2815. [Google Scholar] [CrossRef]
- Terkes, V.; Lisica, K.; Marusic, M.; Verunica, N.; Tolic, A.; Morovic, M. Remdesivir Treatment in Moderately Ill COVID-19 Patients: A Retrospective Single Center Study. J. Clin. Med. 2022, 11, 5066. [Google Scholar] [CrossRef]
- Al-Tannak, N.F.; Novotny, L.; Alhunayan, A. Remdesivir—Bringing Hope for COVID-19 Treatment. Sci. Pharm. 2020, 88, 29. [Google Scholar] [CrossRef]
- Ceramella, J.; Iacopetta, D.; Sinicropi, M.S.; Andreu, I.; Mariconda, A.; Saturnino, C.; Giuzio, F.; Longo, P.; Aquaro, S.; Catalano, A. Drugs for COVID-19: An Update. Molecules 2022, 27, 8562. [Google Scholar] [CrossRef]
- Taieb, F.; Mbaye, K.D.; Tall, B.; Lakhe, N.A.; Talla, C.; Thioub, D.; Ndoye, A.M.; Ka, D.; Gaye, A.; Cissé Diallo, V.M.-P.; et al. Hydroxychloroquine and Azithromycin Treatment of Hospitalized Patients Infected with SARS-CoV-2 in Senegal from March to October. J. Clin. Med. 2021, 10, 2954. [Google Scholar] [CrossRef]
- McAleer, M. Prevention Is Better Than the Cure: Risk Management of COVID-19. J. Risk Financial Manag. 2020, 13, 46. [Google Scholar] [CrossRef] [Green Version]
- Rattanaumpawan, P.; Jirajariyavej, S.; Lerdlamyong, K.; Palavutitotai, N.; Saiyarin, J. Real-World Effectiveness and Optimal Dosage of Favipiravir for Treatment of COVID-19: Results from a Multicenter Observational Study in Thailand. Antibiotics 2022, 11, 805. [Google Scholar] [CrossRef] [PubMed]
- Gentile, I.; Scotto, R.; Schiano Moriello, N.; Pinchera, B.; Villari, R.; Trucillo, E.; Ametrano, L.; Fusco, L.; Castaldo, G.; Buonomo, A.R.; et al. Nirmatrelvir/Ritonavir and Molnupiravir in the Treatment of Mild/Moderate COVID-19: Results of a Real-Life Study. Vaccines 2022, 10, 1731. [Google Scholar] [CrossRef]
- Marino, A.; Munafò, A.; Augello, E.; Bellanca, C.M.; Bonomo, C.; Ceccarelli, M.; Musso, N.; Cantarella, G.; Cacopardo, B.; Bernardini, R. Sarilumab Administration in COVID-19 Patients: Literature Review and Considerations. Infect. Dis. Rep. 2022, 14, 360–371. [Google Scholar] [CrossRef] [PubMed]
- Maraolo, A.E.; Crispo, A.; Piezzo, M.; Di Gennaro, P.; Vitale, M.G.; Mallardo, D.; Ametrano, L.; Celentano, E.; Cuomo, A.; Ascierto, P.A.; et al. The Use of Tocilizumab in Patients with COVID-19: A Systematic Review, Meta-Analysis and Trial Sequential Analysis of Randomized Controlled Studies. J. Clin. Med. 2021, 10, 4935. [Google Scholar] [CrossRef] [PubMed]
- Hussen, J.; Kandeel, M.; Hemida, M.G.; Al-Mubarak, A.I.A. Antibody-Based Immunotherapeutic Strategies for COVID-19. Pathogens 2020, 9, 917. [Google Scholar] [CrossRef] [PubMed]
- Temesgen, Z.; Burger, C.D.; Baker, J.; Polk, C.; Libertin, C.R.; Kelley, C.F.; Marconi, V.C.; Orenstein, R.; Catterson, V.M.; Aronstein, W.S.; et al. Lenzilumab in hospitalised patients with COVID-19 pneumonia (LIVE-AIR): A phase 3, randomised, placebo-controlled trial. Lancet Respir. Med. 2022, 10, 237–246. [Google Scholar] [CrossRef]
- Ho, C.; Lee, P.-C. COVID-19 Treatment—Current Status, Advances, and Gap. Pathogens 2022, 11, 1201. [Google Scholar] [CrossRef]
- Welker, J.; Pulido, J.D.; Catanzaro, A.T.; Malvestutto, C.D.; Li, Z.; Cohen, J.B.; Whitman, E.D.; Byrne, D.; Giddings, O.K.; Lake, J.E.; et al. Efficacy and safety of CD24Fc in hospitalised patients with COVID-19: A randomised, double-blind, placebo-controlled, phase 3 study. Lancet Infect. Dis. 2022, 22, 611–621. [Google Scholar] [CrossRef]
- Gilham, D.; Smith, A.L.; Fu, L.; Moore, D.Y.; Muralidharan, A.; Reid, S.P.M.; Stotz, S.C.; Johansson, J.O.; Sweeney, M.; Wong, N.C.W.; et al. Bromodomain and Extraterminal Protein Inhibitor, Apabetalone (RVX-208), Reduces ACE2 Expression and Attenuates SARS-CoV-2 Infection In Vitro. Biomedicines 2021, 9, 437. [Google Scholar] [CrossRef]
