Surface Engineering of Escherichia coli to Display Its Phytase (AppA) and Functional Analysis of Enzyme Activities
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Strains and Plasmids
2.3. PCR Amplification of AppA
2.4. Construction of pESE-AppA
2.5. Overexpression of the AppA Protein
2.6. Analysis of AppA Expression and Display
2.6.1. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
2.6.2. Immunofluorescence Microscopy (IFM)
2.6.3. Cell-based Acid Phosphatase Assay
2.7. Outer Membrane (OM)-Linked Activity Assays
2.7.1. OM Isolation
2.7.2. Acid Phosphatase Assay
2.7.3. Phytase Assay
2.7.4. MG Colorimetric Assay
2.8. Simulation of Poultry Digestive Tract
2.9. Data Analysis
3. Results and Discussion
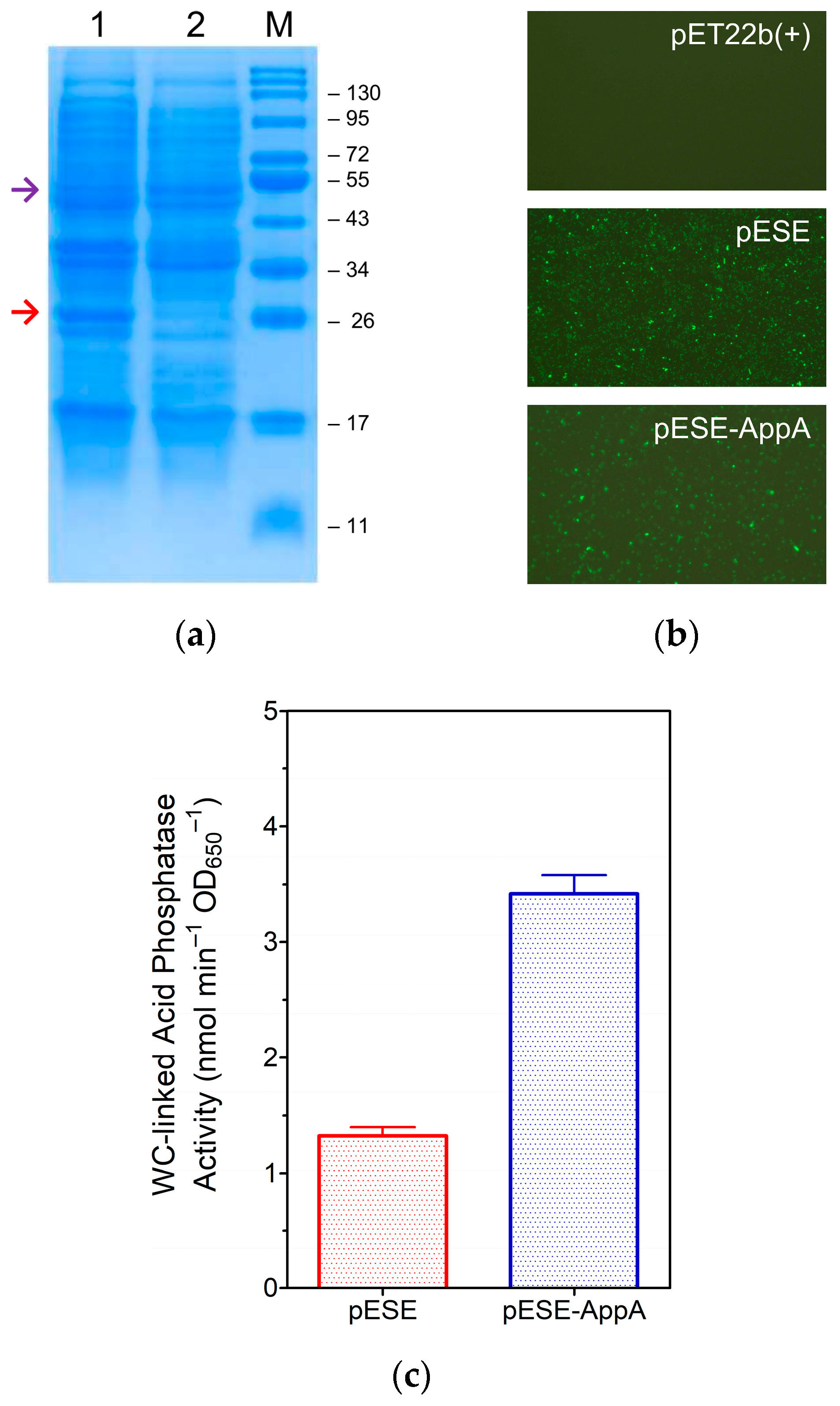
3.1. Production of eLpp-OmpA-AppA and Display of AppA
3.1.1. Construction of the Expression Plasmid, pESE-AppA
3.1.2. Expression of eLpp-OmpA-AppA and Display of AppA
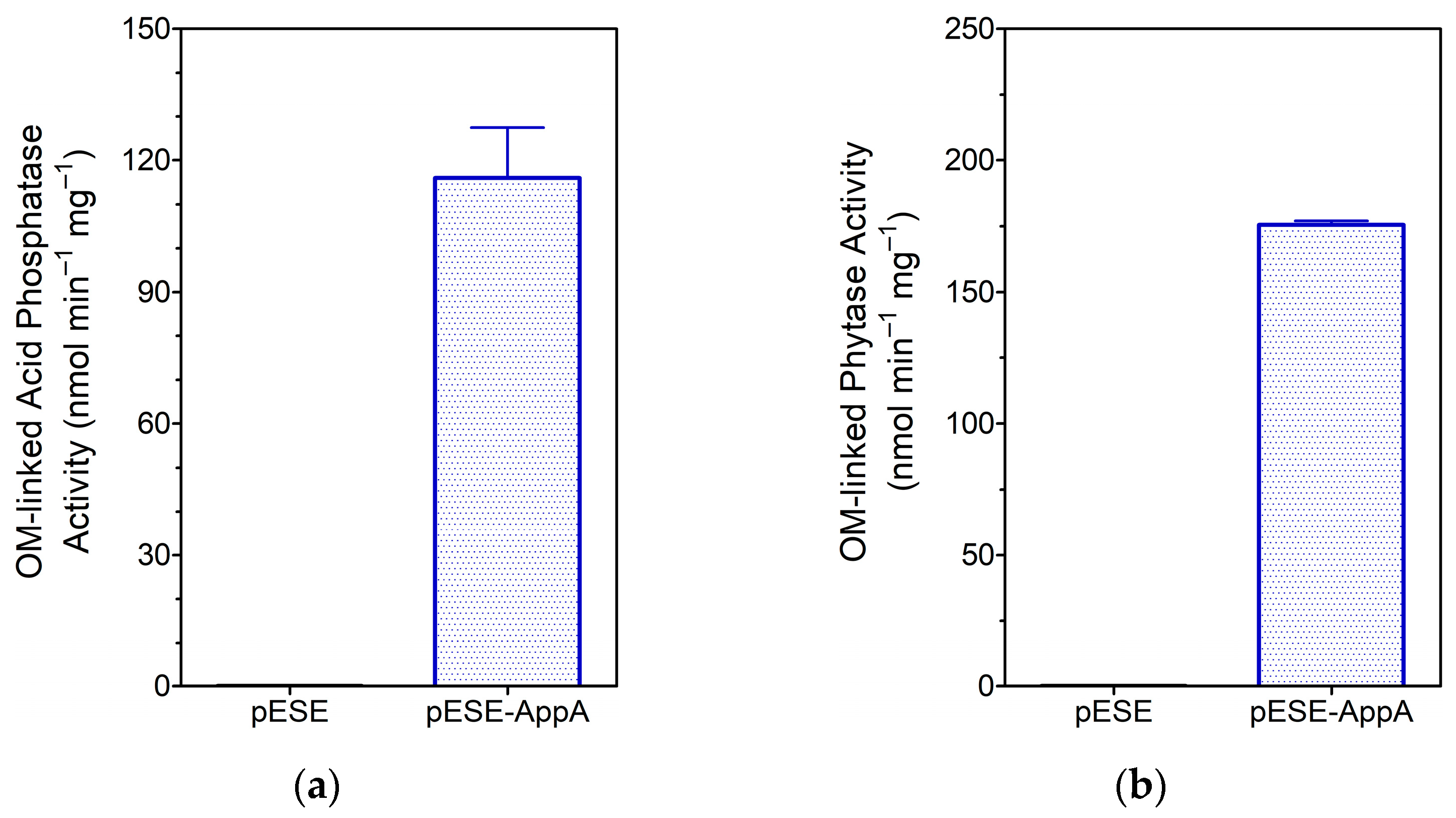
3.1.3. Analysis of OM-Linked AppA Activities
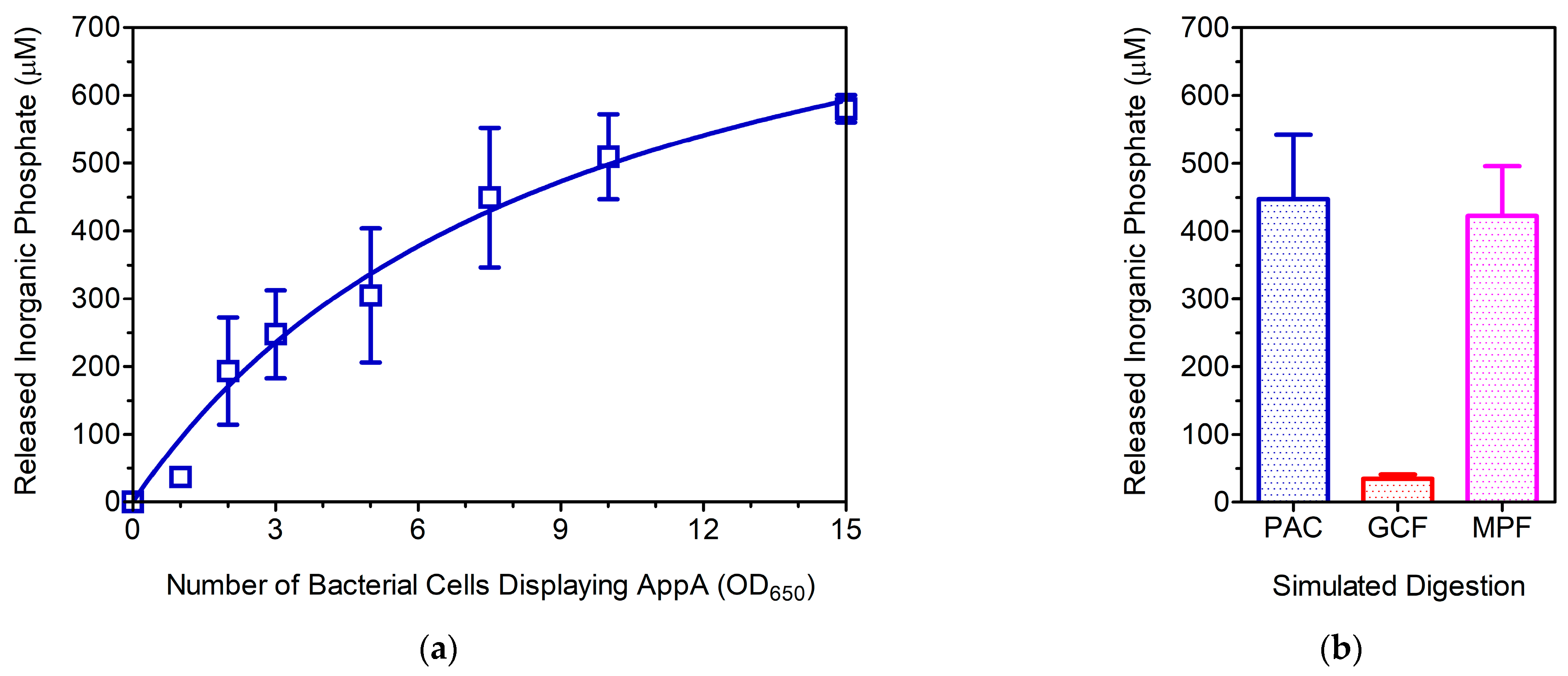
3.2. Evaluation of the WC Biocatalyst as a Poultry Feed Additive
3.3. Is MWCB-Phytase Promising for Monogastric Animal Feed?
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, S.Y.; Choi, J.H.; Xu, Z. Microbial Cell-Surface Display. Trends Biotechnol. 2003, 21, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Cherf, G.M.; Cochran, J.R. Applications of Yeast Surface Display for Protein Engineering. In Yeast Surface Display; Liu, B., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2015; Volume 1319, pp. 155–175. ISBN 978-1-4939-2747-0. [Google Scholar]
- Parwin, S.; Kalan, S.; Srivastava, P. Bacterial Cell Surface Display. In ACS Symposium Series; Rathinam, N.K., Sani, R.K., Eds.; American Chemical Society: Washington, DC, USA, 2019; Volume 1329, pp. 81–108. ISBN 978-0-8412-3500-7. [Google Scholar]
- Zhang, C.; Chen, H.; Zhu, Y.; Zhang, Y.; Li, X.; Wang, F. Saccharomyces Cerevisiae Cell Surface Display Technology: Strategies for Improvement and Applications. Front. Bioeng. Biotechnol. 2022, 10, 1056804. [Google Scholar] [CrossRef] [PubMed]
- Han, M.-J.; Lee, S.H. An Efficient Bacterial Surface Display System Based on a Novel Outer Membrane Anchoring Element from the Escherichia coli Protein YiaT. FEMS Microbiol. Lett. 2015, 362, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Zhao, Y.; Cui, S.; Liang, B. Redesigning of Microbial Cell Surface and Its Application to Whole-Cell Biocatalysis and Biosensors. Appl. Biochem. Biotechnol. 2018, 185, 396–418. [Google Scholar] [CrossRef] [PubMed]
