Contribution of Endothelial Dysfunction to Cancer Susceptibility and Progression: A Comprehensive Narrative Review on the Genetic Risk Component
Abstract
1. Introduction
2. Vascular Homeostasis
Nitric Oxide (NO)
| First Author (Year) | Country/Ethnic Background | Population Characteristics | Study Design | Studied Polymorphisms |
|---|---|---|---|---|
| Choi et al. (2006) [12] | South Korea/Unclear | 1039 BC patients 995 non-cancer controls | Cohort study | rs2070744 rs1799983 |
| Lu et al. (2006) [41] | USA/non-Hispanic Caucasian | 421 BC patients 423 non-cancer controls | Case–control study | rs2070744 rs1799983 rs869109213 |
| Yeh et al. (2009) [54] | Taiwan/Taiwanese | 727 CRC patients 736 healthy controls | Case–control study | rs2070744 rs1799983 rs869109213 |
| Oztürk et al. (2011) [47] | Turkey/Turkish | 89 EMCA patients 60 total hysterectomy controls | Case–control study | rs1799983 rs869109213 |
| Arıkan et al. (2012) [49] | Turkey/Turkish | 84 CRC patients 99 healthy controls | Case–control study | rs1799983 |
| Jang et al. (2013) [43] | South Korea/Korean | 528 CRC patients 509 healthy controls | Case–control study | rs2070744 rs1799983 rs869109213 |
| Ramírez-Patiño et al. (2013) [55] | Mexico/Mexican | 429 BC patients 281 healthy women | Case–control study | rs869109213 |
| Wu et al. (2014) [36] | Mixed | 4169 cancer cases and 4185 controls (rs2070744) 7775 cancer cases and 7817 controls (rs1799983) 3430 cancer cases and 3842 controls (rs869109213) | Meta-analysis | rs2070744 rs1799983 rs869109213 |
| Zhang et al. (2014) [37] | Mixed | 4220 cancer cases and 4016 controls (rs2070744) 8359 cancer cases and 9575 controls (rs1799983) 2873 cancer cases and 3338 controls (rs869109213) | Meta-analysis | rs2070744 rs1799983 rs869109213 |
| Gao et al. (2015) [33] | Mixed (meta-analysis) Han Chinese (case–control) | 873 BC patients 1034 healthy women (case–control) | Meta-analysis Case–control study | rs2070744 rs1799983 rs869109213 |
| Krishnaveni et al. (2015) [34] | India/South Indian | 150 GC patients 150 healthy controls | Case–control study | rs2070744 |
| Polat et al. (2015) [50] | Turkey/Turkish | 75 BLCA patients 143 healthy controls | Case–control study | rs2070744 rs1799983 rs869109213 |
| Diler et al. (2016) [45] | Turkey/Turkish | 84 PCa patients 116 healthy controls | Case–control study | rs2070744 rs1799983 rs869109213 |
| Polat et al. (2016) [39] | Turkey/Turkish | 50 PCa patients 50 healthy controls | Case–control study | rs2070744 rs1799983 rs869109213 |
| Chen et al. (2018) [48] | Taiwan/Taiwanese | 139 premenopausal and 144 postmenopausal BC patients 100 premenopausal and 100 postmenopausal healthy women | Case–control study | rs2070744 rs1799983 rs869109213 |
| Huang et al. (2018) [51] | Taiwan/Taiwanese | 277 LC patients | Cohort study | rs2070744 rs1799983 |
| Su et al. (2018) [42] | Taiwan/Taiwanese | 1044 OSCC patients 1200 healthy controls | Case–control study | rs2070744 rs1799983 |
| Hung et al. (2019) [46] | Taiwan/Taiwanese | 117 UCC patients 95 patients with cervical precancerous lesions 330 healthy controls | Case–control study | rs2070744 rs1799983 |
| Nan et al. (2019) [38] | Mixed | 41 case–control studies | Meta-analysis | rs2070744 rs1799983 rs869109213 |
| Tsay et al. (2019) [52] | Taiwan/Taiwanese | 431 UC patients 862 healthy controls | Case–control study | rs2070744 rs1799983 |
| Abedinzadeh et al. (2020) [40] | Mixed | 4464 cancer cases and 4347 controls (rs1799983) 589 cancer cases and 789 controls (rs869109213) 588 cancer cases and 692 controls (rs2070744) | Meta-analysis | rs2070744 rs1799983 rs869109213 |
| Carkic et al. (2020) [57] | Serbia/Serbian | 50 OSCC patients 110 healthy controls | Case–control study | rs2070744 rs1799983 rs869109213 |
| Koçer et al. (2020) [56] | Turkey/Turkish | 107 LC patients 100 healthy controls | Case–control study | rs1799983 rs869109213 |
| Balci et al. (2023) [44] | Turkey/Unclear | 48 PCa patients 42 biopsy individuals 27 healthy controls | Case–control study | rs2070744 |
3. Consequences of ED
3.1. Endothelin-1 (ET-1)
3.2. Angiotensin II (Ang II)
3.3. Thrombin
| First Author (Year) | Country/Ethnic Background | Population Characteristics | Study Design | Studied Polymorphisms |
|---|---|---|---|---|
| Hajek et al. (2003) [125] | Czech Republic/ Unclear | 25 leukaemia patients | Cohort study | ACE indel |
| Koh et al. (2003) [66] | Singapore/Singaporean | 189 BC patients 671 healthy controls | Nested case–control study | ACE indel ACE rs4291 |
| Tormene et al. (2003) [144] | Italy/Unclear | 52 women operated for gynaecological malignancy 198 women operated for gynaecological non-malignant disease | Case–control study | F2 rs1799963 |
| Freitas-Silva et al. (2004) [67] | Portugal/Portuguese | 70 EMCA patients 101 healthy controls | Case–control study | ACE indel |
| Medeiros et al. (2004) [80] | Portugal/Portuguese | 170 PCa patients 30 healthy controls | Case–control study | ACE indel |
| Chung et al. (2005) [84] | Taiwan/Taiwanese | 61 OPL betel quid chewers 61 asymptomatic betel quid chewers | Case–control study | ACE indel |
| Ebert et al. (2005) [73] | Germany/Caucasian | 88 GC patients 145 healthy controls | Case–control study | ACE indel |
| González-Zuloeta Ladd et al. (2005) [111] | Netherland/Unclear | 4878 female postmenopausal total participants 114 BC patients | Cohort study | ACE indel |
| Goto et al. (2005) [134] | Japan/Japanese | 454 GC patients 202 healthy controls | Case–control study | ACE indel |
| Röcken et al. (2005) [75] | Germany/Unclear | 113 GC patients 189 healthy controls | Case–control study | ACE indel |
| Arima et al. (2006) [98] | Japan/Japanese | 937 total participants 176 subjects died of malignant neoplasm | Cohort study | ACE indel |
| Yaren et al. (2006) [109] | Turkey/Turkish | 44 BC patients 46 healthy premenopausal women | Case–control study | ACE indel |
| Carl-McGrath et al. (2007) [69] | Germany/Unclear | 45 GC patients | Cohort study | ACE indel |
| González-Zuloeta Ladd et (2007) [138] | Netherlands/Unclear | 203 BC cases 3323 controls | Case–control study | AGT rs699 |
| Hsieh et al. (2007) [89] | Taiwan/Taiwanese | 120 UL patients 125 endometriosis patients 128 healthy controls | Case–control study | ACE indel |
| Röcken et al. (2007) [132] | Germany/Unclear | 100 GC patients | Cohort study | ACE indel |
| Röcken et al. (2007) [90] | Germany/Unclear | 141 CRC patients 189 healthy controls | Case–control study | ACE indel |
| Vairaktaris et al. (2007) [86] | Greece/Greek and German | 60 OSCC patients 153 healthy controls | Case–control study | ACE indel |
| Yaren et al. (2007) [108] | Turkey/Turkish | 57 BC patients 52 healthy controls | Case–control study | ACE indel |
| Yigit et al. (2007) [83] | Turkey/Turkish | 48 PCa patients 51 healthy controls | Case–control study | ACE indel |
| van der Knaap et al. (2008) [110] | Netherland/Unclear | 7679 participants * | Cohort study | ACE indel |
| Alves Corrêa et al. (2009) [114] | Brazil/Brazilian | 101 BC patients 307 healthy controls | Case–control study | ACE indel |
| Harman et al. (2009) [100] | Turkey/Turkish | 50 adrenal mass patients 30 healthy controls | Case–control study | ACE indel NOS3 rs1799983 |
| Loh et al. (2009) [71] | Mixed/Asian and Caucasian | 203 case–control studies | Meta-analysis | ACE indel |
| Vairaktaris et al. (2009) [88] | Mixed/Greek and German | 162 OSCC patients 168 healthy controls | Case–control study | ACE indel |
| Vasků et al. (2009) [140] | Czech Republic/Czech | 102 CRC patients 101 healthy controls | Case–control study | AGT rs699 AGT rs5051 |
| Vigano et al. (2009) [129] | Canada/Unclear | 72 GC and NSCLC advanced cancer patients | Cohort study | ACE indel |
| Andreotti et al. (2010) [139] | Mixed | 1035 RCC patients 777 controls | Case–control study | AGT rs7539020 AGT rs3889728 AGT rs3789662 AGT rs1326889 AGT rs2493137 |
| Nacak et al. (2010) [95] | Turkey/Turkish | 25 LC patients 165 healthy controls | Case–control study | ACE indel |
| Namazi et al. (2010) [106] | Iran/Iranian | 70 BC patients 70 healthy controls | Case–control study | ACE indel |
| Srivastava et al. (2010) [116] | India/North Indian | 233 GBC patients 260 non-cancer controls | Case–control study | ACE indel |
| Liu et al. (2011) [92] | China/Chinese | 241 CRC patients 299 non-cancer controls | Case–control study | ACE indel |
| Lukic et al. (2011) [101] | Serbia/Unclear | 45 PC patients 55 chronic pancreatitis patients 128 healthy controls | Case–control study | ACE indel |
| De Martino et al. (2011) [120] | Austria/Unclear | 10 RCC patients 173 healthy controls | Case–control study | ACE indel |
| Mendizábal-Ruiz et al. (2011) [104] | Mexico/Mexican | 65 BC patients 40 benign breast disease patients | Case–control study | ACE indel AGT rs699 |
| Vossen et al. (2011) [147] | Germany/German | 1801 CRC patients 1853 healthy controls | Case–control study | F2 rs1799963 |
| Dević Pavlić et al. (2012) [97] | Croatia/Croatian | 308 LC patients 353 healthy controls | Case–control study | ACE indel |
| Correa-Noronha et al. (2012) [128] | Brazil/Brazilian | 74 EMCA patients and 228 controls 83 EOC patients and 297 controls | Case–control study | ACE indel |
| Huhn et al. (2012) [137] | Mixed/Czech and German | 1025 Czech cancer cases and 787 Czech controls 1798 German cancer cases and 1810 German controls | Case–control study | AGT rs699 |
| Liu et al. (2012) [85] | Taiwan/Taiwanese | 205 male oral cancer patients 88 Oral precancerous lesions patients 120 healthy controls | Case–control study | ACE indel |
| Wang et al. (2012) [79] | China/Han Chinese | 189 PCa patients 290 non-cancer controls | Case–control study | ACE indel |
| Altas et al. (2013) [127] | Turkey/Unclear | 21 hypophyseal adenoma patients 20 healthy controls | Case–control study | ACE indel |