- Wicik, Z.; Eyileten, C.; Jakubik, D.; Simões, S.N.; Martins, D.C., Jr.; Pavão, R.; Siller-Matula, J.M.; Postula, M. ACE2 Interaction Networks in COVID-19: A Physiological Framework for Prediction of Outcome in Patients with Cardiovascular Risk Factors. J. Clin. Med. 2020, 9, 3743. [Google Scholar] [CrossRef]
- Lee, C.-C.; Hsieh, C.-C.; Ko, W.-C. Molnupiravir—A Novel Oral Anti-SARS-CoV-2 Agent. Antibiotics 2021, 10, 1294. [Google Scholar] [CrossRef]
- Jayk Bernal, A.; Gomes da Silva, M.M.; Musungaie, D.B.; Kovalchuk, E.; Gonzalez, A.; Delos Reyes, V.; Martín-Quirós, A.; Caraco, Y.; Williams-Diaz, A.; Brown, M.L.; et al. Molnupiravir for Oral Treatment of COVID-19 in Nonhospitalized Patients. N. Engl. J. Med. 2022, 386, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Corritori, S.; Savchuk, N.; Pauza, C.D. Risk/Benefit Profiles of Currently Approved Oral Antivirals for Treatment of COVID-19: Similarities and Differences. Covid 2022, 2, 1057–1076. [Google Scholar] [CrossRef]
- Alhumaid, S.; Al Mutair, A.; Al Alawi, Z.; Alhmeed, N.; Zaidi, A.R.Z.; Tobaiqy, M. Efficacy and Safety of Lopinavir/Ritonavir for Treatment of COVID-19: A Systematic Review and Meta-Analysis. Trop. Med. Infect. Dis. 2020, 5, 180. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Xie, Y.; Zhu, M.; Yi, D.; Zhao, J.; Guo, S.; Zhang, Y.; Wang, J.; Li, Q.; Wang, Y.; et al. Identification of Darunavir Derivatives for Inhibition of SARS-CoV-2 3CLpro. Int. J. Mol. Sci. 2022, 23, 16011. [Google Scholar] [CrossRef]
- Ahmad, B.; Batool, M.; Ain, Q.U.; Kim, M.S.; Choi, S. Exploring the Binding Mechanism of PF-07321332 SARS-CoV-2 Protease Inhibitor through Molecular Dynamics and Binding Free Energy Simulations. Int. J. Mol. Sci. 2021, 22, 9124. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, H.; Yang, L.; Wang, Q. Progress on COVID-19 Chemotherapeutics Discovery and Novel Technology. Molecules 2022, 27, 8257. [Google Scholar] [CrossRef]
- Mudatsir, M.; Yufika, A.; Nainu, F.; Frediansyah, A.; Megawati, D.; Pranata, A.; Mahdani, W.; Ichsan, I.; Dhama, K.; Harapan, H. Antiviral Activity of Ivermectin Against SARS-CoV-2: An Old-Fashioned Dog with a New Trick—A Literature Review. Sci. Pharm. 2020, 88, 36. [Google Scholar] [CrossRef]
- Samaha, A.A.; Mouawia, H.; Fawaz, M.; Hassan, H.; Salami, A.; Bazzal, A.A.; Saab, H.B.; Al-Wakeel, M.; Alsaabi, A.; Chouman, M.; et al. Retraction: Samaha et al. Effects of a Single Dose of Ivermectin on Viral and Clinical Outcomes in Asymptomatic SARS-CoV-2 Infected Subjects: A Pilot Clinical Trial in Lebanon. Viruses 2021, 13, 989. Viruses 2021, 13, 2154. [Google Scholar] [CrossRef]
- Kumar, S.; Singh, B.; Kumari, P.; Kumar, P.V.; Agnihotri, G.; Khan, S.; Kant Beuria, T.; Syed, G.H.; Dixit, A. Identification of multipotent drugs for COVID-19 therapeutics with the evaluation of their SARS-CoV2 inhibitory activity. Comput. Struct. Biotechnol. J. 2021, 19, 1998–2017. [Google Scholar] [CrossRef] [PubMed]
- Castillejos-López, M.; Torres-Espíndola, L.M.; Huerta-Cruz, J.C.; Flores-Soto, E.; Romero-Martinez, B.S.; Velázquez-Cruz, R.; Higuera-Iglesias, A.; Camarena, Á.; Torres-Soria, A.K.; Salinas-Lara, C.; et al. K.; Salinas-Lara, C.; et al. Ivermectin: A Controversial Focal Point during the COVID-19 Pandemic. Life 2022, 12, 1384. [Google Scholar] [CrossRef] [PubMed]
- Chan-Tack, K.; Sampson, M.; Earp, J.; Arya, V.; Yao, L.; Alexander, J.; Hausman, E.; Belew, Y.; Birnkrant, D.; Struble, K. Considerations and Challenges in the Remdesivir COVID-19 Pediatric Development Program. J. Clin. Pharmacol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Plante, K.S.; Dwivedi, V.; Plante, J.A.; Fernandez, D.; Mirchandani, D.; Bopp, N.; Aguilar, P.V.; Park, J.-G.; Tamayo, P.P.; Delgado, J.; et al. Antiviral activity of oleandrin and a defined extract of Nerium oleander against SARS-CoV-Biomed. Pharmacother. 2021, 138, 111457. [Google Scholar] [CrossRef]
- Kino, T.; Burd, I.; Segars, J.H. Dexamethasone for Severe COVID-19: How Does It Work at Cellular and Molecular Levels? Int. J. Mol. Sci. 2021, 22, 6764. [Google Scholar] [CrossRef]