- Nicchi, S.; Giuliani, M.; Giusti, F.; Pancotto, L.; Maione, D.; Delany, I.; Galeotti, C.L.; Brettoni, C. Decorating the Surface of Escherichia coli with Bacterial Lipoproteins: A Comparative Analysis of Different Display Systems. Microb. Cell Factories 2021, 20, 33. [Google Scholar] [CrossRef] [PubMed]
- Cole, G.B.; Bateman, T.J.; Moraes, T.F. The Surface Lipoproteins of Gram-Negative Bacteria: Protectors and Foragers in Harsh Environments. J. Biol. Chem. 2021, 296, 100147. [Google Scholar] [CrossRef] [PubMed]
- Kjærgaard, K.; Hasman, H.; Schembri, M.A.; Klemm, P. Antigen 43-Mediated Autotransporter Display, a Versatile Bacterial Cell Surface Presentation System. J. Bacteriol. 2002, 184, 4197–4204. [Google Scholar] [CrossRef] [PubMed]
- Tabañag, I.D.F.; Chu, I.-M.; Wei, Y.-H.; Tsai, S.-L. The Role of Yeast-Surface-Display Techniques in Creating Biocatalysts for Consolidated BioProcessing. Catalysts 2018, 8, 94. [Google Scholar] [CrossRef]
- Uchański, T.; Zögg, T.; Yin, J.; Yuan, D.; Wohlkönig, A.; Fischer, B.; Rosenbaum, D.M.; Kobilka, B.K.; Pardon, E.; Steyaert, J. An Improved Yeast Surface Display Platform for the Screening of Nanobody Immune Libraries. Sci. Rep. 2019, 9, 382. [Google Scholar] [CrossRef]
- Lozančić, M.; Sk. Hossain, A.; Mrša, V.; Teparić, R. Surface Display—An Alternative to Classic Enzyme Immobilization. Catalysts 2019, 9, 728. [Google Scholar] [CrossRef]
- Kuroda, K.; Ueda, M. Arming Technology in Yeast—Novel Strategy for Whole-Cell Biocatalyst and Protein Engineering. Biomolecules 2013, 3, 632–650. [Google Scholar] [CrossRef] [PubMed]
- Ueda, M. Establishment of Cell Surface Engineering and Its Development. Biosci. Biotechnol. Biochem. 2016, 80, 1243–1253. [Google Scholar] [CrossRef] [PubMed]
- Hui, C.-Y.; Guo, Y.; Yang, X.-Q.; Zhang, W.; Huang, X.-Q. Surface Display of Metal Binding Domain Derived from PbrR on Escherichia coli Specifically Increases Lead(II) Adsorption. Biotechnol. Lett. 2018, 40, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Erpel, F.; Restovic, F.; Arce-Johnson, P. Development of Phytase-Expressing Chlamydomonas Reinhardtii for Monogastric Animal Nutrition. BMC Biotechnol. 2016, 16, 29. [Google Scholar] [CrossRef] [PubMed]
- Madsen, C.K.; Brinch-Pedersen, H. Molecular Advances on Phytases in Barley and Wheat. Int. J. Mol. Sci. 2019, 20, 2459. [Google Scholar] [CrossRef] [PubMed]
- Mingmongkolchai, S.; Panbangred, W. Display of Escherichia coli Phytase on the Surface of Bacillus Subtilis Spore Using CotG as an Anchor Protein. Appl. Biochem. Biotechnol. 2019, 187, 838–855. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Han, R.; Cao, Y.; Turner, B.L.; Ma, L.Q. Enhancing Phytate Availability in Soils and Phytate-P Acquisition by Plants: A Review. Environ. Sci. Technol. 2022, 56, 9196–9219. [Google Scholar] [CrossRef] [PubMed]