| Fishchuk et al. (2013) [131] | Ukraine/Ukrainian | 131 BC patients 102 healthy women | Case–control study | ACE indel AGT rs699 AGT rs4762 |
| Namazi et al. (2013) [107] | Iran/Iranian | 110 BC patients | Prospective study | ACE indel |
| Vylliotis et al. (2013) [87] | Mixed/Greek and German | 160 OSCC patients 168 healthy controls | Case–control study | ACE indel F2 rs1799963 AGT rs699 |
| Yapijakis et al. (2013) [123] | Greece/Greek | 92 BCC patients 103 healthy controls | Case–control study | ACE indel |
| Yuan et al. (2013) [112] | China/Chinese | 293 HCC patients 384 healthy controls | Case–control study | ACE indel NOS3 rs869109213 |
| Zang et al. (2013) [62] | China/Han Chinese | 260 pulmonary metastatic stage III osteosarcoma patients 260 matched pulmonary metastatic stage IIB osteosarcoma patients | Case–control study | EDN1 rs1800541 EDN1 rs2070699 EDN1 rs5370 |
| Phukan et al. (2014) [94] | India/Northeast Indian | 151 LC patients 151 controls | Case–control study | ACE indel |
| Xie et al. (2014) [82] | Mixed | 7025 cancer cases 34,911 controls | Meta-analysis | ACE indel |
| Zhang et al. (2014) [70] | Mixed | 5007 cancer cases 8173 controls | Meta-analysis | ACE indel |
| Zhou et al. (2014) [58] | China/Han Chinese | 350 Paediatric osteosarcoma patients with <90% tumour necrosis 350 matched osteosarcoma patients with ≥90% tumour necrosis | Case–control study | EDN1 rs1800541 EDN1 rs2070699 EDN1 rs5370 |
| Ding et al. (2015) [130] | China/Han Chinese | 606 BC patients 633 healthy controls | Case–control study | ACE rs4291 ACE rs4343 |
| Gan et al. (2015) [133] | Mixed/Asian and Caucasian | 1480 GC cases 3773 non-cancer controls | Meta-analysis | ACE indel |
| Lian et al. (2015) [119] | China/Chinese | 800 glioma patients 800 healthy controls | Case–control study | ACE indel |
| Pabalan et al. (2015) [74] | Mixed | 1459 cancer cases 2581 controls | Meta-analysis | ACE indel |
| Wei et al. (2015) [64] | Mixed | 1392 cancer cases 2951 controls | Meta-analysis | ACE indel |
| Yang et al. (2015) [76] | Mixed/Asian and White | 2903 GC cases 10,833 controls | Meta-analysis | ACE indel |
| Zha et al. (2015) [113] | China/Dai Chinese | 210 HCC patients 206 healthy controls | Case–control study | ACE indel |
| Hanafy et al. (2016) [145] | Egypt/Egyptian | 280 HCV-infected patients 100 healthy controls | Case–control study | F2 rs1799963 |
| Pringle et al. (2016) [65] | Australia/Mixed | 184 type 1 endometrioid cancer women 153 healthy controls | Case–control study | AGT rs699 ACE rs4291 |
| Ali et al. (2017) [121] | Pakistan/Pakistani | 200 BLCA patients 200 healthy controls | Case–control study | ACE indel |
| Baghad et al. (2017) [146] | Morocco/Moroccan | 76 CRC patients 182 healthy controls | Case–control study | F2 rs1799963 |
| Marques et al. (2017) [91] | Brazil/Admixed Brazilian | 140 CRC patients 140 non-cancer controls | Case–control study | ACE indel |
| Xu et al. (2017) [63] | China/Han Chinese | 234 PCa patients with HRPC within six years after androgen deprivation therapy 234 matched PCa patients without HRPC within six years after androgen deprivation therapy | Case–control study | EDN1 rs1800541 EDN1 rs2070699 EDN1 rs5370 |
| Zheng et al. (2017) [93] | China/Chinese | 146 CRC patients 106 healthy controls | Case–control study | ACE indel |
| Moghimi et al. (2018) [102] | Mixed | 2846 BC cases 9299 controls | Meta-analysis | ACE indel |
| Pandith et al. (2018) [118] | India/Indian | 12 glioma patients 141 non-cancer controls | Case–control study | ACE indel |
| Peddireddy et al. (2018) [99] | India/South Indian | 246 NSCLC patients 250 healthy controls | Case–control study | ACE indel NOS3 rs869109213 |
| Singh et al. (2018) [105] | India/North Indian | 161 BC patients 152 healthy women | Case–control study | ACE indel |
| Wang et al. (2018) [77] | Mixed | 1098 PCa cases 12,960 controls | Meta-analysis | ACE indel |
| Aydin et al. (2019) [61] | Turkey/Unclear | 113 PTC patients 185 healthy controls | Case–control study | EDN1 rs1800541 EDN1 rs5370 |
| Benenemissi et al. (2019) [117] | Algeria/Algerian | 36 glioma patients 195 healthy controls | Case–control study | ACE indel |
| Keshavarzi et al. (2019) [115] | Iran/Iranian | 202 UL patients 211 healthy controls | Case–control study | ACE indel |
| Papaggelopoulos et al. (2019) [135] | Greece/Greek | 190 BCC patients 99 healthy controls | Case–control study | AGT rs699 |
| Xiao et al. (2019) [68] | Mixed | 8 case–control studies | Meta-analysis | ACE rs4291 |
| Banerjee et al. (2021) [96] | India/North Indian | 154 LC patients 205 healthy controls | Case–control study | ACE indel |
| Dastgheib et al. (2021) [103] | Mixed | 35 case–control studies | Meta-analysis | ACE indel ACE rs4291 |
| Koronellos et al. (2021) [124] | Greece/Greek | 104 BCC patients 111 healthy controls | Case–control study | ACE indel |
| Samara et al. (2021) [136] | Greece/Caucasian | 73 BLCA patients 73 healthy controls | Case–control study | AGT rs699 |
| Du et al. (2022) [81] | Mixed | 817 PCa patients 917 controls | Meta-analysis | ACE indel |
| Said et al. (2022) [78] | Tunisia/Tunisian | 124 PCa patients 143 healthy controls | Case–control study | ACE indel |
| Kumbul et al. (2023) [126] | Turkey/Unclear | 44 LaC patients 61 healthy controls | Case–control study | ACE indel |
| Yapijakis et al. (2023) [122] | Greece/Greek | 100 BCC patients 103 healthy controls | Case–control study | AGT rs699 ACE indel |
4. Adhesion Molecules
4.1. P-Selectin
4.2. E-Selectin
4.3. Von Willebrand Factor (vWF)
4.4. ICAM-1
4.5. VCAM-1
| First Author (Year) | Country/Ethnic Background | Population Characteristics | Study Design | Studied Polymorphisms |
|---|---|---|---|---|
| Chen et al. (2006) [185] | USA/African-American | 286 PCa patients 391 healthy controls | Case–control study | ICAM1 rs5498 |
| Theodoropoulos et al. (2006) [177] | Greece/Greek | 222 CRC patients 200 healthy controls | Case–control study | ICAM1 rs5498 ICAM1 rs1799969 |
| Alessandro et al. (2007) [156] | Italy/Caucasian | 172 CRC patients 80 healthy controls | Case–control study | SELE rs5361 |
| Arandi et al. (2008) [167] | Iran/southern Iranian | 276 BC patients and 235 healthy controls 264 BC patients and 200 healthy controls | Case–control study | ICAM1 rs1799969 ICAM1 rs5498 |
| Burim et al. (2009) [187] | Brazil/Unclear | 158 astrocytoma patients and 162 controls | Case–control study | ICAM1 rs5498 ICAM1 rs1799969 |
| Wang et al. (2009) [179] | China/Chinese | 87 CRC patients 102 non-CRC controls | Case–control study | ICAM1 rs5498 ICAM1 rs1799969 |
| Wang et al. (2009) [194] | Jamaica/Jamaican | 395 NHL patients 309 non-NHL controls | Case–control study | VCAM1 rs1041163 |
| Panoussopoulos et al. (2010) [161] | Greece/Unclear | 80 PC patients 160 healthy controls | Case–control study | SELE rs5361 |
| Naidu et al. (2011) [157] | Malaysia/Malaysian | 387 BC patients 252 healthy controls | Case–control study | SELE rs5361 |
| Tan et al. (2012) [152] | Scotland and Canada/Unclear | 775 cancer patients 101 validation cohort patients | Cohort study | SELP rs6136 ICAM1 rs281432 |
| Thanopoulou et al. (2012) [181] | Greece/Unclear | 203 NSCLC patients 175 healthy controls | Case–control study | ICAM1 rs5498 |
| Tian et al. (2012) [180] | China/Chinese | 332 GC patients 380 healthy controls | Case–control study | ICAM1 rs5498 |
| Xia et al. (2012) [159] | China/Chinese | 311 GC patients 425 controls | Case–control study | SELE rs5361 |
| Kontogianni et al. (2013) [163] | Greece/Unclear | 261 BC patients 480 healthy controls | Case–control study | SELE rs5361 |
| Liarmakopoulos et al. (2013) [160] | Greece/Greek | 88 GC patients 480 healthy controls | Case–control study | SELE rs5361 |
| Lin et al. (2013) [174] | Taiwan/Unclear | 595 OSCC patients 561 healthy controls | Case–control study | ICAM1 rs5498 |
| Miaskowski et al. (2013) [193] | Mixed | 155 BC patients with lymphedema 387 BC patients without lymphedema | Case–control study | VCAM1 rs3176861 |
| Yilmaz et al. (2013) [168] | Turkey/Turkish | 92 primary brain tumour patients 92 healthy controls | Case–control study | ICAM1 rs5498 ICAM1 rs1799969 |
| Avan et al. (2014) [153] | Italy/Unclear | 303 locally advanced or metastatic PC | Cohort study | SELP rs6136 |
| Cai et al. (2014) [182] | China/Northern Han Chinese | 408 OC patients 520 healthy controls | Case–control study | ICAM1 rs5498 |
| Cheng et al. (2014) [164] | Mixed/Asian and Caucasian | 1675 cancer patients 2285 controls | Meta-analysis | SELE rs5361 |
| Wang et al. (2014) [183] | Taiwan/Taiwanese | 279 UC patients 279 healthy controls | Case–control study | ICAM1 rs5498 |
| Andrew et al. (2015) [196] | USA/Caucasian | 783 UC patients | Cohort study | VCAM1 rs3176879 |
| Cheng et al. (2015) [172] | Mixed | 4844 cancer patients 5618 healthy controls | Meta-analysis | ICAM1 rs5498 ICAM1 rs1799969 |
| Tang et al. (2015) [173] | Mixed | 5528 cancer patients and 6173 controls for rs5498 3138 cancer cases and 3699 controls for rs3093030 | Meta-analysis | ICAM1 rs5498 ICAM1 rs3093030 |
| Chen et al. (2016) [184] | Taiwan/Taiwanese | 305 HCC patients 613 healthy controls | Case–control study | ICAM1 rs5498 |
| Ghazy et al. (2016) [191] | Egypt/Unclear | 60 mixed-type OC patients 20 healthy controls | Case–control study | ICAM1 rs1437 |
| Golnarnik et al. (2016) [158] | Iran/Northern Iranian | 100 BC patients 120 healthy controls | Case–control study | SELE rs5361 |
| Lu et al. (2016) [162] | China/Chinese | 687 OC patients 687 healthy controls | Case–control study | SELE rs5361 |
| Novikov et al. (2016) [178] | Russia/unclear | 49 CRC patients 30 BC patients 33 controls | Case–control study | ICAM1 rs5498 |
| Sun et al. (2016) [186] | Taiwan/Taiwanese | 91 UCC patients 63 patients with precancerous lesions 290 healthy controls | Case–control study | ICAM1 rs5498 ICAM1 rs3093030 ICAM1 rs281432 |