- Romanou, V.; Koukaki, E.; Chantziara, V.; Stamou, P.; Kote, A.; Vasileiadis, I.; Koutsoukou, A.; Rovina, N. Dexamethasone in the Treatment of COVID-19: Primus Inter Pares? J. Pers. Med. 2021, 11, 556. [Google Scholar] [CrossRef]
- Calzetta, L.; Aiello, M.; Frizzelli, A.; Rogliani, P.; Chetta, A. Dexamethasone in Patients Hospitalized with COVID-19: Whether, When and to Whom. J. Clin. Med. 2021, 10, 1607. [Google Scholar] [CrossRef]
- Maláska, J.; Stašek, J.; Duška, F.; Balík, M.; Máca, J.; Hruda, J.; Vymazal, T.; Klementová, O.; Zatloukal, J.; Gabrhelík, T.; et al. Effect of dexamethasone in patients with ARDS and COVID-19 (REMED trial)—Study protocol for a prospective, multi-centre, open-label, parallel-group, randomized controlled trial. Trials 2022, 23, 1–15. [Google Scholar] [CrossRef]
- Mahdi, M.; Hermán, L.; Réthelyi, J.M.; Bálint, B.L. Potential Role of the Antidepressants Fluoxetine and Fluvoxamine in the Treatment of COVID-19. Int. J. Mol. Sci. 2022, 23, 3812. [Google Scholar] [CrossRef]
- Sukhatme, V.P.; Reiersen, A.M.; Vayttaden, S.J.; Sukhatme, V.V. Fluvoxamine: A Review of Its Mechanism of Action and Its Role in COVID-19. Front. Pharmacol. 2021, 12, 652688. [Google Scholar] [CrossRef]
- Aldén, M.; Olofsson Falla, F.; Yang, D.; Barghouth, M.; Luan, C.; Rasmussen, M.; De Marinis, Y. Intracellular Reverse Transcription of Pfizer BioNTech COVID-19 mRNA Vaccine BNT162b2 In Vitro in Human Liver Cell Line. Curr. Issues Mol. Biol. 2022, 44, 1115–1126. [Google Scholar] [CrossRef] [PubMed]
- Balasubramaniyam, A.; Ryan, E.; Brown, D.; Hamza, T.; Harrison, W.; Gan, M.; Sankhala, R.S.; Chen, W.-H.; Martinez, E.J.; Jensen, J.L.; et al. Unglycosylated Soluble SARS-CoV-2 Receptor Binding Domain (RBD) Produced in E. coli Combined with the Army Liposomal Formulation Containing QS21 (ALFQ) Elicits Neutralizing Antibodies against Mismatched Variants. Vaccines 2023, 11, 42. [Google Scholar] [CrossRef]
- Ghimire, D.; Han, Y.; Lu, M. Structural Plasticity and Immune Evasion of SARS-CoV-2 Spike Variants. Viruses 2022, 14, 1255. [Google Scholar] [CrossRef] [PubMed]
- McGill, A.R.; Kahlil, R.; Dutta, R.; Green, R.; Howell, M.; Mohapatra, S.; Mohapatra, S.S. SARS-CoV-2 Immuno-Pathogenesis and Potential for Diverse Vaccines and Therapies: Opportunities and Challenges. Infect. Dis. Rep. 2021, 13, 102–125. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.-Y.; Liu, W.-C.; Zheng, J.-Q.; Lu, C.-L.; Hou, Y.-C.; Zheng, C.-M.; Song, J.-Y.; Lu, K.-C.; Chao, Y.-C. Immunological Aspects of SARS-CoV-2 Infection and the Putative Beneficial Role of Vitamin-D. Int. J. Mol. Sci. 2021, 22, 5251. [Google Scholar] [CrossRef] [PubMed]
- Coban, M.A.; Morrison, J.; Maharjan, S.; Hernandez Medina, D.; Li, W.; Zhang, Y.S.; Freeman, W.D.; Radisky, E.S.; Le Roch, K.G.; Weisend, C.M.; et al. Attacking COVID-19 Progression Using Multi-Drug Therapy for Synergetic Target Engagement. Biomolecules 2021, 11, 787. [Google Scholar] [CrossRef]
- Holms, R.D. Long COVID (PASC) Is Maintained by a Self-Sustaining Pro-Inflammatory TLR4/RAGE-Loop of S100A8/A9 > TLR4/RAGE Signalling, Inducing Chronic Expression of IL-1b, IL-6 and TNFa: Anti-Inflammatory Ezrin Peptides as Potential Therapy. Immuno 2022, 2, 512–533. [Google Scholar] [CrossRef]
- Saber-Ayad, M.; Hammoudeh, S.; Abu-Gharbieh, E.; Hamoudi, R.; Tarazi, H.; Al-Tel, T.; Hamid, Q. Current Status of Baricitinib as a Repurposed Therapy for COVID-19. Pharmaceuticals 2021, 14, 680. [Google Scholar] [CrossRef]
- A García-García, J.; Pérez-Quintana, M.; Ramos-Giráldez, C.; Cebrián-González, I.; Martín-Ponce, M.L.; del Valle-Villagrán, J.; A Navarro-Puerto, M.; Sánchez-Villegas, J.; Gómez-Herreros, R.; Manoja-Bustos, I.; et al. Anakinra versus Baricitinib: Different Strategies for Patients Hospitalized with COVID-19. J. Clin. Med. 2021, 10, 4019. [Google Scholar] [CrossRef]