- Humer, E.; Schwarz, C.; Schedle, K. Phytate in Pig and Poultry Nutrition. J. Anim. Physiol. Anim. Nutr. 2015, 99, 605–625. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.K.; Gangoliya, S.S.; Singh, N.K. Reduction of Phytic Acid and Enhancement of Bioavailable Micronutrients in Food Grains. J. Food Sci. Technol. 2015, 52, 676–684. [Google Scholar] [CrossRef]
- Samtiya, M.; Aluko, R.E.; Dhewa, T. Plant Food Anti-Nutritional Factors and Their Reduction Strategies: An Overview. Food Prod. Process. Nutr. 2020, 2, 6. [Google Scholar] [CrossRef]
- Kumar, V.; Singh, D.; Sangwan, P.; Gill, P.K. Management of Environmental Phosphorus Pollution Using Phytases: Current Challenges and Future Prospects. In Applied Environmental Biotechnology: Present Scenario and Future Trends; Kaushik, G., Ed.; Springer: New Delhi, India, 2015; pp. 97–114. ISBN 978-81-322-2122-7. [Google Scholar]
- Joudaki, H.; Aria, N.; Moravej, R.; Rezaei Yazdi, M.; Emami-Karvani, Z.; Hamblin, M.R. Microbial Phytases: Properties and Applications in the Food Industry. Curr. Microbiol. 2023, 80, 374. [Google Scholar] [CrossRef] [PubMed]
- Rizwanuddin, S.; Kumar, V.; Naik, B.; Singh, P.; Mishra, S.; Rustagi, S.; Kumar, V. Microbial Phytase: Their Sources, Production, and Role in the Enhancement of Nutritional Aspects of Food and Feed Additives. J. Agric. Food Res. 2023, 12, 100559. [Google Scholar] [CrossRef]
- Golovan, S.; Wang, G.; Zhang, J.; Forsberg, C.W. Characterization and Overproduction of the Escherichia coli appA Encoded Bifunctional Enzyme That Exhibits Both Phytase and Acid Phosphatase Activities. Can. J. Microbiol. 1999, 46, 59–71. [Google Scholar] [CrossRef]
- Dassa, E.; Cahu, M.; Desjoyaux-Cherel, B.; Boquet, P.L. The Acid Phosphatase with Optimum pH of 2.5 of Escherichia coli. Physiological and Biochemical Study. J. Biol. Chem. 1982, 257, 6669–6676. [Google Scholar] [CrossRef] [PubMed]
- Greiner, R.; Carlsson, N.-G.; Alminger, M.L. Stereospecificity of Myo-Inositol Hexakisphosphate Dephosphorylation by a Phytate-Degrading Enzyme of Escherichia coli. J. Biotechnol. 2000, 84, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Acquistapace, I.M.; Thompson, E.J.; Kühn, I.; Bedford, M.R.; Brearley, C.A.; Hemmings, A.M. Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA. Int. J. Mol. Sci. 2022, 23, 6346. [Google Scholar] [CrossRef] [PubMed]
- Dersjant-Li, Y.; Awati, A.; Schulze, H.; Partridge, G. Phytase in Non-ruminant Animal Nutrition: A Critical Review on Phytase Activities in the Gastrointestinal Tract and Influencing Factors. J. Sci. Food Agric. 2015, 95, 878–896. [Google Scholar] [CrossRef] [PubMed]