| Zhang et al. (2016) [188] | Mixed | 4608 cancer patients 4913 controls | Meta-analysis | ICAM1 rs1799969 ICAM1 rs3093030 |
| Liu et al. (2017) [175] | China/Chinese | 195 CRC patients 188 healthy controls | Case–control study | ICAM1 rs5498 |
| Powrózek et al. (2019) [151] | Poland/Unclear | 62 HNC patients | Cohort study | SELP rs3917647 SELP rs6136 |
| Qian et al. (2019) [166] | Mixed European/Caucasian | 948 NSCLC patients | Cohort study | VWF rs73049469 |
| Ghazy et al. (2020) [190] | Egypt/Egyptian | 40 BC patients 40 healthy controls | Case–control study | ICAM1 rs281437 |
| Feng et al. (2021) [169] | China/Northern Chinese Han | 488 UCC patients 684 patients with cervical precancerous lesions 510 healthy females | Case–control study | ICAM1 rs5498 ICAM1 rs3093030 ICAM1 rs281432 |
| He et al. (2021) [189] | China/Unclear | 290 HCC patients 290 healthy controls | Case–control study | ICAM1 rs281437 ICAM1 rs923366 ICAM1 rs3093030 |
| Qiu et al. (2021) [176] | Mixed | 1003 CRC patients 1303 healthy controls | Case–control study | ICAM1 rs5498 ICAM1 rs3093030 |
| Zakariya et al. (2022) [154] | Iraq/Iraqi | 60 BC patients 40 healthy controls | Case–control study | SELE rs5361 SELE rs5368 SELE rs5362 |
5. ED-Related Proteins and Cancer Hallmarks
| Gene | Polymorphism | Substitution | Ancestral Allele | Global MAF (MA) | Most Severe Consequence |
|---|---|---|---|---|---|
| NOS3 | rs2070744 | C>G | C | 23% (C) | Intron variant |
| rs1799983 | T>G/A | G | 18% (T) | Missense variant | |
| rs869109213 | VNTR | NA | NA | Intron variant | |
| EDN1 | rs5370 | G>T | G | 25% (T) | Missense variant |
| rs1800541 | T>G | T | 28% (G) | Regulatory region variant | |
| rs2070699 | G>C/T | G | 36% (T) | Intron variant | |
| ACE | Indel | Indel | NA | NA | - |
| rs4291 | T>A/G | A | 35% (T) | Regulatory region variant | |
| rs4343 | G>A | A | 36% (G) | Synonymous variant | |
| AGT | rs699 | A>G | G | 29% (A) | Missense variant |
| rs4762 | G>A | G | 10% (A) | Missense variant | |
| rs1326889 | C>T/A | T | 22% (C) | Intron variant | |
| rs281432 | C>G | G | 48% (C) | Intron variant | |
| rs2493137 | T>C | T | 48% (T) | Intron variant | |
| rs5050 | T>C/G | G | 18% (G) | 5 prime UTR variant | |
| rs5051 | C>G/A/T | T | 29%(C) | 5 prime UTR variant | |
| rs7539020 | C>T | C | 49%(C) | Intron variant | |
| rs3889728 | C>G/T | C | 30%(T) | Intron variant | |
| rs3789662 | A>G | A | 34%(G) | 3 prime UTR variant | |
| F2 | rs1799963 | G>A | G | <1% (A) | 3 prime UTR variant |
| SELP | rs3917647 | G>A | G | 46% (A) | Intergenic variant |
| rs6136 | T>C/G | T | 4% (G) | Missense variant | |
| SELE | rs5361 | T>G/A | T | 5% (G) | Missense variant |
| rs5362 | A>G | G | 5% (G) | Non-coding transcript exon variant | |
| rs5367 | A>G | A | 5% (G) | Splice region variant | |
| rs5368 | G>A | G | 15% (A) | Missense variant | |
| VWF | rs73049469 | C>A | C | 13% (A) | Intron variant |
| ICAM1 | rs1437 | A>G/T | G | 37% (G) | 3 prime UTR variant |
| rs5498 | A>G | A | 36% (G) | Missense variant | |
| rs1799969 | G>A | G | 6% (A) | Missense variant | |
| rs281437 | C>G/T | C | 26% (T) | 3 prime UTR variant | |
| rs923366 | C>T/A | T | 35% (T) | 3 prime UTR variant | |
| rs3093030 | C>T | C | 32% (T) | Non-coding transcript exon variant | |
| VCAM1 | rs3176861 | C>T | C | 20% (T) | Intron variant |
| rs3176879 | G>A | A | 13% (G) | Synonymous variant | |
| rs1041163 | T>C | T | 18% (C) | Intergenic variant |
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marques, I.S.; Tavares, V.; Neto, B.V.; Mota, I.N.R.; Pereira, D.; Medeiros, R. Long Non-Coding RNAs in Venous Thromboembolism: Where Do We Stand? Int. J. Mol. Sci. 2023, 24, 12103. [Google Scholar] [CrossRef] [PubMed]
- Lutsey, P.L.; Zakai, N.A. Epidemiology and prevention of venous thromboembolism. Nat. Rev. Cardiol. 2023, 20, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Tavares, V.; Neto, B.V.; Vilas-Boas, M.I.; Pereira, D.; Medeiros, R. Impact of hereditary thrombophilia on cancer-associated thrombosis, tumour susceptibility and progression: A review of existing evidence. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188778. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, C.J.; Morinaga, L.T.K.; Alves, J.L.J.; Castro, M.A.; Calderaro, D.; Jardim, C.V.P.; Souza, R. Cancer-associated thrombosis: The when, how and why. Eur. Respir. Rev. 2019, 28, 180119. [Google Scholar] [CrossRef] [PubMed]
- Kacimi, S.E.O.; Moeinafshar, A.; Haghighi, S.S.; Saghazadeh, A.; Rezaei, N. Venous thromboembolism in cancer and cancer immunotherapy. Crit. Rev. Oncol. Hematol. 2022, 178, 103782. [Google Scholar] [CrossRef]
- Poredos, P.; Jezovnik, M.K. Endothelial Dysfunction and Venous Thrombosis. Angiology 2018, 69, 564–567. [Google Scholar] [CrossRef]
- Zhang, J. Biomarkers of endothelial activation and dysfunction in cardiovascular diseases. Rev. Cardiovasc. Med. 2022, 23, 73. [Google Scholar] [CrossRef] [PubMed]
- Migliacci, R.; Becattini, C.; Pesavento, R.; Davi, G.; Vedovati, M.C.; Guglielmini, G.; Falcinelli, E.; Ciabattoni, G.; Dalla Valle, F.; Prandoni, P.; et al. Endothelial dysfunction in patients with spontaneous venous thromboembolism. Haematologica 2007, 92, 812–818. [Google Scholar] [CrossRef]
- Rajendran, P.; Rengarajan, T.; Thangavel, J.; Nishigaki, Y.; Sakthisekaran, D.; Sethi, G.; Nishigaki, I. The vascular endothelium and human diseases. Int. J. Biol. Sci. 2013, 9, 1057–1069. [Google Scholar] [CrossRef]
- Cyr, A.R.; Huckaby, L.V.; Shiva, S.S.; Zuckerbraun, B.S. Nitric Oxide and Endothelial Dysfunction. Crit. Care Clin. 2020, 36, 307–321. [Google Scholar] [CrossRef]
- Franses, J.W.; Drosu, N.C.; Gibson, W.J.; Chitalia, V.C.; Edelman, E.R. Dysfunctional endothelial cells directly stimulate cancer inflammation and metastasis. Int. J. Cancer 2013, 133, 1334–1344. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.Y.; Lee, K.M.; Noh, D.Y.; Ahn, S.H.; Lee, J.E.; Han, W.; Jang, I.J.; Shin, S.G.; Yoo, K.Y.; Hayes, R.B.; et al. Genetic polymorphisms of eNOS, hormone receptor status, and survival of breast cancer. Breast Cancer Res. Treat. 2006, 100, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Trent, R.J. Molecular Medicine, 4th ed.; Academic Press: Cambridge, MA, USA, 2012; pp. 81–115. [Google Scholar]
- Chiarella, P.; Capone, P.; Sisto, R. Contribution of Genetic Polymorphisms in Human Health. Int. J. Environ. Res. Public Health 2023, 20, 912. [Google Scholar] [CrossRef] [PubMed]
- Leaché, A.D.; Oaks, J.R. The utility of single nucleotide polymorphism (SNP) data in phylogenetics. Annu. Rev. Ecol. Evol. Syst. 2017, 48, 69–84. [Google Scholar] [CrossRef]
- Hovhannisyan, G.; Harutyunyan, T.; Aroutiounian, R.; Liehr, T. DNA copy number variations as markers of mutagenic impact. Int. J. Mol. Sci. 2019, 20, 4723. [Google Scholar] [CrossRef] [PubMed]
- Sehn, J.K. Insertions and deletions (indels). In Clinical Genomics; Elsevier: Amsterdam, The Netherlands, 2015; pp. 129–150. [Google Scholar]
- Gemayel, R.; Vinces, M.D.; Legendre, M.; Verstrepen, K.J. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu. Rev. Genet. 2010, 44, 445–477. [Google Scholar] [CrossRef] [PubMed]
- Theofilis, P.; Sagris, M.; Oikonomou, E.; Antonopoulos, A.S.; Siasos, G.; Tsioufis, C.; Tousoulis, D. Inflammatory mechanisms contributing to endothelial dysfunction. Biomedicines 2021, 9, 781. [Google Scholar] [CrossRef] [PubMed]
- Verhamme, P.; Hoylaerts, M. The pivotal role of the endothelium in haemostasis and thrombosis. Acta Clin. Belg. 2006, 61, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Neubauer, K.; Zieger, B. Endothelial cells and coagulation. Cell Tissue Res. 2022, 387, 391–398. [Google Scholar] [CrossRef]
- Shibata, S.; Harpel, P.; Bona, C.; Fillit, H. Monoclonal antibodies to heparan sulfate inhibit the formation of thrombin-antithrombin III complexes. Clin. Immunol. Immunopathol. 1993, 67, 264–272. [Google Scholar] [CrossRef]
- Weiler, H.; Isermann, B. Thrombomodulin. J. Thromb. Haemost. 2003, 1, 1515–1524. [Google Scholar] [CrossRef]
- Sandset, P.M. Tissue factor pathway inhibitor (TFPI)–an update. Pathophysiol. Haemost. Thromb. 1996, 26, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Durand, M.J.; Gutterman, D.D. Diversity in mechanisms of endothelium-dependent vasodilation in health and disease. Microcirculation 2013, 20, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Incalza, M.A.; D’Oria, R.; Natalicchio, A.; Perrini, S.; Laviola, L.; Giorgino, F. Oxidative stress and reactive oxygen species in endothelial dysfunction associated with cardiovascular and metabolic diseases. Vascul Pharmacol. 2018, 100, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Tavares, V.; Pinto, R.; Assis, J.; Pereira, D.; Medeiros, R. Venous thromboembolism GWAS reported genetic makeup and the hallmarks of cancer: Linkage to ovarian tumour behaviour. Biochim. Biophys. Acta-Rev. Cancer 2020, 1873, 188331. [Google Scholar] [CrossRef]
- Endemann, D.H.; Schiffrin, E.L. Endothelial dysfunction. J. Am. Soc. Nephrol. 2004, 15, 1983–1992. [Google Scholar] [CrossRef]
- Vanhoutte, P.M.; Shimokawa, H.; Feletou, M.; Tang, E.H. Endothelial dysfunction and vascular disease—A 30th anniversary update. Acta Physiol. 2017, 219, 22–96. [Google Scholar] [CrossRef] [PubMed]
- Dardi, P.; Dos Reis Costa, D.E.F.; Assunção, H.C.R.; Rossoni, L.V. Venous endothelial function in cardiovascular disease. Biosci. Rep. 2022, 42, BSR20220285. [Google Scholar] [CrossRef]
- Vannini, F.; Kashfi, K.; Nath, N. The dual role of iNOS in cancer. Redox Biol. 2015, 6, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Napoli, C.; Paolisso, G.; Casamassimi, A.; Al-Omran, M.; Barbieri, M.; Sommese, L.; Infante, T.; Ignarro, L.J. Effects of nitric oxide on cell proliferation: Novel insights. J. Am. Coll. Cardiol. 2013, 62, 89–95. [Google Scholar] [CrossRef]