- Kojima, Y.; Nakakubo, S.; Takei, N.; Kamada, K.; Yamashita, Y.; Nakamura, J.; Matsumoto, M.; Horii, H.; Sato, K.; Shima, H.; et al. Comparative Efficacy of Tocilizumab and Baricitinib Administration in COVID-19 Treatment: A Retrospective Cohort Study. Medicina 2022, 58, 513. [Google Scholar] [CrossRef]
- Richardson, P.J.; Robinson, B.W.S.; Smith, D.P.; Stebbing, J. The AI-Assisted Identification and Clinical Efficacy of Baricitinib in the Treatment of COVID-19. Vaccines 2022, 10, 951. [Google Scholar] [CrossRef]
- Atluri, K.; Aimlin, I.; Arora, S. Current Effective Therapeutics in Management of COVID-19. J. Clin. Med. 2022, 11, 3838. [Google Scholar] [CrossRef] [PubMed]
- Vannucchi, A.M.; Mortara, A.; D’Alessio, A.; Morelli, M.; Tedeschi, A.; Festuccia, M.B.; Monforte, A.D.; Capochiani, E.; Selleri, C.; Simonetti, F.; et al. JAK Inhibition with Ruxolitinib in Patients with COVID-19 and Severe Pneumonia: Multicenter Clinical Experience from a Compassionate Use Program in Italy. J. Clin. Med. 2021, 10, 3752. [Google Scholar] [CrossRef]
- Gatti, M.; Turrini, E.; Raschi, E.; Sestili, P.; Fimognari, C. Janus Kinase Inhibitors and Coronavirus Disease (COVID)-19: Rationale, Clinical Evidence and Safety Issues. Pharmaceuticals 2021, 14, 738. [Google Scholar] [CrossRef] [PubMed]
- Atzeni, F.; Masala, I.F.; Rodríguez-Carrio, J.; Ríos-Garcés, R.; Gerratana, E.; La Corte, L.; Giallanza, M.; Nucera, V.; Riva, A.; Espinosa, G.; et al. The Rheumatology Drugs for COVID-19 Management: Which and When? J. Clin. Med. 2021, 10, 783. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.W.X.; Ren, D.; Lee, P.H.; Sutjipto, S.; Dugan, C.; Khoo, B.Y.; Tay, J.X.; Vasoo, S.; Young, B.E.; Lye, D.C. Real-World Use of Sotrovimab for Pre-Emptive Treatment in High-Risk Hospitalized COVID-19 Patients: An Observational Cross-Sectional Study. Antibiotics 2022, 11, 345. [Google Scholar] [CrossRef] [PubMed]
- Biscarini, S.; Villa, S.; Genovese, C.; Tomasello, M.; Tonizzo, A.; Fava, M.; Iannotti, N.; Bolis, M.; Mariani, B.; Valzano, A.G.; et al. Safety Profile and Outcomes of Early COVID-19 Treatments in Immunocompromised Patients: A Single-Centre Cohort Study. Biomedicines 2022, 10, 2002. [Google Scholar] [CrossRef] [PubMed]
- Quiros-Roldan, E.; Amadasi, S.; Zanella, I.; Degli Antoni, M.; Storti, S.; Tiecco, G.; Castelli, F. Monoclonal Antibodies against SARS-CoV-2: Current Scenario and Future Perspectives. Pharmaceuticals 2021, 14, 1272. [Google Scholar] [CrossRef] [PubMed]
- Cicchitto, G.; Cardillo, L.; de Martinis, C.; Sabatini, P.; Marchitiello, R.; Abate, G.; Rovetti, A.; Cavallera, A.; Apuzzo, C.; Ferrigno, F.; et al. Effects of Casirivimab/Imdevimab Monoclonal Antibody Treatment among Vaccinated Patients Infected by SARS-CoV-2 Delta Variant. Viruses 2022, 14, 650. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.; Salama, M. Survival Benefit of Tocilizumab in COVID-19 May Be Greater in Patients with Higher Measured Interleukin 6 Levels. COVID 2022, 2, 578–585. [Google Scholar] [CrossRef]
- Conti, V.; Corbi, G.; Sellitto, C.; Sabbatino, F.; Maci, C.; Bertini, N.; De Bellis, E.; Iuliano, A.; Davinelli, S.; Pagliano, P.; et al. Effect of Tocilizumab in Reducing the Mortality Rate in COVID-19 Patients: A Systematic Review with Meta-Analysis. J. Pers. Med. 2021, 11, 628. [Google Scholar] [CrossRef] [PubMed]
- Vena, A.; Cenderello, G.; Balletto, E.; Mezzogori, L.; Santagostino Barbone, A.; Berruti, M.; Ball, L.; Battaglini, D.; Bonsignore, A.; Dentone, C.; et al. Early Administration of Bamlanivimab in Combination with Etesevimab Increases the Benefits of COVID-19 Treatment: Real-World Experience from the Liguria Region. J. Clin. Med. 2021, 10, 4682. [Google Scholar] [CrossRef]
- Focosi, D.; Tuccori, M. Prescription of Anti-Spike Monoclonal Antibodies in COVID-19 Patients with Resistant SARS-CoV-2 Variants in Italy. Pathogens 2022, 11, 823. [Google Scholar] [CrossRef] [PubMed]