- Nezhad, N.G.; Raja Abd Rahman, R.N.Z.; Normi, Y.M.; Oslan, S.N.; Shariff, F.M.; Leow, T.C. Integrative Structural and Computational Biology of Phytases for the Animal Feed Industry. Catalysts 2020, 10, 844. [Google Scholar] [CrossRef]
- Mo, A.-Y.; Park, S.-M.; Kim, Y.-S.; Yang, M.-S.; Kim, D.-H. Expression of Fungal Phytase on the Cell Surface ofSaccharomyces Cerevisiae. Biotechnol. Bioprocess Eng. 2005, 10, 576. [Google Scholar] [CrossRef]
- Harnpicharnchai, P.; Sornlake, W.; Tang, K.; Eurwilaichitr, L.; Tanapongpipat, S. Cell-Surface Phytase on Pichia Pastoris Cell Wall Offers Great Potential as a Feed Supplement. FEMS Microbiol. Lett. 2010, 302, 8–14. [Google Scholar] [CrossRef]
- Li, C.; Lin, Y.; Huang, Y.; Liu, X.; Liang, S. Citrobacter Amalonaticus Phytase on the Cell Surface of Pichia Pastoris Exhibits High pH Stability as a Promising Potential Feed Supplement. PLoS ONE 2014, 9, e114728. [Google Scholar] [CrossRef]
- Gallus, S.; Mittmann, E.; Rabe, K.S. A Modular System for the Rapid Comparison of Different Membrane Anchors for Surface Display on Escherichia coli. ChemBioChem 2022, 23, e202100472. [Google Scholar] [CrossRef] [PubMed]
- Nanudorn, P.; Thiengmag, S.; Whangsuk, W.; Mongkolsuk, S.; Loprasert, S. Potential Use of Two Aryl Sulfotransferase Cell-Surface Display Systems to Detoxify the Endocrine Disruptor Bisphenol A. Biochem. Biophys. Res. Commun. 2020, 528, 691–697. [Google Scholar] [CrossRef]
- Francisco, J.A.; Earhart, C.F.; Georgiou, G. Transport and Anchoring of Beta-Lactamase to the External Surface of Escherichia coli. Proc. Natl. Acad. Sci. USA 1992, 89, 2713–2717. [Google Scholar] [CrossRef]
- Jo, J.-H.; Han, C.-W.; Kim, S.-H.; Kwon, H.-J.; Lee, H.-H. Surface Display Expression of Bacillus Licheniformis Lipase in Escherichia coli Using Lpp’OmpA Chimera. J. Microbiol. 2014, 52, 856–862. [Google Scholar] [CrossRef] [PubMed]
- Hantke, K.; Braun, V. Covalent Binding of Lipid to Protein: Diglyceride and Amide-Linked Fatty Acid at the N-Terminal End of the Murein-Lipoprotein of the Escherichia coli Outer Membrane. Eur. J. Biochem. 1973, 34, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, G.; Stephens, D.L.; Stathopoulos, C.; Poetschke, H.L.; Mendenhall, J.; Earhart, C.F. Display of β-Lactamase on the Escherichia coli Surface: Outer Membrane Phenotypes Conferred by Lpp′–OmpA′–β-Lactamase Fusions. Protein Eng. Des. Sel. 1996, 9, 239–247. [Google Scholar] [CrossRef]
- Torres-Bañaga, R.; Mares-Alejandre, R.E.; Terán-Ramírez, C.; Estrada-González, A.L.; Muñoz-Muñoz, P.L.A.; Meléndez-López, S.G.; Rivero, I.A.; Ramos-Ibarra, M.A. Functional Display of an Amoebic Chitinase in Escherichia coli Expressing the Catalytic Domain of EhCHT1 on the Bacterial Cell Surface. Appl. Biochem. Biotechnol. 2020, 192, 1255–1269. [Google Scholar] [CrossRef]