- Gao, X.; Wang, J.; Wang, W.; Wang, M.; Zhang, J. eNOS Genetic Polymorphisms and Cancer Risk: A Meta-Analysis and a Case-Control Study of Breast Cancer. Medicine 2015, 94, e972. [Google Scholar] [CrossRef] [PubMed]
- Krishnaveni, D.; Amar Chand, B.; Shravan Kumar, P.; Uma Devi, M.; Ramanna, M.; Jyothy, A.; Pratibha, N.; Balakrishna, N.; Venkateshwari, A. Association of endothelial nitric oxide synthase gene T-786C promoter polymorphism with gastric cancer. World J. Gastrointest. Oncol. 2015, 7, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Sun, H.; Qi, X.; Zhou, M. eNOS rs2070744 polymorphism might influence predisposition to hemorrhagic cerebral vascular diseases in East Asians: A meta-analysis. Brain Behav. 2020, 10, e01538. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wang, Z.F.; Xu, Y.; Ren, R.; Heng, B.L.; Su, Z.X. Association between three eNOS polymorphisms and cancer risk: A meta-analysis. Asian Pac. J. Cancer Prev. 2014, 15, 5317–5324. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, L.M.; Wang, M.N.; Chen, X.J.; Li, N.; Huang, Y.D.; Chen, M. The G894t, T-786c and 4b/a polymorphisms in Enos gene and cancer risk: A meta-analysis. J. Evid. Based Med. 2014, 7, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Nan, J.; Liu, Y.; Xu, C.; Ge, D. Effects of eNOS gene polymorphisms on individual susceptibility to cancer: A meta-analysis. Nitric Oxide 2019, 84, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Polat, F.; Turaçlar, N.; Yilmaz, M.; Bingöl, G.; Cingilli Vural, H. eNOS gene polymorphisms in paraffin-embedded tissues of prostate cancer patients. Turk. J. Med. Sci. 2016, 46, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Abedinzadeh, M.; Dastgheib, S.A.; Maleki, H.; Heiranizadeh, N.; Zare, M.; Jafari-Nedooshan, J.; Kargar, S.; Neamatzadeh, H. Association of Endothelial Nitric Oxide Synthase Gene Polymorphisms with Susceptibility to Prostate Cancer: A Comprehensive Systematic Review and Meta-Analysis. Urol. J. 2020, 17, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Wei, Q.; Bondy, M.L.; Yu, T.K.; Li, D.; Brewster, A.; Shete, S.; Sahin, A.; Meric-Bernstam, F.; Wang, L.E. Promoter polymorphism (-786t>C) in the endothelial nitric oxide synthase gene is associated with risk of sporadic breast cancer in non-Hispanic white women age younger than 55 years. Cancer 2006, 107, 2245–2253. [Google Scholar] [CrossRef]
- Su, C.W.; Chien, M.H.; Lin, C.W.; Chen, M.K.; Chow, J.M.; Chuang, C.Y.; Chou, C.H.; Liu, Y.C.; Yang, S.F. Associations of genetic variations of the endothelial nitric oxide synthase gene and environmental carcinogens with oral cancer susceptibility and development. Nitric Oxide 2018, 79, 1–7. [Google Scholar] [CrossRef]
- Jang, M.J.; Jeon, Y.J.; Kim, J.W.; Chong, S.Y.; Hong, S.P.; Oh, D.; Cho, Y.K.; Chung, K.W.; Kim, N.K. Association of eNOS polymorphisms (-786T>C, 4a4b, 894G>T) with colorectal cancer susceptibility in the Korean population. Gene 2013, 512, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Balci, S.; Akbayir, S.; Bozlu, M.; Tamer, L. Investigation of the relationship between endothelial nitric oxide synthase T786C polymorphism and PSA, PSA derivatives, and prostate cancer in the Turkish population. J. Med. Biochem. 2023, 42, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Diler, S.B.; Öden, A. The T-786C, G894T, and Intron 4 VNTR (4a/b) Polymorphisms of the Endothelial Nitric Oxide Synthase Gene in Prostate Cancer Cases. Genetika 2016, 52, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Hung, W.C.; Wu, T.F.; Ng, S.C.; Lee, Y.C.; Shen, H.P.; Yang, S.F.; Wang, P.H. Involvement of endothelial nitric oxide synthase gene variants in the aggressiveness of uterine cervical cancer. J. Cancer 2019, 10, 2594–2600. [Google Scholar] [CrossRef] [PubMed]
- Oztürk, E.; Dikensoy, E.; Balat, O.; Uğur, M.G.; Balcı, S.O.; Aydın, A.; Kazancı, U.; Pehlivan, S. Association of endothelial nitric oxide synthase gene polymorphisms with endometrial carcinoma: A preliminary study. J. Turk. Ger. Gynecol. Assoc. 2011, 12, 229–233. [Google Scholar] [CrossRef]
- Chen, C.H.; Wu, S.H.; Tseng, Y.M.; Hou, M.F.; Tsai, L.Y.; Tsai, S.M. Distinct role of endothelial nitric oxide synthase gene polymorphisms from menopausal status in the patients with sporadic breast cancer in Taiwan. Nitric Oxide 2018, 72, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Arıkan, S.; Cacina, C.; Guler, E.; Çulcu, S.; Tuna, G.; Yaylım-Eraltan, I. The effects of NOS3 Glu298Asp variant on colorectal cancer risk and progression in Turkish population. Mol. Biol. Rep. 2012, 39, 3245–3249. [Google Scholar] [CrossRef] [PubMed]
- Polat, F.; Diler, S.B.; Azazi, İ.; Öden, A. T-786C, G894T, and intron 4 VNTR (4a/b) polymorphisms of the endothelial nitric oxide synthase gene in bladder cancer cases. Asian Pac. J. Cancer Prev. 2015, 16, 2199–2202. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, C.Y.; Hsieh, M.J.; Wu, W.J.; Chiang, W.L.; Liu, T.C.; Yang, S.F.; Tsao, T.C. Association of endothelial nitric oxide synthase (eNOS) polymorphisms with EGFR-mutated lung adenocarcinoma in Taiwan. J. Cancer 2018, 9, 2518–2524. [Google Scholar] [CrossRef]
- Tsay, M.D.; Hsieh, M.J.; Wang, S.S.; Wang, W.C.; Chou, Y.Y.; Shih, C.H.; Yang, S.F.; Chou, Y.E. Impact of endothelial nitric oxide synthase polymorphisms on urothelial cell carcinoma development. Urol. Oncol. 2019, 37, e291–e293. [Google Scholar] [CrossRef]
- Raina, P.; Sikka, R.; Gupta, H.; Matharoo, K.; Bali, S.K.; Singh, V.; Bhanwer, A. Association of eNOS and MCP-1 Genetic Variants with Type 2 Diabetes and Diabetic Nephropathy Susceptibility: A Case–Control and Meta-Analysis Study. Biochem. Genet. 2021, 59, 966–996. [Google Scholar] [CrossRef]
- Yeh, C.C.; Santella, R.M.; Hsieh, L.L.; Sung, F.C.; Tang, R. An intron 4 VNTR polymorphism of the endothelial nitric oxide synthase gene is associated with early-onset colorectal cancer. Int. J. Cancer 2009, 124, 1565–1571. [Google Scholar] [CrossRef]
- Ramírez-Patiño, R.; Figuera, L.E.; Puebla-Pérez, A.M.; Delgado-Saucedo, J.I.; Legazpí-Macias, M.M.; Mariaud-Schmidt, R.P.; Ramos-Silva, A.; Gutiérrez-Hurtado, I.A.; Gómez Flores-Ramos, L.; Zúñiga-González, G.M.; et al. Intron 4 VNTR (4a/b) polymorphism of the endothelial nitric oxide synthase gene is associated with breast cancer in Mexican women. J. Korean Med. Sci. 2013, 28, 1587–1594. [Google Scholar] [CrossRef] [PubMed]
- Koçer, C.; Benlier, N.; Balci, S.O.; Pehlivan, S.; Şanli, M.; Nacak, M. The role of endothelial nitric oxide synthase gene polymorphisms in patients with lung cancer. Clin. Respir. J. 2020, 14, 948–955. [Google Scholar] [CrossRef] [PubMed]
- Carkic, J.; Nikolic, N.; Nisevic, J.; Lazarevic, M.; Kuzmanovic-Pficer, J.; Jelovac, D.; Milasin, J. Endothelial nitric oxide synthase polymorphisms/haplotypes are strong modulators of oral cancer risk in Serbian population. J. Oral Sci. 2020, 62, 322–326. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, B.; Wang, M.; Ni, J. Endothelin-1 gene polymorphisms and risk of chemoresistant pediatric osteosarcoma. Pediatr. Blood Cancer 2014, 61, 612–617. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Rghigh, A. Polymorphism in Endothelin-1 Gene: An Overview. Curr. Clin. Pharmacol. 2016, 11, 191–210. [Google Scholar] [CrossRef] [PubMed]
- Kirshbom, P.M.; Mahle, W.T.; Joyner, R.W.; Leong, T.; Wilson, M.; Kogon, B.E.; Kanter, K.R.; Bouzyk, M.M. The endothelin-1 G5665T polymorphism impacts transplant-free survival for single ventricle patients. J. Thorac. Cardiovasc. Surg. 2008, 136, 117–122. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Aydin, A.F.; Vural, P.; Doğru-Abbasoğlu, S.; Çil, E. The endothelin 1 and endothelin receptor A gene polymorphisms increase the risk of developing papillary thyroid cancer. Mol. Biol. Rep. 2019, 46, 199–205. [Google Scholar] [CrossRef]
- Zang, X.; Zhou, Y.; Huang, Z.; Zhang, C. Endothelin-1 single nucleotide polymorphisms and risk of pulmonary metastatic osteosarcoma. PLoS ONE 2013, 8, e73349. [Google Scholar] [CrossRef][Green Version]
- Xu, D.; Wang, X.; Lou, Y. Association of endothelin-1 gene single-nucleotide polymorphisms and haplotypes with risk of hormone refractory prostate cancer. Pharmazie 2017, 72, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.T.; Chen, N.; He, Y.Z.; Wang, J.R.; Yang, Y.; Guo, X.J.; Wang, Z.Q. Angiotensin-converting enzyme insertion/deletion polymorphism and gastric cancer: A systematic review and meta-analysis. Clin. Res. Hepatol. Gastroenterol. 2015, 39, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Pringle, K.G.; Delforce, S.J.; Wang, Y.; Ashton, K.A.; Proietto, A.; Otton, G.; Blackwell, C.C.; Scott, R.J.; Lumbers, E.R. Renin-angiotensin system gene polymorphisms and endometrial cancer. Endocr. Connect. 2016, 5, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Koh, W.P.; Yuan, J.M.; Sun, C.L.; van den Berg, D.; Seow, A.; Lee, H.P.; Yu, M.C. Angiotensin I-converting enzyme (ACE) gene polymorphism and breast cancer risk among Chinese women in Singapore. Cancer Res. 2003, 63, 573–578. [Google Scholar] [PubMed]
- Freitas-Silva, M.; Pereira, D.; Coelho, C.; Bicho, M.; Lopes, C.; Medeiros, R. Angiotensin I-converting enzyme gene insertion/deletion polymorphism and endometrial human cancer in normotensive and hypertensive women. Cancer Genet. Cytogenet. 2004, 155, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Dong, Z.; Zhu, J.; You, J.; Fan, J. Association between ACE A240T polymorphism and cancer risk: A meta-analysis. J. Int. Med. Res. 2019, 47, 5917–5925. [Google Scholar] [CrossRef] [PubMed]
- Carl-McGrath, S.; Ebert, M.P.; Lendeckel, U.; Röcken, C. Expression of the local angiotensin II system in gastric cancer may facilitate lymphatic invasion and nodal spread. Cancer Biol. Ther. 2007, 6, 1218–1226. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, K.; Cheng, D.; Yi, L.; Shi, H.; Zhen, G. Association between angiotensin I-converting enzyme gene polymorphism and susceptibility to cancer: A meta analysis. Int. J. Clin. Exp. Pathol. 2014, 7, 6291–6300. [Google Scholar] [PubMed]