- Plichta, J.; Kuna, P.; Panek, M. Monoclonal Antibodies as Potential COVID-19 Therapeutic Agents. COVID 2022, 2, 599–620. [Google Scholar] [CrossRef]
- Drouin, A.C.; Theberge, M.W.; Liu, S.Y.; Smither, A.R.; Flaherty, S.M.; Zeller, M.; Geba, G.P.; Reynaud, P.; Rothwell, W.B.; Luk, A.P.; et al. Successful Clearance of 300 Day SARS-CoV-2 Infection in a Subject with B-Cell Depletion Associated Prolonged (B-DEAP) COVID by REGEN-COV Anti-Spike Monoclonal Antibody Cocktail. Viruses 2021, 13, 1202. [Google Scholar] [CrossRef] [PubMed]
- Focosi, D.; Casadevall, A. A Critical Analysis of the Use of Cilgavimab plus Tixagevimab Monoclonal Antibody Cocktail (Evusheld™) for COVID-19 Prophylaxis and Treatment. Viruses 2022, 14, 1999. [Google Scholar] [CrossRef]
- Awadasseid, A.; Yin, Q.; Wu, Y.; Zhang, W. Potential protective role of the anti-PD-1 blockade against SARS-CoV-2 infection. Biomed. Pharmacother. 2021, 142, 111957. [Google Scholar] [CrossRef]
- Félix, L.; Correia, R.; Sequeira, R.; Ribeiro, C.; Monteiro, S.; Antunes, L.; Silva, J.; Venâncio, C.; Valentim, A. MS-222 and Propofol Sedation during and after the Simulated Transport of Nile tilapia (Oreochromis niloticus). Biology 2021, 10, 1309. [Google Scholar] [CrossRef]
- Beghini, D.G.; Horita, S.I.; Henriques-Pons, A. Mesenchymal Stem Cells in the Treatment of COVID-19, a Promising Future. Cells 2021, 10, 2588. [Google Scholar] [CrossRef]
- Karakaş, N.; Üçüncüoğlu, S.; Uludağ, D.; Karaoğlan, B.S.; Shah, K.; Öztürk, G. Mesenchymal Stem Cell-Based COVID-19 Therapy: Bioengineering Perspectives. Cells 2022, 11, 465. [Google Scholar] [CrossRef]
- Câmara, D.A.D.; Porcacchia, A.S.; Lizier, N.F.; De-Sá-Júnior, P.L. A COVID-19 Overview and Potential Applications of Cell Therapy. Biologics 2021, 1, 177–188. [Google Scholar] [CrossRef]
- De Masi, L.; Argenio, M.A.; Giordano, D.; Facchiano, A. Molecular Aspects of Spike–ACE2 Interaction. Encyclopedia 2022, 2, 96–108. [Google Scholar] [CrossRef]
- Yalcin, H.C.; Sukumaran, V.; Al-Ruweidi, M.K.A.A.; Shurbaji, S. Do Changes in ACE-2 Expression Affect SARS-CoV-2 Virulence and Related Complications: A Closer Look into Membrane-Bound and Soluble Forms. Int. J. Mol. Sci. 2021, 22, 6703. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.C.; Zhang, H.X.; Zhang, Z.; Rinkiko, S.; Cui, Y.M.; Zhu, Y.Z. The Two-Way Switch Role of ACE2 in the Treatment of Novel Coronavirus Pneumonia and Underlying Comorbidities. Molecules 2021, 26, 142. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.; Chen, C.; Yin, T.; Fang, Q.; Hong, Z.; Zhou, R.; Tang, H.; Dong, H. ACE2 and Innate Immunity in the Regulation of SARS-CoV-2-Induced Acute Lung Injury: A Review. Int. J. Mol. Sci. 2021, 22, 11483. [Google Scholar] [CrossRef] [PubMed]
- Triposkiadis, F.; Xanthopoulos, A.; Giamouzis, G.; Boudoulas, K.D.; Starling, R.C.; Skoularigis, J.; Boudoulas, H.; Iliodromitis, E. ACE2, the Counter-Regulatory Renin–Angiotensin System Axis and COVID-19 Severity. J. Clin. Med. 2021, 10, 3885. [Google Scholar] [CrossRef]
- Tan, M.I.; Alfarafisa, N.M.; Septiani, P.; Barlian, A.; Firmansyah, M.; Faizal, A.; Melani, L.; Nugrahapraja, H. Potential Cell-Based and Cell-Free Therapy for Patients with COVID-19. Cells 2022, 11, 2319. [Google Scholar] [CrossRef]
- Rapin, A.; Noujaim, P.-J.; Taiar, R.; Carazo-Mendez, S.; Deslee, G.; Jolly, D.; Boyer, F.C. Characteristics of COVID-19 Inpatients in Rehabilitation Units during the First Pandemic Wave: A Cohort Study from a Large Hospital in Champagne Region. Biology 2022, 11, 937. [Google Scholar] [CrossRef]
- Crum, R.J.; Capella-Monsonís, H.; Badylak, S.F.; Hussey, G.S. Extracellular Vesicles for Regenerative Medicine Applications. Appl. Sci. 2022, 12, 7472. [Google Scholar] [CrossRef]