- Pocanschi, C.L.; Popot, J.-L.; Kleinschmidt, J.H. Folding and Stability of Outer Membrane Protein A (OmpA) from Escherichia coli in an Amphipathic Polymer, Amphipol A8-35. Eur. Biophys. J. 2013, 42, 103–118. [Google Scholar] [CrossRef]
- Maurya, S.R.; Chaturvedi, D.; Mahalakshmi, R. Modulating Lipid Dynamics and Membrane Fluidity to Drive Rapid Folding of a Transmembrane Barrel. Sci. Rep. 2013, 3, 1989. [Google Scholar] [CrossRef]
- Guzman, L.M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight Regulation, Modulation, and High-Level Expression by Vectors Containing the Arabinose PBAD Promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [CrossRef] [PubMed]
- Mares, R.E.; Minchaca, A.Z.; Villagrana, S.; Meléndez-López, S.G.; Ramos, M.A. Analysis of the Isomerase and Chaperone-Like Activities of an Amebic PDI (Eh PDI). BioMed Res. Int. 2015, 2015, 286972. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Westermeier, R. Sensitive, Quantitative, and Fast Modifications for Coomassie Blue Staining of Polyacrylamide Gels. Proteomics 2006, 6, 61–64. [Google Scholar] [CrossRef]
- Dassa, E.; Tetu, C.; Boquet, P.-L. Identification of the Acid Phosphatase (Optimum pH 2.5) of Escherichia coli. FEBS Lett. 1980, 113, 275–278. [Google Scholar] [CrossRef] [PubMed]
- Dassa, E.; Boquet, P.L. Identification of the Gene appA for the Acid Phosphatase (pH Optimum 2.5) of Escherichia coli. Mol. Gen. Genet. MGG 1985, 200, 68. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Yoo, G.; Bong, J.-H.; Jose, J.; Kang, M.-J.; Pyun, J.-C. Isolation and Characterization of the Outer Membrane of Escherichia coli with Autodisplayed Z-Domains. Biochim. Biophys. Acta BBA Biomembr. 2015, 1848, 842–847. [Google Scholar] [CrossRef]
- Baykov, A.A.; Evtushenko, O.A.; Avaeva, S.M. A Malachite Green Procedure for Orthophosphate Determination and Its Use in Alkaline Phosphatase-Based Enzyme Immunoassay. Anal. Biochem. 1988, 171, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Menezes-Blackburn, D.; Gabler, S.; Greiner, R. Performance of Seven Commercial Phytases in an in Vitro Simulation of Poultry Digestive Tract. J. Agric. Food Chem. 2015, 63, 6142–6149. [Google Scholar] [CrossRef]
- Studier, F.W.; Moffatt, B.A. Use of Bacteriophage T7 RNA Polymerase to Direct Selective High-Level Expression of Cloned Genes. J. Mol. Biol. 1986, 189, 113–130. [Google Scholar] [CrossRef]