- Loh, M.; Koh, K.X.; Yeo, B.H.; Song, C.M.; Chia, K.S.; Zhu, F.; Yeoh, K.G.; Hill, J.; Iacopetta, B.; Soong, R. Meta-analysis of genetic polymorphisms and gastric cancer risk: Variability in associations according to race. Eur. J. Cancer 2009, 45, 2562–2568. [Google Scholar] [CrossRef]
- Gard, P.R. Implications of the angiotensin converting enzyme gene insertion/deletion polymorphism in health and disease: A snapshot review. Int. J. Mol. Epidemiol. Genet. 2010, 1, 145–157. [Google Scholar]
- Ebert, M.P.; Lendeckel, U.; Westphal, S.; Dierkes, J.; Glas, J.; Folwaczny, C.; Roessner, A.; Stolte, M.; Malfertheiner, P.; Röcken, C. The angiotensin I-converting enzyme gene insertion/deletion polymorphism is linked to early gastric cancer. Cancer Epidemiol. Biomark. Prev. 2005, 14, 2987–2989. [Google Scholar] [CrossRef] [PubMed]
- Pabalan, N.; Jarjanazi, H.; Ozcelik, H. Associations of the Insertion/Deletion Polymorphism in the ACE Gene and Risk of Gastric Cancer: A Meta-Analysis. J. Gastrointest. Cancer 2015, 46, 370–379. [Google Scholar] [CrossRef]
- Röcken, C.; Lendeckel, U.; Dierkes, J.; Westphal, S.; Carl-McGrath, S.; Peters, B.; Krüger, S.; Malfertheiner, P.; Roessner, A.; Ebert, M.P. The number of lymph node metastases in gastric cancer correlates with the angiotensin I-converting enzyme gene insertion/deletion polymorphism. Clin. Cancer Res. 2005, 11, 2526–2530. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Cai, C.; Ye, L.; Rao, Y.; Wang, Q.; Hu, D.; Huang, X. The relationship between angiotensin-converting enzyme gene insertion/deletion polymorphism and digestive cancer risk: Insights from a meta-analysis. J. Renin Angiotensin Aldosterone Syst. 2015, 16, 1306–1313. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Li, H.Y.; Jiang, Z.P.; Zhou, T.B. Relationship between angiotensin-converting enzyme insertion/deletion gene polymorphism and prostate cancer susceptibility. J. Cancer Res. Ther. 2018, 14, S375–S380. [Google Scholar] [CrossRef] [PubMed]
- Said, R.; Jenni, R.; Boussetta, S.; Ammous, F.; Zouari, S.; Zaghbib, S.; Chakroun, M.; Derouiche, A.; Chebil, M.; Ouerhani, S. Association of a common genetic variant (insertion/deletion) in ACE gene with prostate cancer susceptibility in a Tunisian population. J. Clin. Lab. Anal. 2022, 36, e24129. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, S.; Lin, Y.W.; Wu, J.; Chen, H.; Mao, Y.Q.; Zheng, X.Y.; Zhou, C.; Xie, L.P. Angiotensin-converting enzyme insertion/deletion polymorphism and the risk of prostate cancer in the Han population of China. Med. Oncol. 2012, 29, 1964–1971. [Google Scholar] [CrossRef] [PubMed]
- Medeiros, R.; Vasconcelos, A.; Costa, S.; Pinto, D.; Lobo, F.; Morais, A.; Oliveira, J.; Lopes, C. Linkage of angiotensin I-converting enzyme gene insertion/deletion polymorphism to the progression of human prostate cancer. J. Pathol. 2004, 202, 330–335. [Google Scholar] [CrossRef]
- Du, J.; Lan, J.; Yang, H.; Ying, Q.; Huang, G.; Mou, J.; Long, J.; Qiao, Z.; Hu, Q. Association of angiotensin-converting enzyme insertion/deletion (ACE I/D) gene polymorphism with susceptibility to prostate cancer: An updated meta-analysis. World J. Surg. Oncol. 2022, 20, 354. [Google Scholar] [CrossRef]
- Xie, Y.; You, C.; Chen, J. An updated meta-analysis on association between angiotensin I-converting enzyme gene insertion/deletion polymorphism and cancer risk. Tumour Biol. 2014, 35, 6567–6579. [Google Scholar] [CrossRef]
- Yigit, B.; Bozkurt, N.; Narter, F.; Yilmaz, H.; Yucebas, E.; Isbir, T. Effects of ACE I/D polymorphism on prostate cancer risk, tumor grade and metastatis. Anticancer Res. 2007, 27, 933–936. [Google Scholar] [PubMed]
- Chung, F.M.; Yang, Y.H.; Chen, C.H.; Lin, C.C.; Shieh, T.Y. Angiotensin-converting enzyme gene insertion/deletion polymorphism is associated with risk of oral precancerous lesion in betel quid chewers. Br. J. Cancer 2005, 93, 602–606. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.T.; Lin, L.W.; Chen, C.Y.; Wang, C.P.; Liu, H.P.; Houng, J.Y.; Chung, F.M.; Shieh, T.Y. Polymorphism of angiotensin I-converting enzyme gene is related to oral cancer and lymph node metastasis in male betel quid chewers. Oral Oncol. 2012, 48, 1257–1262. [Google Scholar] [CrossRef] [PubMed]
- Vairaktaris, E.; Yapijakis, C.; Tsigris, C.; Vassiliou, S.; Derka, S.; Nkenke, E.; Spyridonidou, S.; Vylliotis, A.; Vorris, E.; Ragos, V.; et al. Association of angiotensin-converting enzyme gene insertion/deletion polymorphism with increased risk for oral cancer. Acta Oncol. 2007, 46, 1097–1102. [Google Scholar] [CrossRef] [PubMed]
- Vylliotis, A.; Yapijakis, C.; Nkenke, E.; Nisyrios, T.; Avgoustidis, D.; Adamopoulou, M.; Ragos, V.; Vassiliou, S.; Koronellos, N.; Vairaktaris, E. Effect of thrombosis-related gene polymorphisms upon oral cancer: A regression analysis. Anticancer Res. 2013, 33, 4033–4039. [Google Scholar]
- Vairaktaris, E.; Serefoglou, Z.; Avgoustidis, D.; Yapijakis, C.; Critselis, E.; Vylliotis, A.; Spyridonidou, S.; Derka, S.; Vassiliou, S.; Nkenke, E.; et al. Gene polymorphisms related to angiogenesis, inflammation and thrombosis that influence risk for oral cancer. Oral Oncol. 2009, 45, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.Y.; Lee, C.C.; Chang, C.C.; Wang, Y.K.; Yeh, L.S.; Lin, C.S. Angiotensin I-converting enzyme insertion-related genotypes and allele are associated with higher susceptibility of endometriosis and leiomyoma. Mol. Reprod. Dev. 2007, 74, 808–814. [Google Scholar] [CrossRef] [PubMed]
- Röcken, C.; Neumann, K.; Carl-McGrath, S.; Lage, H.; Ebert, M.P.; Dierkes, J.; Jacobi, C.A.; Kalmuk, S.; Neuhaus, P.; Neumann, U. The gene polymorphism of the angiotensin I-converting enzyme correlates with tumor size and patient survival in colorectal cancer patients. Neoplasia 2007, 9, 716–722. [Google Scholar] [CrossRef] [PubMed]
- Marques, D.; Ferreira-Costa, L.R.; Ferreira-Costa, L.L.; Correa, R.D.S.; Borges, A.M.P.; Ito, F.R.; Ramos, C.C.O.; Bortolin, R.H.; Luchessi, A.D.; Ribeiro-Dos-Santos, Â.; et al. Association of insertion-deletions polymorphisms with colorectal cancer risk and clinical features. World J. Gastroenterol. 2017, 23, 6854–6867. [Google Scholar] [CrossRef]
- Liu, S.Y.; Sima, X.; Wang, C.H.; Gao, M. The association between ACE polymorphism and risk of colorectal cancer in a Chinese population. Clin. Biochem. 2011, 44, 1223–1226. [Google Scholar] [CrossRef]
- Zheng, X.; Liu, G.; Cui, G.; Cheng, M.; Zhang, N.; Hu, S. Angiotensin-Converting Enzyme Gene Deletion Polymorphism is Associated with Lymph Node Metastasis in Colorectal Cancer Patients in a Chinese Population. Med. Sci. Monit. 2017, 23, 4926–4931. [Google Scholar] [CrossRef] [PubMed]
- Phukan, R.K.; Borah, P.K.; Saikia, B.J.; Das, M.; Sekhon, G.S.; Mahanta, J. Interaction of tobacco smoking and chewing with Angiotensin converting enzyme (insertion/deletion) gene polymorphisms and risk of lung cancer in a high risk area from northeast India. Asian Pac. J. Cancer Prev. 2014, 15, 10691–10695. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nacak, M.; Nacak, I.; Sanli, M.; Ozkur, M.; Pektaş, M.; Aynacioğlu, A.S. Association of angiotensin converting enzyme gene insertion/deletion polymorphism with lung cancer in Turkey. Cancer Genet. Cytogenet. 2010, 198, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, J.; Gupta, A.; Agnihotri, V.; Pradhan, R.; Kandel, R.; Upadhyay, A.D.; Dwivedi, S.; Kumar, L.; Dey, S.; Dey, A.B. Lung cancer in the older population: Interactive effects of angiotensin converting enzyme gene polymorphism (rs 4340 ID) and tobacco addiction in risk assessment. Indian J. Cancer 2021, 60, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Dević Pavlić, S.; Ristić, S.; Flego, V.; Kapović, M.; Radojčić Badovinac, A. Angiotensin-converting enzyme insertion/deletion gene polymorphism in lung cancer patients. Genet. Test. Mol. Biomark. 2012, 16, 722–725. [Google Scholar] [CrossRef]
- Arima, H.; Kiyohara, Y.; Tanizaki, Y.; Nakabeppu, Y.; Kubo, M.; Kato, I.; Sueishi, K.; Tsuneyoshi, M.; Fujishima, M.; Iida, M. Angiotensin I-converting enzyme gene polymorphism modifies the smoking-cancer association: The Hisayama Study. Eur. J. Cancer Prev. 2006, 15, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Peddireddy, V.; Badabagni, S.P.; Gundimeda, S.D.; Mundluru, H.P. Association of eNOS and ACE gene polymorphisms and plasma nitric oxide with risk of non-small cell lung cancer in South India. Clin. Respir. J. 2018, 12, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Harman, E.; Karadeniz, M.; Biray, C.; Zengi, A.; Cetinkalp, S.; Ozgen, A.G.; Saygili, F.; Berdeli, A.; Gündüz, C.; Yilmaz, C. The relation of adiponectin and tumor necrosis factor alpha levels between endothelial nitric oxide synthase, angiotensin-converting enzyme, transforming growth factor beta, and tumor necrosis factor alpha gene polymorphism in adrenal incidentalomas. J. Endocrinol. Investig. 2009, 32, 881–888. [Google Scholar] [CrossRef]
- Lukic, S.; Nikolic, A.; Alempijevic, T.; Popovic, D.; Sokic Milutinovic, A.; Ugljesic, M.; Knezevic, S.; Milicic, B.; Dinic, D.; Radojkovic, D. Angiotensin-converting enzyme gene insertion/deletion polymorphism in patients with chronic pancreatitis and pancreatic cancer. Dig. Surg. 2011, 28, 258–262. [Google Scholar] [CrossRef]
- Moghimi, M.; Kargar, S.; Jafari, M.A.; Ahrar, H.; Jarahzadeh, M.H.; Neamatzadeh, H.; Sadeghizadeh- Yazdi, J. Angiotensin Converting Enzyme Insertion/Deletion Polymorphism is Associated with Breast Cancer Risk: A Meta-Analysis. Asian Pac. J. Cancer Prev. 2018, 19, 3225–3231. [Google Scholar] [CrossRef][Green Version]