- Nalesso, F.; Garzotto, F.; Cattarin, L.; Gobbi, L.; Qassim, L.; Sgarabotto, L.; Tiberio, I.; Calò, L.A. A Continuous Renal Replacement Therapy Protocol for Patients with Acute Kidney Injury in Intensive Care Unit with COVID-19. J. Clin. Med. 2020, 9, 1529. [Google Scholar] [CrossRef]
- Chávez-Valencia, V.; Orizaga-De-La-Cruz, C.; Lagunas-Rangel, F.A. Acute Kidney Injury in COVID-19 Patients: Pathogenesis, Clinical Characteristics, Therapy, and Mortality. Diseases 2022, 10, 53. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, V.; Aquino, R.P.; Del Gaudio, P.; Campiglia, P.; Russo, P. Post-COVID Syndrome: The Research Progress in the Treatment of Pulmonary sequelae after COVID-19 Infection. Pharmaceutics 2022, 14, 1135. [Google Scholar] [CrossRef] [PubMed]
- Souri, M.; Chiani, M.; Farhangi, A.; Mehrabi, M.R.; Nourouzian, D.; Raahemifar, K.; Soltani, M. Anti-COVID-19 Nanomaterials: Directions to Improve Prevention, Diagnosis, and Treatment. Nanomaterials 2022, 12, 783. [Google Scholar] [CrossRef] [PubMed]
- Ramezani, Z.; Dayer, M.R.; Noorizadeh, S.; Thompson, M. Deactivation of SARS-CoV-2 via Shielding of Spike Glycoprotein Using Carbon Quantum Dots: Bioinformatic Perspective. COVID 2021, 1, 120–129. [Google Scholar] [CrossRef]
- He, Q.; Lu, J.; Liu, N.; Lu, W.; Li, Y.; Shang, C.; Li, X.; Hu, L.; Jiang, G. Antiviral Properties of Silver Nanoparticles against SARS-CoV-2: Effects of Surface Coating and Particle Size. Nanomaterials 2022, 12, 990. [Google Scholar] [CrossRef] [PubMed]
- Merkl, P.; Long, S.; McInerney, G.M.; Sotiriou, G.A. Antiviral Activity of Silver, Copper Oxide and Zinc Oxide Nanoparticle Coatings against SARS-CoV-2. Nanomaterials 2021, 11, 1312. [Google Scholar] [CrossRef] [PubMed]
- Mosselhy, D.A.; Kareinen, L.; Kivistö, I.; Aaltonen, K.; Virtanen, J.; Ge, Y.; Sironen, T. Copper-Silver Nanohybrids: SARS-CoV-2 Inhibitory Surfaces. Nanomaterials 2021, 11, 1820. [Google Scholar] [CrossRef]
- Al-Hindawi, A.; AlDallal, U.; Waly, Y.M.; Hussain, M.H.; Shelig, M.; Saleh ElMitwalli, O.S.M.M.; Deen, G.R.; Henari, F.Z. An Exploration of Nanoparticle-Based Diagnostic Approaches for Coronaviruses: SARS-CoV-2, SARS-CoV and MERS-CoV. Nanomaterials 2022, 12, 3550. [Google Scholar] [CrossRef]
- Ardekani, L.S.; Thulstrup, P.W. Gold Nanoparticle-Mediated Lateral Flow Assays for Detection of Host Antibodies and COVID-19 Proteins. Nanomaterials 2022, 12, 1456. [Google Scholar] [CrossRef]
- Pranskuniene, Z.; Balciunaite, R.; Simaitiene, Z.; Bernatoniene, J. Herbal Medicine Uses for Respiratory System Disorders and Possible Trends in New Herbal Medicinal Recipes during COVID-19 in Pasvalys District, Lithuania. Int. J. Environ. Res. Public Health 2022, 19, 8905. [Google Scholar] [CrossRef]
- Mamedov, T.; Yuksel, D.; Ilgın, M.; Gürbüzaslan, I.; Gulec, B.; Mammadova, G.; Ozdarendeli, A.; Yetiskin, H.; Kaplan, B.; Islam Pavel, S.T.; et al. Production and Characterization of Nucleocapsid and RBD Cocktail Antigens of SARS-CoV-2 in Nicotiana benthamiana Plant as a Vaccine Candidate against COVID-19. Vaccines 2021, 9, 1337. [Google Scholar] [CrossRef] [PubMed]
- Ruocco, V.; Strasser, R. Transient Expression of Glycosylated SARS-CoV-2 Antigens in Nicotiana benthamiana. Plants 2022, 11, 1093. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.L.; Raju, C.S.; Mahboob, T.; Kayesth, S.; Gupta, K.K.; Jain, G.K.; Dhobi, M.; Nawaz, M.; Wilairatana, P.; de Lourdes Pereira, M.; et al. Precision and Advanced Nano-Phytopharmaceuticals for Therapeutic Applications. Nanomaterials 2022, 12, 238. [Google Scholar] [CrossRef] [PubMed]
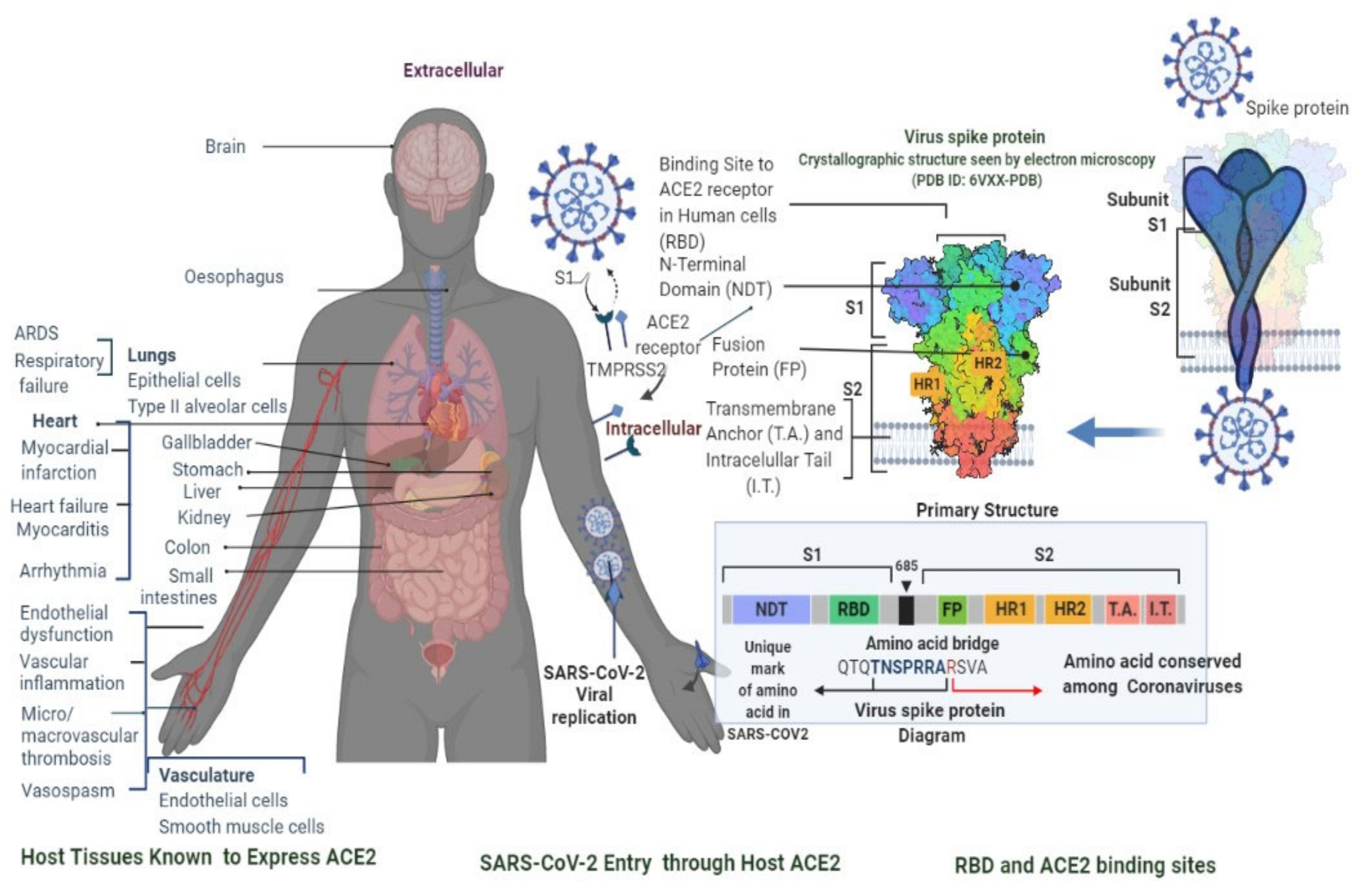
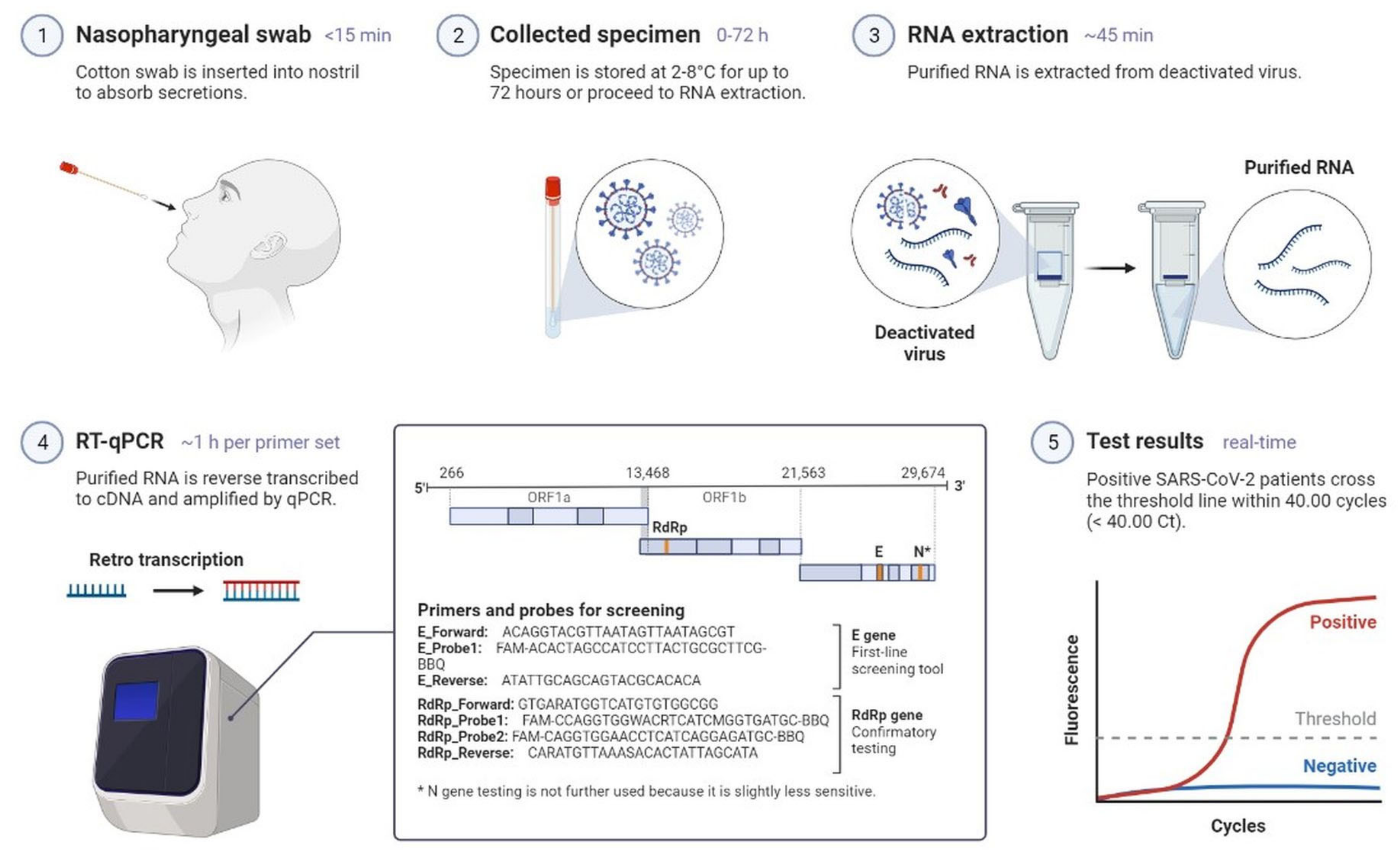
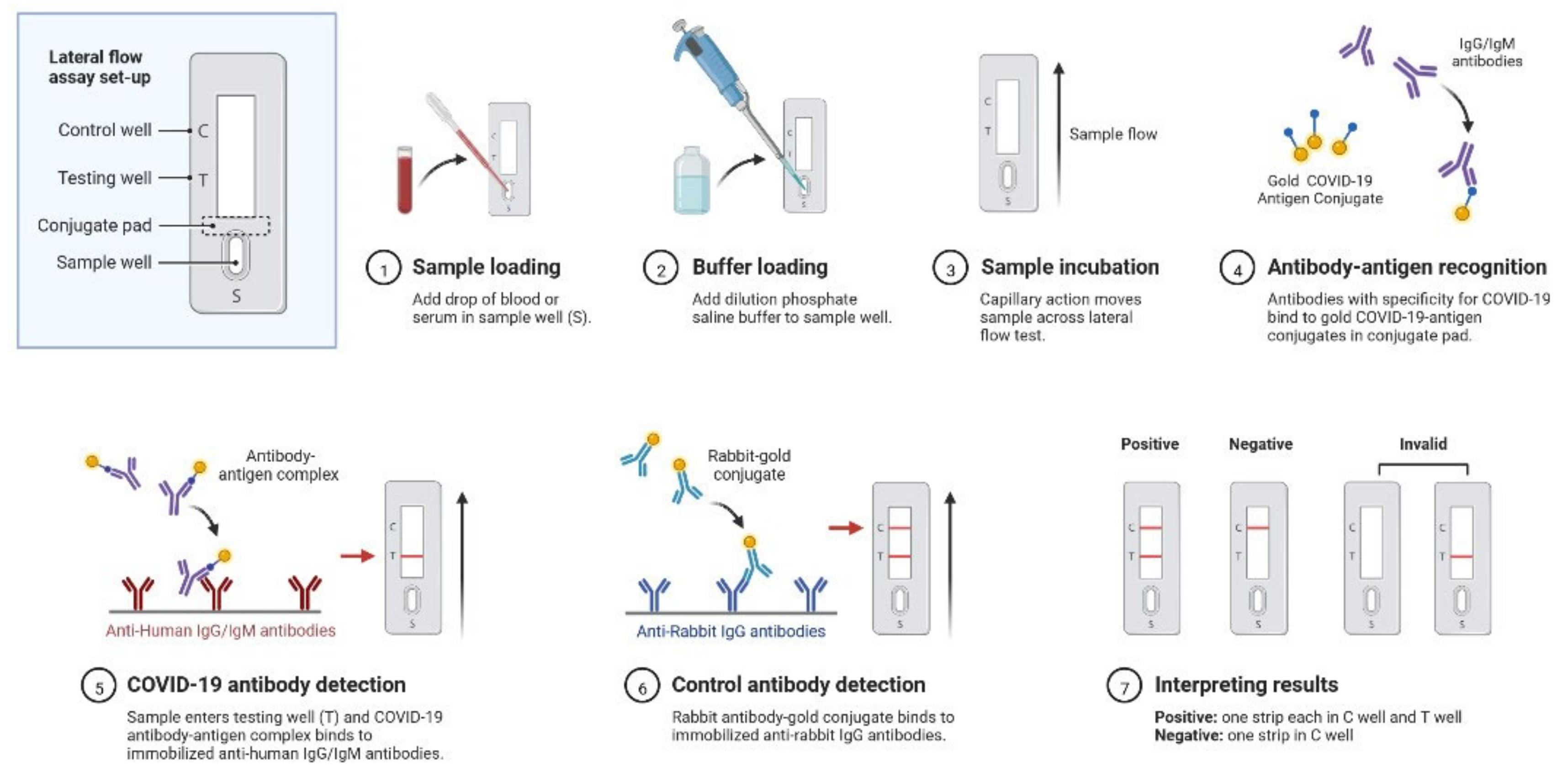

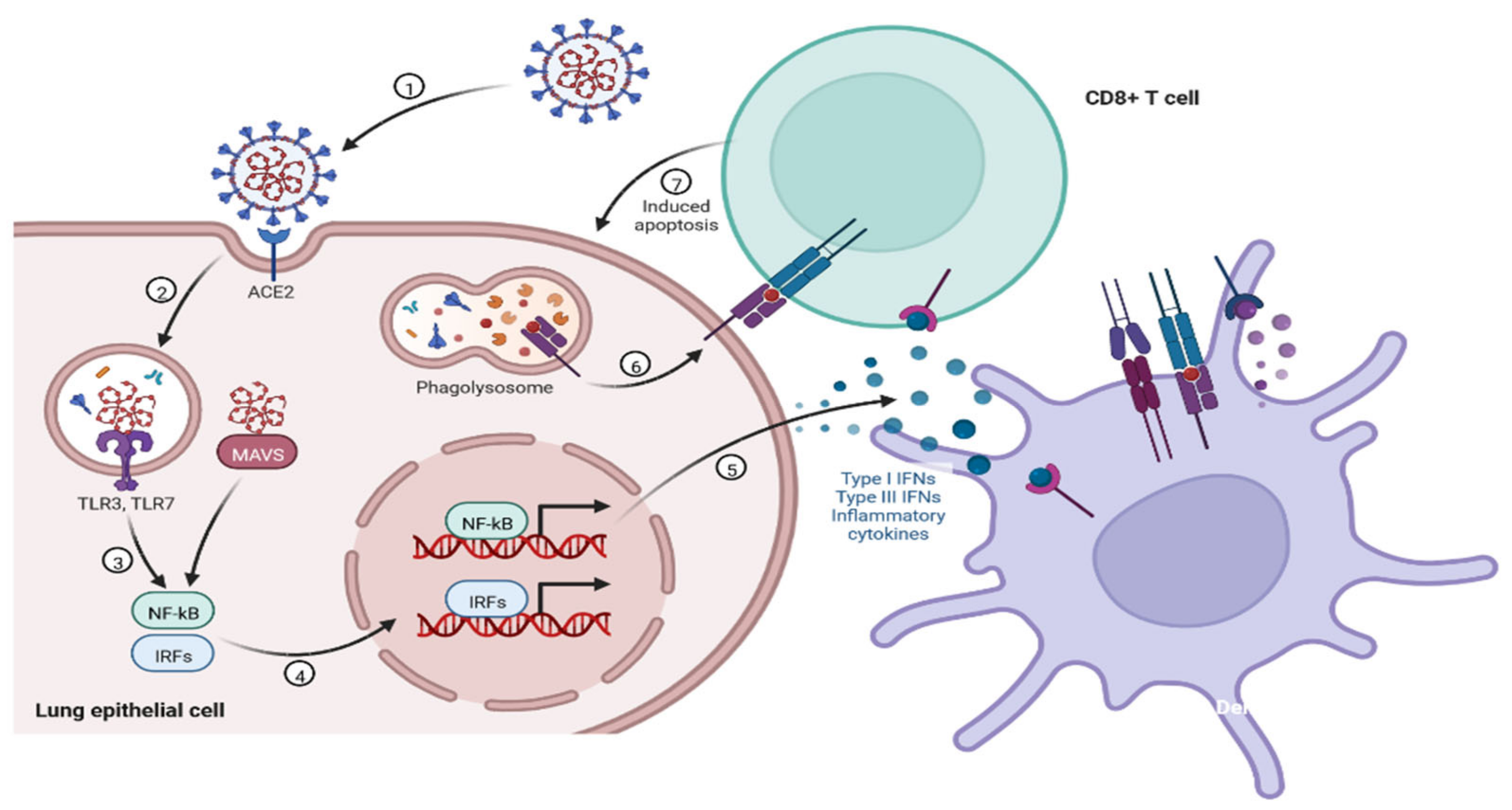





| Characteristics | SARS-CoV-2 | MERS-CoV | SARS-CoV |
|---|---|---|---|
| Outbreak Beginning Date | December 2019 [18] | April 2012 [20] | November 2002 [20] |
| Location of the First Case | Wuhan, China [18] | Saudi Arabia [20] | Guangdong, China [20] |