- Dubendorff, J.W.; Studier, F.W. Creation of a T7 Autogene. J. Mol. Biol. 1991, 219, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant Protein Expression in Escherichia coli: Advances and Challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Liu, Y.-Q.; Xu, Y.-S.; Li, Z.-J.; Wang, Y.-Z.; Zhang, Z.-X.; Sun, X.-M. Regulating the T7 RNA Polymerase Expression in E. coli BL21 (DE3) to Provide More Host Options for Recombinant Protein Production. Microb. Cell Factories 2021, 20, 189. [Google Scholar] [CrossRef]
- Schlegel, S.; Löfblom, J.; Lee, C.; Hjelm, A.; Klepsch, M.; Strous, M.; Drew, D.; Slotboom, D.J.; De Gier, J.-W. Optimizing Membrane Protein Overexpression in the Escherichia coli Strain Lemo21(DE3). J. Mol. Biol. 2012, 423, 648–659. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Klepsch, M.M.; Schlegel, S.; Appel, A.; Draheim, R.; Tarry, M.; Högbom, M.; Van Wijk, K.J.; Slotboom, D.J.; Persson, J.O.; et al. Tuning Escherichia coli for Membrane Protein Overexpression. Proc. Natl. Acad. Sci. USA 2008, 105, 14371–14376. [Google Scholar] [CrossRef]
- Studier, F.W. Use of Bacteriophage T7 Lysozyme to Improve an Inducible T7 Expression System. J. Mol. Biol. 1991, 219, 37–44. [Google Scholar] [CrossRef]
- Sugawara, E.; Nikaido, H. Pore-Forming Activity of OmpA Protein of Escherichia coli. J. Biol. Chem. 1992, 267, 2507–2511. [Google Scholar] [CrossRef] [PubMed]
- Park, M. Surface Display Technology for Biosensor Applications: A Review. Sensors 2020, 20, 2775. [Google Scholar] [CrossRef] [PubMed]
- Moraskie, M.; Roshid, M.H.O.; O’Connor, G.; Dikici, E.; Zingg, J.-M.; Deo, S.; Daunert, S. Microbial Whole-Cell Biosensors: Current Applications, Challenges, and Future Perspectives. Biosens. Bioelectron. 2021, 191, 113359. [Google Scholar] [CrossRef]
- Song, Y.; Rampley, C.P.N.; Chen, X.; Du, F.; Thompson, I.P.; Huang, W.E. Application of Bacterial Whole-Cell Biosensors in Health. In Handbook of Cell Biosensors; Thouand, G., Ed.; Springer International Publishing: Cham, Switzerland, 2022; pp. 945–961. ISBN 978-3-030-23216-0. [Google Scholar]
- Schüürmann, J.; Quehl, P.; Festel, G.; Jose, J. Bacterial Whole-Cell Biocatalysts by Surface Display of Enzymes: Toward Industrial Application. Appl. Microbiol. Biotechnol. 2014, 98, 8031–8046. [Google Scholar] [CrossRef]
- De Carvalho, C.C.C.R. Whole Cell Biocatalysts: Essential Workers from Nature to the Industry. Microb. Biotechnol. 2017, 10, 250–263. [Google Scholar] [CrossRef] [PubMed]
- Salema, V.; Fernández, L.Á. Escherichia coli Surface Display for the Selection of Nanobodies. Microb. Biotechnol. 2017, 10, 1468–1484. [Google Scholar] [CrossRef] [PubMed]