- Dastgheib, S.A.; Asadian, F.; Farbod, M.; Karimi-Zarchi, M.; Meibodi, B.; Akbarian, E.; Neamatzadeh, H. Association of ACE I/D, -240A > T and AT1R A1166C polymorphisms with susceptibility to breast cancer: A systematic review and meta-analysis based on 35 case-control studies. Nucleosides Nucleotides Nucleic Acids 2021, 40, 117–135. [Google Scholar] [CrossRef]
- Mendizábal-Ruiz, A.P.; Morales, J.; Castro Martinez, X.; Gutierrez Rubio, S.A.; Valdez, L.; Vásquez-Camacho, J.G.; Sanchez Corona, J.; Moran Moguel, M.C. RAS polymorphisms in cancerous and benign breast tissue. J. Renin Angiotensin Aldosterone Syst. 2011, 12, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Srivastava, N.; Amit, S.; Prasad, S.N.; Misra, M.P.; Ateeq, B. Association of AGTR1 (A1166C) and ACE (I/D) Polymorphisms with Breast Cancer Risk in North Indian Population. Transl. Oncol. 2018, 11, 233–242. [Google Scholar] [CrossRef]
- Namazi, S.; Monabati, A.; Ardeshir-Rouhani-Fard, S.; Azarpira, N. Association of angiotensin I converting enzyme (insertion/deletion) and angiotensin II type 1 receptor (A1166C) polymorphisms with breast cancer prognostic factors in Iranian population. Mol. Carcinog. 2010, 49, 1022–1030. [Google Scholar] [CrossRef] [PubMed]
- Namazi, S.; Daneshian, A.; Mohammadianpanah, M.; Jafari, P.; Ardeshir-Rouhani-Fard, S.; Nasirabadi, S. The impact of renin-angiotensin system, angiotensin I converting enzyme (insertion/deletion), and angiotensin II type 1 receptor (A1166C) polymorphisms on breast cancer survival in Iran. Gene 2013, 532, 125–131. [Google Scholar] [CrossRef]
- Yaren, A.; Turgut, S.; Kursunluoglu, R.; Oztop, I.; Turgut, G.; Degirmencioglu, S.; Kelten, C.; Erdem, E. Insertion/deletion polymorphism of the angiotensin I-converting enzyme gene in patients with breast cancer and effects on prognostic factors. J. Investig. Med. 2007, 55, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Yaren, A.; Turgut, S.; Kursunluoglu, R.; Oztop, I.; Turgut, G.; Kelten, C.; Erdem, E. Association between the polymorphism of the angiotensin-converting enzyme gene and tumor size of breast cancer in premenopausal patients. Tohoku J. Exp. Med. 2006, 210, 109–116. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Van der Knaap, R.; Siemes, C.; Coebergh, J.W.; van Duijn, C.M.; Hofman, A.; Stricker, B.H. Renin-angiotensin system inhibitors, angiotensin I-converting enzyme gene insertion/deletion polymorphism, and cancer: The Rotterdam Study. Cancer 2008, 112, 748–757. [Google Scholar] [CrossRef]
- González-Zuloeta Ladd, A.M.; Arias Vásquez, A.; Sayed-Tabatabaei, F.A.; Coebergh, J.W.; Hofman, A.; Njajou, O.; Stricker, B.; van Duijn, C. Angiotensin-converting enzyme gene insertion/deletion polymorphism and breast cancer risk. Cancer Epidemiol. Biomark. Prev. 2005, 14, 2143–2146. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yuan, F.; Zhang, L.S.; Li, H.Y.; Liao, M.; Lv, M.; Zhang, C. Influence of angiotensin I-converting enzyme gene polymorphism on hepatocellular carcinoma risk in China. DNA Cell Biol. 2013, 32, 268–273. [Google Scholar] [CrossRef]
- Zha, Y.; Gan, P.; Liu, Q.; Tan, J. Relationship between polymorphism of angiotensin-converting enzyme gene insertion/deletion and risk of hepatocellular carcinoma in a Chinese Dai population. J. Renin Angiotensin Aldosterone Syst. 2015, 16, 695–699. [Google Scholar] [CrossRef] [PubMed]
- Alves Corrêa, S.A.; Ribeiro de Noronha, S.M.; Nogueira-de-Souza, N.C.; Valleta de Carvalho, C.; Massad Costa, A.M.; Juvenal Linhares, J.; Vieira Gomes, M.T.; Guerreiro da Silva, I.D. Association between the angiotensin-converting enzyme (insertion/deletion) and angiotensin II type 1 receptor (A1166C) polymorphisms and breast cancer among Brazilian women. J. Renin Angiotensin Aldosterone Syst. 2009, 10, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Keshavarzi, F.; Teimoori, B.; Farzaneh, F.; Mokhtari, M.; Najafi, D.; Salimi, S. Association of ACE I/D and AGTR1 A1166C Gene Polymorphisms and Risk of Uterine Leiomyoma: A Case-Control Study. Asian Pac. J. Cancer Prev. 2019, 20, 2595–2599. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, K.; Srivastava, A.; Mittal, B. Angiotensin I-converting enzyme insertion/deletion polymorphism and increased risk of gall bladder cancer in women. DNA Cell Biol. 2010, 29, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Benenemissi, I.H.; Sifi, K.; Sahli, L.K.; Semmam, O.; Abadi, N.; Satta, D. Angiotensin-converting enzyme insertion/deletion gene polymorphisms and the risk of glioma in an Algerian population. Pan Afr. Med. J. 2019, 32, 197. [Google Scholar] [CrossRef]
- Pandith, A.A.; Qasim, I.; Zahoor, W.; Shah, P.; Bhat, A.R. ACE I/D sequence variants but not MTHFR C677T, is strongly linked to malignant glioma risk and its variant DD genotype may act as a promising predictive biomarker for overall survival of glioma patients. Gene 2018, 639, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Lian, M.; Jiang, H.; Wang, H.; Guo, S. Angiotensin-converting enzyme insertion/deletion gene polymorphisms is associated with risk of glioma in a Chinese population. J. Renin Angiotensin Aldosterone Syst. 2015, 16, 443–447. [Google Scholar] [CrossRef]
- De Martino, M.; Klatte, T.; Schatzl, G.; Waldert, M.; Remzi, M.; Haitel, A.; Kramer, G.; Marberger, M. Insertion/deletion polymorphism of angiotensin I-converting enzyme gene is linked with chromophobe renal cell carcinoma. Urology 2011, 77, e1005.e9–e1005.e13. [Google Scholar] [CrossRef]
- Ali, S.H.B.; Bangash, K.S.; Rauf, A.; Younis, M.; Anwar, K.; Khurram, R.; Khawaja, M.A.; Azam, M.; Qureshi, A.A.; Akhter, S.; et al. Identification of novel potential genetic predictors of urothelial bladder carcinoma susceptibility in Pakistani population. Fam. Cancer 2017, 16, 577–594. [Google Scholar] [CrossRef]
- Yapijakis, C.; Gintoni, I.; Charalampidou, S.; Angelopoulou, A.; Papakosta, V.; Vassiliou, S.; Chrousos, G.P. Angiotensinogen, Angiotensin-Converting Enzyme, and Chymase Gene Polymorphisms as Biomarkers for Basal Cell Carcinoma Susceptibility. Adv. Exp. Med. Biol. 2023, 1423, 175–180. [Google Scholar] [CrossRef]
- Yapijakis, C.; Koronellos, N.; Spyridonidou, S.; Vylliotis, A.; Avgoustidis, D.; Goutas, N.; Vlachodimitropoulos, D.; Vairaktaris, E. Association of angiotensin-converting enzyme gene insertion/deletion polymorphism with decreased risk for basal cell carcinoma. Arch. Dermatol. Res. 2013, 305, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Koronellos, N.; Yapijakis, C.; Katoulis, A.; Avgoustidis, D.; Vylliotis, A.; Papakosta, V.; Diamantopoulou, S.; Zografos, O.; Vairaktari, G.; Vairaktaris, E.; et al. Association study indicates combined effect of interleukin-10 and angiotensin-converting enzyme in basal cell carcinoma development. Arch. Dermatol. Res. 2021, 313, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Hajek, D.; Tomiska, M.; Krahulcova, E.; Druckmuller, M.; Florianova, M.; Izakovicova-Holla, L.; Vacha, J. I/D ACE gene polymorphism in survival of leukemia patients—Hypothesis and pilot study. Med. Hypotheses 2003, 61, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Kumbul, Y.; Hekimler Öztürk, K.; Yasan, H.; Akın, V.; Sivrice, M.E.; Caner, F. Angiotensin-converting enzyme insertion/deletion gene polymorphism in patients with laryngeal cancer. Acta Otorhinolaryngol. Ital. 2023, 43, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Altas, M.; Bayrak, O.F.; Serefhan, A.; Silav, G.; Coskun, K.K.; Cerci, A.; Isik, N.; Elmaci, I. Investigation of ACE genome insertion/deletion correlation with immunohistochemical profile in pituitary adenomas. Turk. Neurosurg. 2013, 23, 464–469. [Google Scholar] [CrossRef]
- Correa-Noronha, S.A.; Noronha, S.M.; Alecrim, C.; Mesquita Ade, C.; Brito, G.S.; Junqueira, M.G.; Leite, D.B.; Carvalho, C.V.; Silva, I.D. Association of angiotensin-converting enzyme I gene I/D polymorphism with endometrial but not with ovarian cancer. Gynecol. Endocrinol. 2012, 28, 889–891. [Google Scholar] [CrossRef] [PubMed]
- Vigano, A.; Trutschnigg, B.; Kilgour, R.D.; Hamel, N.; Hornby, L.; Lucar, E.; Foulkes, W.; Tremblay, M.L.; Morais, J.A. Relationship between angiotensin-converting enzyme gene polymorphism and body composition, functional performance, and blood biomarkers in advanced cancer patients. Clin. Cancer Res. 2009, 15, 2442–2447. [Google Scholar] [CrossRef]
- Ding, P.; Yang, Y.; Ding, S.; Sun, B. Synergistic association of six well-characterized polymorphisms in three genes of the renin-angiotensin system with breast cancer among Han Chinese women. J. Renin Angiotensin Aldosterone Syst. 2015, 16, 1232–1239. [Google Scholar] [CrossRef]
- Fishchuk, L.E.; Gorovenko, N.G. Genetic polymorphisms of the renin-angiotensin system in breast cancer patients. Exp. Oncol. 2013, 35, 101–104. [Google Scholar]
- Röcken, C.; Röhl, F.W.; Diebler, E.; Lendeckel, U.; Pross, M.; Carl-McGrath, S.; Ebert, M.P. The angiotensin II/angiotensin II receptor system correlates with nodal spread in intestinal type gastric cancer. Cancer Epidemiol. Biomark. Prev. 2007, 16, 1206–1212. [Google Scholar] [CrossRef][Green Version]
- Gan, L.; Liu, X.; Wu, Z.; Huang, M.; Zhang, X.; Guo, W. Angiotensin-converting enzyme insertion/deletion polymorphism and gastric cancer: A systematic review and meta-analysis. Int. J. Clin. Exp. Med. 2015, 8, 5788–5793. [Google Scholar] [PubMed]
- Goto, Y.; Ando, T.; Nishio, K.; Ishida, Y.; Kawai, S.; Goto, H.; Hamajima, N. The ACE gene polymorphism is associated with the incidence of gastric cancer among H. pylori seropositive subjects with atrophic gastritis. Asian Pac. J. Cancer Prev. 2005, 6, 464–467. [Google Scholar] [PubMed]
- Papaggelopoulos, J.; Angelopoulou, A.; Avgoustidis, D.; Koronellos, N.; Derka, S.; Vassiliou, S.; Yapijakis, C. Association of Polymorphisms in the Genes of Angiotensinogen and Angiotensin Receptors with Risk for Basal Cell Carcinoma. Anticancer Res. 2019, 39, 5525–5530. [Google Scholar] [CrossRef]