| Confirmed Cases | 651,918,402 [From December 2019 to 23 December 2022] [22] | 2519 (From 2012 until 31 January 2020) [20] | 8096 [20] |
| Latency | Between 5 and 14 days [18,19] | Between 2 and 14 days [20] | Between 2 and 7 days [20] |
| Contagious Period | 5 to 20 days after onset of disease, depends on severity of disease | 5–14 days after onset of disease [20] | 10 days after onset of disease [20] |
| Fatality Rate | ~2.3% [19] | ~36% [20] | ~10% [20] |
| Incidental Host | Malayan pangolin [19,20] | Dromedary camels [20] | Masked palm civets [20] |
| Transmission | Respiratory droplets Possibly faecal-oral Close contact with diseased patients Possibly aerosol [18,19] | Respiratory droplets Ingestion of camel milk Close contact with diseased patients/camels [20] | Respiratory droplets Faecal-oral Close contact with diseased patients Aerosol [20] |
| Radiologic Features | Various, from focal faint patchy ground-glass opacity to bilateral ambiguous consolidation of the air space of plain chest radiographs. Not sufficiently unique to differentiate between the three diseases [20]. | ||
| Clinical Presentation | From asymptomatic or moderate illness to severe upper-respiratory distress and death-related multiorgan failure, varying from person to person. Diarrhoea is recorded as well [20]. | ||
| Lineage | Strain | Mutations (Spike Protein) | First Identified | Effects on Clinical Outcome | References |
|---|---|---|---|---|---|
| B.1.1.7 | 20I/501Y.V1 | N501Y, A570D, D614G, P681H, Δ69/70, Δ144Y. | United Kingdom | Increased severity based on hospitalizations and case fatality rates Limited effect of monoclonal antibody treatments on neutralising | [10,33] |
| P.1 | 20J/501Y.V3 | K417N/T E484K N501Y D614G | Japan/Brazil | Significant effect of some monoclonal antibody therapy on neutralising Bamlanivimab–etesevimab: reduced susceptibility. Casirivimab-imdevimab: No change in susceptibility | [33,35] |
| B.1.351 | 20H/501.V2 | N501Y D614G K417N E484K | South Africa | Increased transmission (50%). Bamlanivimab–etesevimab: >45-fold decrease in susceptibility | [35] |
| B.1.427 and B.1.429 | 20C/S:452R | S13I W152C L452R D614G | US-California | Increased transmission (20%). Bamlanivimab–etesevimab: Active but 7.4-fold reduced the susceptibility | [33] |
| Name of Emerged Variants | Lineage | Name of Country (First Identified) | Mutation of Interest in Spike Region | Year and Month First Detected | Effect on Disease Severity | Rate of Transmission | References |
|---|---|---|---|---|---|---|---|
| Omicron | BA.2 | South Africa | T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q954H, N969K, G142D, N211I, Δ212, V213G, G339D, S371F, S373P, S375F, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y | In November 2021 | High | Community level | [32] |
| Omicron | BA.4 | South Africa | L452R, F486V, R493Q | In January 2022 | High | Community level | [38] |
| Omicron | BA.5 | South Africa | L452R, F486V, R493Q | In February 2022 | High | Community level | [37] |
| Name of Emerged Variants | Lineage | Name of Country (First Identified) | Mutation of Interest in Spike Region | Year and Month First Detected | Effect on Disease Severity | Rate of Transmission | References |
|---|---|---|---|---|---|---|---|
| Omicron | BA.2.75 | India | W152R, F157L, I210V, G257S, D339H, G446S, N460K, Q493 (reversion) | In May 2022 | High | Not Known | [32] |
| Omicron | BQ.1 | No evidence | K444T, N460K | No evidence | High | No evidence | [36] |
| Name of Emerged Variants | Lineage | Name of Country (First Identified) | Mutation of Interest in Spike Region | Year and Month First Detected | Effect on Disease Severity | Rate of Transmission | References |
|---|---|---|---|---|---|---|---|
| Omicron | B.1.1.529+R346X Any amino-acid substitution | No evidence | R346X | No evidence | No evidence | [33] | |