- Valldorf, B.; Hinz, S.C.; Russo, G.; Pekar, L.; Mohr, L.; Klemm, J.; Doerner, A.; Krah, S.; Hust, M.; Zielonka, S. Antibody Display Technologies: Selecting the Cream of the Crop. Biol. Chem. 2022, 403, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Mahdavi, S.Z.B.; Oroojalian, F.; Eyvazi, S.; Hejazi, M.; Baradaran, B.; Pouladi, N.; Tohidkia, M.R.; Mokhtarzadeh, A.; Muyldermans, S. An Overview on Display Systems (Phage, Bacterial, and Yeast Display) for Production of Anticancer Antibodies; Advantages and Disadvantages. Int. J. Biol. Macromol. 2022, 208, 421–442. [Google Scholar] [CrossRef]
- Celi, P.; Cowieson, A.J.; Fru-Nji, F.; Steinert, R.E.; Kluenter, A.-M.; Verlhac, V. Gastrointestinal Functionality in Animal Nutrition and Health: New Opportunities for Sustainable Animal Production. Anim. Feed Sci. Technol. 2017, 234, 88–100. [Google Scholar] [CrossRef]
- Mourand, G.; Le Devendec, L.; Delannoy, S.; Fach, P.; Keita, A.; Amelot, M.; Jaunet, H.; Dia, M.E.H.; Kempf, I. Variations of the Escherichia coli Population in the Digestive Tract of Broilers. Avian Pathol. 2020, 49, 678–688. [Google Scholar] [CrossRef]
- Richards, J.D.; Gong, J.; De Lange, C.F.M. The Gastrointestinal Microbiota and Its Role in Monogastric Nutrition and Health with an Emphasis on Pigs: Current Understanding, Possible Modulations, and New Technologies for Ecological Studies. Can. J. Anim. Sci. 2005, 85, 421–435. [Google Scholar] [CrossRef]
- Van Elsas, J.D.; Semenov, A.V.; Costa, R.; Trevors, J.T. Survival of Escherichia coli in the Environment: Fundamental and Public Health Aspects. ISME J. 2011, 5, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Jin, D.; Wu, S.; Yang, J.; Lan, R.; Bai, X.; Liu, S.; Meng, Q.; Yuan, X.; Zhou, J.; et al. Insights into the Evolution of Pathogenicity of Escherichia coli from Genomic Analysis of Intestinal E. coli of Marmota himalayana in Qinghai–Tibet Plateau of China. Emerg. Microbes Infect. 2016, 5, 1–9. [Google Scholar] [CrossRef]
- Ramos, S.; Silva, V.; Dapkevicius, M.D.L.E.; Caniça, M.; Tejedor-Junco, M.T.; Igrejas, G.; Poeta, P. Escherichia coli as Commensal and Pathogenic Bacteria among Food-Producing Animals: Health Implications of Extended Spectrum β-Lactamase (ESBL) Production. Animals 2020, 10, 2239. [Google Scholar] [CrossRef]
- Amalia, L.; Tsai, S.-L. Functionalization of OMVs for Biocatalytic Applications. Membranes 2023, 13, 459. [Google Scholar] [CrossRef] [PubMed]
- Renault, P. Genetically Modified Lactic Acid Bacteria: Applications to Food or Health and Risk Assessment. Biochimie 2002, 84, 1073–1087. [Google Scholar] [CrossRef] [PubMed]
- Deckers, M.; Deforce, D.; Fraiture, M.-A.; Roosens, N.H.C. Genetically Modified Micro-Organisms for Industrial Food Enzyme Production: An Overview. Foods 2020, 9, 326. [Google Scholar] [CrossRef]