- Samara, M.; Papathanassiou, M.; Farmakioti, I.; Anagnostou, M.; Satra, M.; Mitrakas, L.; Anastasiou, D.; Chasiotis, G.; Christopoulos, A.; Anagnostou, A.; et al. Renin-Angiotensin System Single Nucleotide Polymorphisms Are Associated with Bladder Cancer Risk. Curr. Oncol. 2021, 28, 4702–4708. [Google Scholar] [CrossRef]
- Huhn, S.; Bevier, M.; Rudolph, A.; Pardini, B.; Naccarati, A.; Hein, R.; Hoffmeister, M.; Vodickova, L.; Novotny, J.; Brenner, H.; et al. Shared ancestral susceptibility to colorectal cancer and other nutrition related diseases. BMC Med. Genet. 2012, 13, 94. [Google Scholar] [CrossRef] [PubMed]
- González-Zuloeta Ladd, A.M.; Arias Vásquez, A.; Siemes, C.; Yazdanpanah, M.; Coebergh, J.W.; Hofman, A.; Stricker, B.H.; van Duijn, C.M. Differential roles of Angiotensinogen and Angiotensin Receptor type 1 polymorphisms in breast cancer risk. Breast Cancer Res. Treat. 2007, 101, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Andreotti, G.; Boffetta, P.; Rosenberg, P.S.; Berndt, S.I.; Karami, S.; Menashe, I.; Yeager, M.; Chanock, S.J.; Zaridze, D.; Matteev, V.; et al. Variants in blood pressure genes and the risk of renal cell carcinoma. Carcinogenesis 2010, 31, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Vasků, A.; Vokurka, J.; Bienertová-Vasků, J. Obesity-related genes variability in Czech patients with sporadic colorectal cancer: Preliminary results. Int. J. Color. Dis. 2009, 24, 289–294. [Google Scholar] [CrossRef] [PubMed]
- League, S.; Hooper, W.C. Molecular diagnostics of inherited thrombosis. Clin. Lab. Sci. 2005, 18, 271–279. [Google Scholar]
- Lim, M.Y.; Deal, A.M.; Kim, S.; Musty, M.D.; Conard, J.; Simioni, P.; Dutrillaux, F.; Eid, S.S.; Middeldorp, S.; Halbmayer, W.M.; et al. Thrombophilic risk of individuals with rare compound factor V Leiden and prothrombin G20210A polymorphisms: An international case series of 100 individuals. Eur. J. Haematol. 2016, 97, 353–360. [Google Scholar] [CrossRef]
- Poort, S.R.; Rosendaal, F.R.; Reitsma, P.H.; Bertina, R.M. A common genetic variation in the 3′-untranslated region of the prothrombin gene is associated with elevated plasma prothrombin levels and an increase in venous thrombosis. Blood 1996, 88, 3698–3703. [Google Scholar] [CrossRef]
- Tormene, D.; Beltramello, P.; Perlati, M.; Brandolin, B.; Barbar, S.; De Toffoli, G.; Simioni, P. The risk of cancer progression in women with gynecological malignancies and thrombophilic polymorphisms: A pilot case-control study. Clin. Appl. Thromb. Hemost. 2009, 15, 535–539. [Google Scholar] [CrossRef] [PubMed]
- Hanafy, A.S.; Alaa, F.A.; Randa, M.H. Association of thrombogenic genes polymorphisms with hepatocellular carcinoma in HCV Egyptian patients. Gene 2016, 580, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Baghad, I.; Erguibi, D.; Chehab, F.; Nadifi, S. Risk of colorectal cancer and clotting factor gene polymorphisms in Moroccan Population. Int. J. Adv. Res. 2017, 5, 1141–1146. [Google Scholar] [CrossRef] [PubMed]
- Vossen, C.Y.; Hoffmeister, M.; Chang-Claude, J.C.; Rosendaal, F.R.; Brenner, H. Clotting factor gene polymorphisms and colorectal cancer risk. J. Clin. Oncol. 2011, 29, 1722–1727. [Google Scholar] [CrossRef] [PubMed]
- Leite, A.R.; Borges-Canha, M.; Cardoso, R.; Neves, J.S.; Castro-Ferreira, R.; Leite-Moreira, A. Novel Biomarkers for Evaluation of Endothelial Dysfunction. Angiology 2020, 71, 397–410. [Google Scholar] [CrossRef] [PubMed]
- Deanfield, J.E.; Halcox, J.P.; Rabelink, T.J. Endothelial function and dysfunction: Testing and clinical relevance. Circulation 2007, 115, 1285–1295. [Google Scholar] [CrossRef] [PubMed]
- Chia, P.Y.; Teo, A.; Yeo, T.W. Overview of the Assessment of Endothelial Function in Humans. Front. Med. 2020, 7, 542567. [Google Scholar] [CrossRef] [PubMed]
- Powrózek, T.; Mlak, R.; Brzozowska, A.; Mazurek, M.; Gołębiowski, P.; Małecka-Massalska, T. Relationship Between -2028 C/T SELP Gene Polymorphism, Concentration of Plasma P-Selectin and Risk of Malnutrition in Head and Neck Cancer Patients. Pathol. Oncol. Res. 2019, 25, 741–749. [Google Scholar] [CrossRef]
- Tan, B.H.; Fladvad, T.; Braun, T.P.; Vigano, A.; Strasser, F.; Deans, D.A.; Skipworth, R.J.; Solheim, T.S.; Damaraju, S.; Ross, J.A.; et al. P-selectin genotype is associated with the development of cancer cachexia. EMBO Mol. Med. 2012, 4, 462–471. [Google Scholar] [CrossRef]
- Avan, A.; Avan, A.; Le Large, T.Y.; Mambrini, A.; Funel, N.; Maftouh, M.; Ghayour-Mobarhan, M.; Cantore, M.; Boggi, U.; Peters, G.J.; et al. AKT1 and SELP polymorphisms predict the risk of developing cachexia in pancreatic cancer patients. PLoS ONE 2014, 9, e108057. [Google Scholar] [CrossRef] [PubMed]
- Zakariya, B.F.; Almohaidi, A.M.S.; Şimşek, S.A.; Al-Waysi, S.A.; Al-Dabbagh, W.H.; Kamal, A.M. The relationship of E-selectin singlenucleotide polymorphisms with breast cancer in Iraqi Arab women. Genom. Inform. 2022, 20, e42. [Google Scholar] [CrossRef] [PubMed]
- Turpin, A.; Labreuche, J.; Fléjou, J.F.; Andre, T.; de Gramont, A.; Hebbar, M. Prognostic factors in patients with stage II colon cancer: Role of E-selectin gene polymorphisms. Dig. Liver Dis. 2019, 51, 1198–1201. [Google Scholar] [CrossRef] [PubMed]
- Alessandro, R.; Seidita, G.; Flugy, A.M.; Damiani, F.; Russo, A.; Corrado, C.; Colomba, P.; Gullotti, L.; Buettner, R.; Bruno, L.; et al. Role of S128R polymorphism of E-selectin in colon metastasis formation. Int. J. Cancer 2007, 121, 528–535. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Naidu, R.; Har, Y.C.; Taib, N.A. Polymorphic variant Ser128Arg of E-Selectin is associated with breast cancer risk and high grade tumors. Onkologie 2011, 34, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Golnarnik, G.; Mashayekhi, F.; Saedi, H.S. The E-selectin S149R polymorphisms in breast cancer in a northern Iran population. Cell. Mol. Biol. 2016, 62, 34–37. [Google Scholar] [PubMed]
- Xia, H.Z.; Du, W.D.; Wu, Q.; Chen, G.; Zhou, Y.; Tang, X.F.; Tang, H.Y.; Liu, Y.; Yang, F.; Ruan, J.; et al. E-selectin rs5361 and FCGR2A rs1801274 variants were associated with increased risk of gastric cancer in a Chinese population. Mol. Carcinog. 2012, 51, 597–607. [Google Scholar] [CrossRef]
- Liarmakopoulos, E.; Gazouli, M.; Aravantinos, G.; Theodoropoulos, G.; Rizos, S.; Vaiopoulou, A.; Kouraklis, G.; Nikiteas, N. E-Selectin S128R gene polymorphism in gastric cancer. Int. J. Biol. Markers 2013, 28, 38–42. [Google Scholar] [CrossRef]
- Panoussopoulos, G.S.; Theodoropoulos, G.; Michalopoulos, N.V.; Gazouli, M.; Flessas, J.; Taka, S.; Stamopoulos, P.; Manouras, A.; Zografos, G.C. Analysis of E-Selectin S128R gene polymorphism in pancreatic cancer. J. Surg. Oncol. 2010, 102, 604–607. [Google Scholar] [CrossRef]
- Lu, Z.H.; Gu, X.J.; Shi, K.Z.; Li, X.; Chen, D.D.; Chen, L. Association between genetic polymorphisms of inflammatory response genes and the risk of ovarian cancer. J. Formos. Med. Assoc. 2016, 115, 31–37. [Google Scholar] [CrossRef][Green Version]
- Kontogianni, P.; Zambirinis, C.P.; Theodoropoulos, G.; Gazouli, M.; Michalopoulos, N.V.; Flessas, J.; Liberi, M.; Zografos, G.C. The impact of the stromal cell-derived factor-1-3′A and E-selectin S128R polymorphisms on breast cancer. Mol. Biol. Rep. 2013, 40, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.Y.; Hao, Y.W.; Zhou, W.L.; Ma, Y.R. E-selectin S128R polymorphism is associated with cancer risk: A meta-analysis. Asian Pac. J. Cancer Prev. 2014, 15, 3247–3252. [Google Scholar] [CrossRef] [PubMed]
- Calabrò, P.; Gragnano, F.; Golia, E.; Grove, E.L. von Willebrand Factor and Venous Thromboembolism: Pathogenic Link and Therapeutic Implications. Semin. Thromb. Hemost. 2018, 44, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Qian, D.; Liu, H.; Wang, X.; Ge, J.; Luo, S.; Patz, E.F., Jr.; Moorman, P.G.; Su, L.; Shen, S.; Christiani, D.C.; et al. Potentially functional genetic variants in the complement-related immunity gene-set are associated with non-small cell lung cancer survival. Int. J. Cancer 2019, 144, 1867–1876. [Google Scholar] [CrossRef]
- Arandi, N.; Talei, A.; Erfani, N.; Ghaderi, A. Intercellular adhesion molecule-1 genetic markers (+241G/A and +469A/G) in Iranian women with breast cancer. Cancer Genet. Cytogenet. 2008, 183, 9–13. [Google Scholar] [CrossRef]
- Yilmaz, U.; Zeybek, U.; Kahraman, O.T.; Kafadar, A.M.; Toptas, B.; Yamak, N.; Celik, F.; Yaylim, I. Investigation of ICAM-1 and β3 integrin gene variations in patients with brain tumors. Asian Pac. J. Cancer Prev. 2013, 14, 5929–5934. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Feng, Y.; Li, X.; Ma, Q.; Zhang, S.; Zhu, M.; Li, S.; Fang, L.; Tian, J.; Sun, L. Intercellular Adhesion Molecule-1 Gene Polymorphisms and Susceptibility to Cervical Cancer in the Northern Chinese Han Population. Front. Genet. 2021, 12, 668539. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Wan, A.; Feng, A.; Rui, R.; Zhou, B. ICAM-1 gene rs5498 polymorphism decreases the risk of coronary artery disease. Medicine 2018, 97, e12523. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Wang, Y.; Zhang, Z.; Qin, R.; Peng, Y.; Tang, W.; Xi, Y.; Tian, G.; Zhang, Y. Roles of intercellular cell adhesion molecule-1 (ICAM-1) in colorectal cancer: Expression, functions, prognosis, tumorigenesis, polymorphisms and therapeutic implications. Front. Oncol. 2022, 12, 1052672. [Google Scholar] [CrossRef]
- Cheng, D.; Liang, B. Intercellular Adhesion Molecule-1 (ICAM-1) Polymorphisms and Cancer Risk: A Meta-Analysis. Iran. J. Public Health 2015, 44, 615–624. [Google Scholar]
- Tang, W.; Wang, Y.; Chen, Y.; Gu, H.; Chen, S.; Kang, M. Polymorphisms in the intercellular adhesion molecule 1 gene and cancer risk: A meta-analysis. Int. J. Clin. Exp. Med. 2015, 8, 11996–12008. [Google Scholar] [PubMed]