| Omicron | B.1.1.529+K444X, N460X | No evidence | K444X, N460X | No evidence | High | No evidence | [37] |
| Omicron | B.1.1.529+N460X, F490X, including XBB and other sub-lineages | No evidence | N460X, F490X | No evidence | High | No evidence | [38] |
| Name of the Drug | Potential | Description | References |
|---|---|---|---|
| Remdesivir | Used as antiviral, inhibiting RNA synthesis in coronaviruses | Selectively provided in 2020 for COVID-19 emergency use, both promising and negative effects reported; Trial was sponsored by Gilead, WHO, INSERM, NIAID. | [49,50,51] |
| Hydroxychloroquine or chloroquine | Used for malaria, lupus (international), rheumatoid arthritis | Selectively provided in 2020 for COVID-19 emergency use, both promising and negative effects reported; Trial was sponsored by CEPI, WHO, INSERM; Discontinued by WHO. | [52] |
| Favipiravir | Used as antiviral against influenza | Used against COVID-19 in 2020; Favipiravir reduced mortality, but the finding was not statistically significant; Trial was sponsored by Fujifilm, China. | [53] |
| Lopinavir/ritonavir without or with interferon beta-1a | Used as antiviral, immune suppression. | Ritonavir has been proposed as a treatment for COVID-19 based on in vitro activity, preclinical studies, and observational studies; Trial was sponsored by CEPI, WHO, UK Government, Univ. of Oxford, INSERM | [54] |
| Sarilumab | Human monoclonal antibody against interleukin-6 receptor | Clinical efficacy of sarilumab relative to the control arm in adult participants hospitalized with severe or critical coronavirus disease 2019; Clinical trial sponsored by Regeneron-Sanofi. | [55] |
| Ritonavir+ ASC-09 | Antiviral | Clinical trial determined whether PF-07321332/ritonavir is safe and effective for the treatment of adults; Trial was performed at multiple sites in China; Combination not approved; ritonavir approved for HIV. | [56] |
| Tocilizumab | Used in rheumatoid arthritis, immune suppression (US, Europe). | Ritonavir has been proposed as a treatment for COVID-19 based on Phase III trial and sponsored by Tocilizumab Genentech-Hoffmann-La Roche | [57] |
| Lenzilumab | New drug candidate; Humanized monoclonal antibody for relieving pneumonia | Tested against COVID-19 in 2020; Trial was sponsored by Humanigen, Inc. | [58] |
| Dapagliflozin | Used as hypoglycaemia agent | Dapagliflozin did not significantly reduce the severity of infection; Trial was sponsored in 2020 by Saint Luke’s Mid America Heart Institute, AstraZeneca. | [59] |
| CD24Fc | Used as antiviral immunomodulator against inflammatory response | Phase III study to evaluate the safety and efficacy of CD24Fc in COVID-19 treatment; Trial was sponsored by OncoImmune, Inc. in 2021 | [60] |
| Apabetalone | Selective BET inhibitor | Tested against COVID-19 in 2020; Trial was sponsored by Resverlogix Corp | [61] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, D.D.; Han, I.; Choi, E.-H.; Yadav, D.K. A Clinical Update on SARS-CoV-2: Pathology and Development of Potential Inhibitors. Curr. Issues Mol. Biol. 2023, 45, 400-433. https://doi.org/10.3390/cimb45010028
Singh DD, Han I, Choi E-H, Yadav DK. A Clinical Update on SARS-CoV-2: Pathology and Development of Potential Inhibitors. Current Issues in Molecular Biology. 2023; 45(1):400-433. https://doi.org/10.3390/cimb45010028
Chicago/Turabian StyleSingh, Desh Deepak, Ihn Han, Eun-Ha Choi, and Dharmendra Kumar Yadav. 2023. "A Clinical Update on SARS-CoV-2: Pathology and Development of Potential Inhibitors" Current Issues in Molecular Biology 45, no. 1: 400-433. https://doi.org/10.3390/cimb45010028
APA StyleSingh, D. D., Han, I., Choi, E.-H., & Yadav, D. K. (2023). A Clinical Update on SARS-CoV-2: Pathology and Development of Potential Inhibitors. Current Issues in Molecular Biology, 45(1), 400-433. https://doi.org/10.3390/cimb45010028