- Hanlon, P.; Sewalt, V. GEMs: Genetically Engineered Microorganisms and the Regulatory Oversight of Their Uses in Modern Food Production. Crit. Rev. Food Sci. Nutr. 2021, 61, 959–970. [Google Scholar] [CrossRef] [PubMed]


| Strain | Genotype | Source |
|---|---|---|
| ER2728 | F′ proA+B+ lacIq Δ(lacZ)M15 zzf::Tn10(TetR)/fhuA2 glnV Δ(lac-proAB) thi-1 Δ(hsdS-mcrB)5 | New England Biolabs |
| BL21(DE3) | fhuA2 [lon] ompT gal (λ DE3) [dcm] ∆hsdS λ DE3 = λ sBamHIo ∆EcoRI-B int::(lacI::PlacUV5::T7 gene1)i21 ∆nin5 | Novagen |
| Lemo21(DE3) | BL21(DE3)/pLemo pLemo = pACYC184-PrhaBAD-lysY (CamR) | New England Biolabs |
| MWC-22b | Lemo21(DE3)/pET22b(+) | This study |
| MWC-ESE | Lemo21(DE3)/pESE | This study |
| MWCB-phytase | Lemo21(DE3)/pESE-AppA | This study |
| Plasmid | Features | Source |
| pBAD33 | araBAD promoter (arabinose regulation), p15A origin, CamR | ATCC 1 [44] |
| pBAD-AppA | pBAD33-based, periplasmic expression of AppA | Lab Stock [45] |
| pET22b(+) | T7lacO promoter (lactose regulation), ColE1 origin, AmpR | Novagen |
| pESE | pET22b(+)-based, OM-linked expression of eLpp-OmpA | Lab Stock [41] |
| pESE-AppA | pESE-derived, OM-linked expression of eLpp-OmpA-AppA | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muñoz-Muñoz, P.L.A.; Terán-Ramírez, C.; Mares-Alejandre, R.E.; Márquez-González, A.B.; Madero-Ayala, P.A.; Meléndez-López, S.G.; Ramos-Ibarra, M.A. Surface Engineering of Escherichia coli to Display Its Phytase (AppA) and Functional Analysis of Enzyme Activities. Curr. Issues Mol. Biol. 2024, 46, 3424-3437. https://doi.org/10.3390/cimb46040215
Muñoz-Muñoz PLA, Terán-Ramírez C, Mares-Alejandre RE, Márquez-González AB, Madero-Ayala PA, Meléndez-López SG, Ramos-Ibarra MA. Surface Engineering of Escherichia coli to Display Its Phytase (AppA) and Functional Analysis of Enzyme Activities. Current Issues in Molecular Biology. 2024; 46(4):3424-3437. https://doi.org/10.3390/cimb46040215
Chicago/Turabian StyleMuñoz-Muñoz, Patricia L. A., Celina Terán-Ramírez, Rosa E. Mares-Alejandre, Ariana B. Márquez-González, Pablo A. Madero-Ayala, Samuel G. Meléndez-López, and Marco A. Ramos-Ibarra. 2024. "Surface Engineering of Escherichia coli to Display Its Phytase (AppA) and Functional Analysis of Enzyme Activities" Current Issues in Molecular Biology 46, no. 4: 3424-3437. https://doi.org/10.3390/cimb46040215
APA StyleMuñoz-Muñoz, P. L. A., Terán-Ramírez, C., Mares-Alejandre, R. E., Márquez-González, A. B., Madero-Ayala, P. A., Meléndez-López, S. G., & Ramos-Ibarra, M. A. (2024). Surface Engineering of Escherichia coli to Display Its Phytase (AppA) and Functional Analysis of Enzyme Activities. Current Issues in Molecular Biology, 46(4), 3424-3437. https://doi.org/10.3390/cimb46040215