- Lin, C.W.; Chuang, C.Y.; Tang, C.H.; Chang, J.L.; Lee, L.M.; Lee, W.J.; Chow, J.M.; Yang, S.F.; Chien, M.H. Combined effects of icam-1 single-nucleotide polymorphisms and environmental carcinogens on oral cancer susceptibility and clinicopathologic development. PLoS ONE 2013, 8, e72940. [Google Scholar] [CrossRef]
- Liu, L.B.; Liu, T.; Xin, F.Z. Correlations of ICAM-1 gene polymorphisms with susceptibility and multidrug resistance in colorectal cancer in a Chinese population. Medicine 2017, 96, e7481. [Google Scholar] [CrossRef]
- Qiu, Z.; Xie, Z.; Qin, R.; Chen, M.; He, H.; Zhang, Z.; Wang, Y.; Hong, M.; Tang, W.; Xi, Y.; et al. Evaluation of ICAM-1 rs5498 and rs3093030 Polymorphisms in Chinese Patients with Colorectal Cancer. DNA Cell Biol. 2021, 40, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Theodoropoulos, G.; Papaconstantinou, I.; Felekouras, E.; Nikiteas, N.; Karakitsos, P.; Panoussopoulos, D.; Lazaris, A.; Patsouris, E.; Bramis, J.; Gazouli, M. Relation between common polymorphisms in genes related to inflammatory response and colorectal cancer. World J. Gastroenterol. 2006, 12, 5037–5043. [Google Scholar] [CrossRef]
- Novikov, V.V.; Shumilova, S.V.; Novikov, D.V.; Kalugin, A.V.; Fomina, S.G.; Karaulov, A.V. Genetic Instability in Locus rs5498 E469K (A/G) of ICAM-1 Gene in Patients with Colorectal Cancer and Breast Cancer. Bull. Exp. Biol. Med. 2016, 160, 811–813. [Google Scholar] [CrossRef]
- Wang, Q.L.; Li, B.H.; Liu, B.; Liu, Y.B.; Liu, Y.P.; Miao, S.B.; Han, Y.; Wen, J.K.; Han, M. Polymorphisms of the ICAM-1 exon 6 (E469K) are associated with differentiation of colorectal cancer. J. Exp. Clin. Cancer Res. 2009, 28, 139. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.M.; Sun, Y.; Li, Z.W.; Wu, Y.; Zhao, A.L.; Li, J.Y. Polymorphisms of ICAM-1 are associated with gastric cancer risk and prognosis. World J. Gastroenterol. 2012, 18, 368–374. [Google Scholar] [CrossRef]
- Thanopoulou, E.; Kotzamanis, G.; Pateras, I.S.; Ziras, N.; Papalambros, A.; Mariolis-Sapsakos, T.; Sigala, F.; Johnson, E.; Kotsinas, A.; Scorilas, A.; et al. The single nucleotide polymorphism g.1548A > G (K469E) of the ICAM-1 gene is associated with worse prognosis in non-small cell lung cancer. Tumor Biol. 2012, 33, 1429–1436. [Google Scholar] [CrossRef]
- Cai, G.; Ma, X.; Zou, W.; Huang, Y.; Zhang, J.; Wang, D.; Chen, B. Prediction value of intercellular adhesion molecule-1 gene polymorphisms for epithelial ovarian cancer risk, clinical features, and prognosis. Gene 2014, 546, 117–123. [Google Scholar] [CrossRef]
- Wang, S.S.; Hsieh, M.J.; Ou, Y.C.; Chen, C.S.; Li, J.R.; Hsiao, P.C.; Yang, S.F. Impacts of ICAM-1 gene polymorphisms on urothelial cell carcinoma susceptibility and clinicopathologic characteristics in Taiwan. Tumor Biol. 2014, 35, 7483–7490. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.P.; Lee, H.L.; Huang, Y.H.; Hsieh, M.J.; Chiang, W.L.; Kuo, W.H.; Chou, M.C.; Yang, S.F.; Yeh, C.B. Association of intercellular adhesion molecule-1 single nucleotide polymorphisms with hepatocellular carcinoma susceptibility and clinicopathologic development. Tumor Biol. 2016, 37, 2067–2074. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Hernandez, W.; Shriver, M.D.; Ahaghotu, C.A.; Kittles, R.A. ICAM gene cluster SNPs and prostate cancer risk in African Americans. Hum. Genet. 2006, 120, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.H.; Yang, S.F.; Liu, Y.F.; Ko, J.L.; Wu, C.H.; Wu, T.F.; Wang, P.H. Single-Nucleotide Polymorphisms and Haplotypes of Intercellular Adhesion Molecule-1 in Uterine Cervical Carcinogenesis in Taiwanese Women. Reprod. Sci. 2016, 23, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Burim, R.V.; Teixeira, S.A.; Colli, B.O.; Peria, F.M.; Tirapelli, L.F.; Marie, S.K.; Malheiros, S.M.; Oba-Shinjo, S.M.; Gabbai, A.A.; Lotufo, P.A.; et al. ICAM-1 (Lys469Glu) and PECAM-1 (Leu125Val) polymorphisms in diffuse astrocytomas. Clin. Exp. Med. 2009, 9, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Huang, J.; Bai, J.; Lu, W.; Zhang, M.; Mei, H. Association of Polymorphisms in Intercellular Adhesion Molecule 1 (ICAM-1) Gene with Cancer Susceptibility: A Meta-Analysis of 14 Case-Control Studies. Med. Sci. Monit. 2016, 22, 569–579. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Wang, S.; Ma, X. The Influence of ICAM1 3′UTR Gene Polymorphism on the Occurrence and Metastasis of Primary Liver Cancer. Biomed. Res. Int. 2021, 2021, 7377299. [Google Scholar] [CrossRef] [PubMed]
- Ghazy, A.A.; Elsheredy, H.G.; Abouelella, A.M.; Rashwan, E.K.; Khaled, B.E.A.; Elsheredy, A.G. Significance of Intracellular Adhesion Molecule-1 Polymorphism and IP-10 among Breast Cancer Patients. Egypt. J. Immunol. 2020, 27, 187–195. [Google Scholar] [PubMed]
- Ghazy, A.A.; El-Etreby, N.M. Relevance of HLA-DP/DQ and ICAM-1 SNPs among Ovarian Cancer Patients. Front. Immunol. 2016, 7, 202. [Google Scholar] [CrossRef]
- Dadgar Pakdel, F.; Keramatipour, M.; Noorbakhsh, F.; Talebi, S.; Vodjgani, M. Investigating the Effect of rs3783605 Single-nucleotide Polymorphism on the Activity of VCAM-1 Promoter in Human Umbilical Vein Endohelial Cells. Iran. J. Allergy Asthma Immunol. 2015, 14, 179–187. [Google Scholar]
- Miaskowski, C.; Dodd, M.; Paul, S.M.; West, C.; Hamolsky, D.; Abrams, G.; Cooper, B.A.; Elboim, C.; Neuhaus, J.; Schmidt, B.L.; et al. Lymphatic and angiogenic candidate genes predict the development of secondary lymphedema following breast cancer surgery. PLoS ONE 2013, 8, e60164. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.S.; Carreon, J.D.; Hanchard, B.; Chanock, S.; Hisada, M. Common genetic variants and risk for non-Hodgkin lymphoma and adult T-cell lymphoma/leukemia in Jamaica. Int. J. Cancer 2009, 125, 1479–1482. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.I.; Jun, S.E.; Shin, D.H. Associations of VCAM-1 gene polymorphisms with obesity and inflammation markers. Inflamm. Res. 2017, 66, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Andrew, A.S.; Gui, J.; Hu, T.; Wyszynski, A.; Marsit, C.J.; Kelsey, K.T.; Schned, A.R.; Tanyos, S.A.; Pendleton, E.M.; Ekstrom, R.M.; et al. Genetic polymorphisms modify bladder cancer recurrence and survival in a USA population-based prognostic study. BJU Int. 2015, 115, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Ehrenfeld, P.; Cordova, F.; Duran, W.N.; Sanchez, F.A. S-nitrosylation and its role in breast cancer angiogenesis and metastasis. Nitric Oxide 2019, 87, 52–59. [Google Scholar] [CrossRef]
- Nastri, C.O.; Martins Wde, P.; Reis, F.J.; Ferriani, R.A. Breast cancer and endothelial dysfunction. Rev. Assoc. Med. Bras. 2008, 54, 467–470. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Choudhari, S.K.; Chaudhary, M.; Bagde, S.; Gadbail, A.R.; Joshi, V. Nitric oxide and cancer: A review. World J. Surg. Oncol. 2013, 11, 118. [Google Scholar] [CrossRef] [PubMed]
- Knowles, J.; Loizidou, M.; Taylor, I. Endothelin-1 and angiogenesis in cancer. Curr. Vasc. Pharmacol. 2005, 3, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Cazaubon, S.; Deshayes, F.; Couraud, P.O.; Nahmias, C. Endothelin-1, angiotensin II and cancer. Med. Sci. 2006, 22, 416–422. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, X.; Cai, L.; Yang, Y.; Sui, G.; Fu, S. Angiotensin II/angiotensin II type I receptor (AT1R) signaling promotes MCF-7 breast cancer cells survival via PI3-kinase/Akt pathway. J. Cell. Physiol. 2010, 225, 168–173. [Google Scholar] [CrossRef]
- Deshayes, F.; Nahmias, C. Angiotensin receptors: A new role in cancer? Trends Endocrinol. Metab. 2005, 16, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Franchini, M.; Mannucci, P.M. Thrombin and cancer: From molecular basis to therapeutic implications. Semin. Thromb. Hemost. 2012, 38, 95–101. [Google Scholar] [CrossRef]
- Rickles, F.R.; Patierno, S.; Fernandez, P.M. Tissue factor, thrombin, and cancer. Chest 2003, 124, 58s–68s. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Wei, W. Angiotensin II in inflammation, immunity and rheumatoid arthritis. Clin. Exp. Immunol. 2015, 179, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Harjunpää, H.; Llort Asens, M.; Guenther, C.; Fagerholm, S.C. Cell Adhesion Molecules and Their Roles and Regulation in the Immune and Tumor Microenvironment. Front. Immunol. 2019, 10, 1078. [Google Scholar] [CrossRef]
- Kobayashi, H.; Boelte, K.C.; Lin, P.C. Endothelial cell adhesion molecules and cancer progression. Curr. Med. Chem. 2007, 14, 377–386. [Google Scholar] [CrossRef]
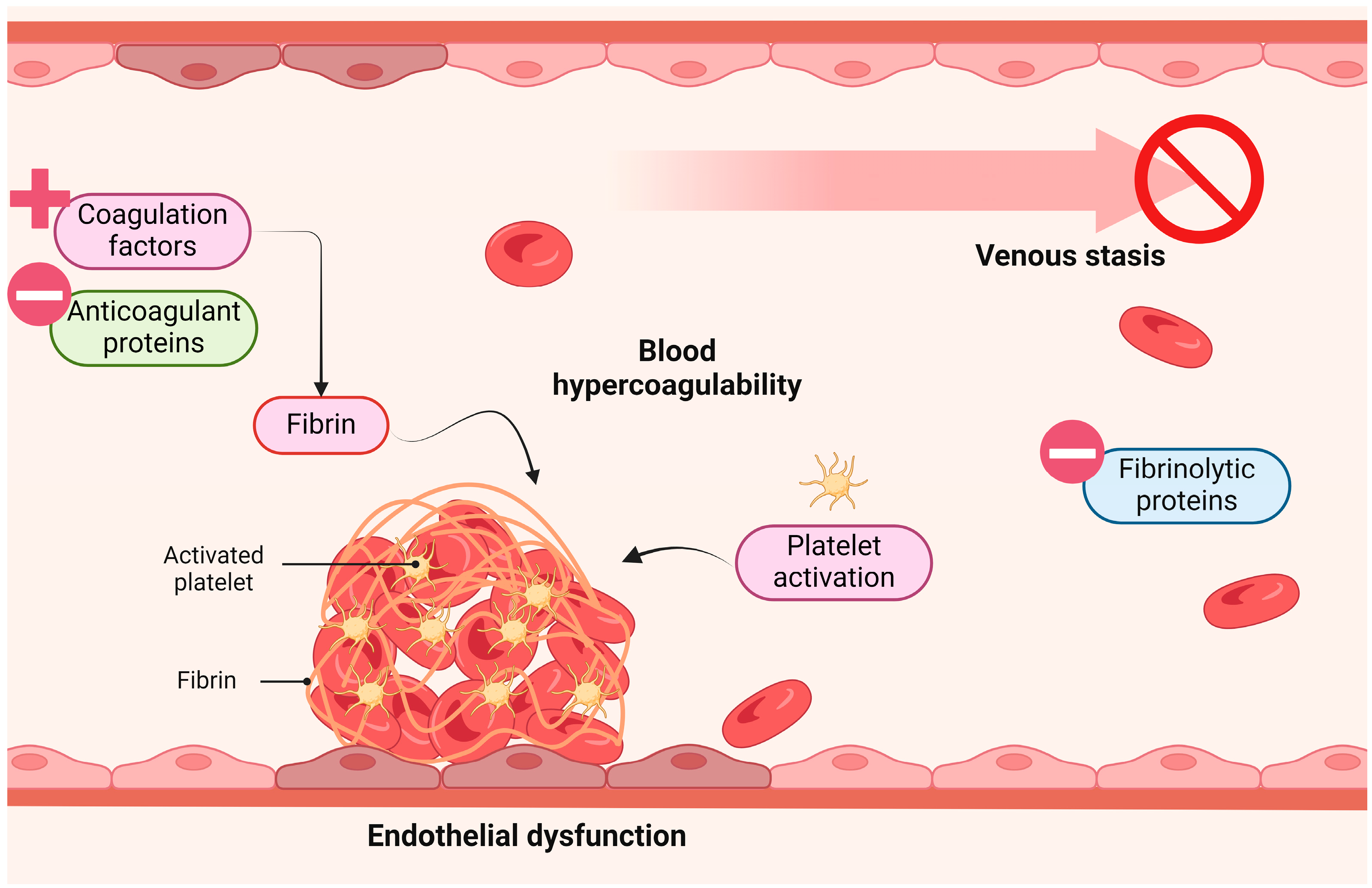
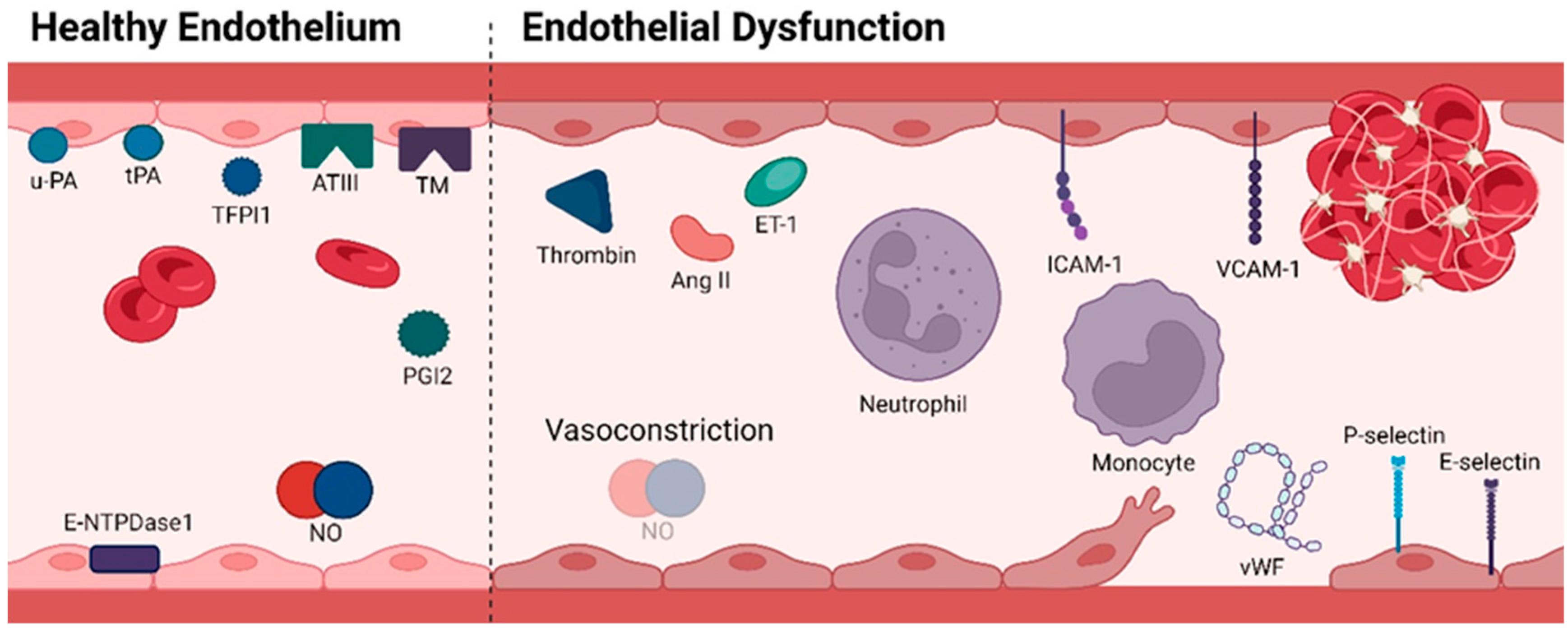

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Melo, I.G.; Tavares, V.; Pereira, D.; Medeiros, R. Contribution of Endothelial Dysfunction to Cancer Susceptibility and Progression: A Comprehensive Narrative Review on the Genetic Risk Component. Curr. Issues Mol. Biol. 2024, 46, 4845-4873. https://doi.org/10.3390/cimb46050292
de Melo IG, Tavares V, Pereira D, Medeiros R. Contribution of Endothelial Dysfunction to Cancer Susceptibility and Progression: A Comprehensive Narrative Review on the Genetic Risk Component. Current Issues in Molecular Biology. 2024; 46(5):4845-4873. https://doi.org/10.3390/cimb46050292
Chicago/Turabian Stylede Melo, Inês Guerra, Valéria Tavares, Deolinda Pereira, and Rui Medeiros. 2024. "Contribution of Endothelial Dysfunction to Cancer Susceptibility and Progression: A Comprehensive Narrative Review on the Genetic Risk Component" Current Issues in Molecular Biology 46, no. 5: 4845-4873. https://doi.org/10.3390/cimb46050292
APA Stylede Melo, I. G., Tavares, V., Pereira, D., & Medeiros, R. (2024). Contribution of Endothelial Dysfunction to Cancer Susceptibility and Progression: A Comprehensive Narrative Review on the Genetic Risk Component. Current Issues in Molecular Biology, 46(5), 4845-4873. https://doi.org/10.3390/cimb46050292






