Multiomics Analysis of the PHLDA Gene Family in Different Cancers and Their Clinical Prognostic Value
Abstract
1. Introduction
2. Materials and Methods
2.1. Oncomine Analysis
2.2. Analysis of the Expression of Gene and Mutation by Using cBioPortal
2.3. Prognosis Analysis Using PrognoScan
2.4. mRNA Expression Profiling with GEPIA
2.5. Survival Data Analysis Applying Kaplan–Meier Plotter
2.6. R2 Platform for Correlation and Survival Analysis
2.7. Protein–Protein Interaction (PPI) Analysis Using GeneMANIA
2.8. Statistical Analysis
3. Results
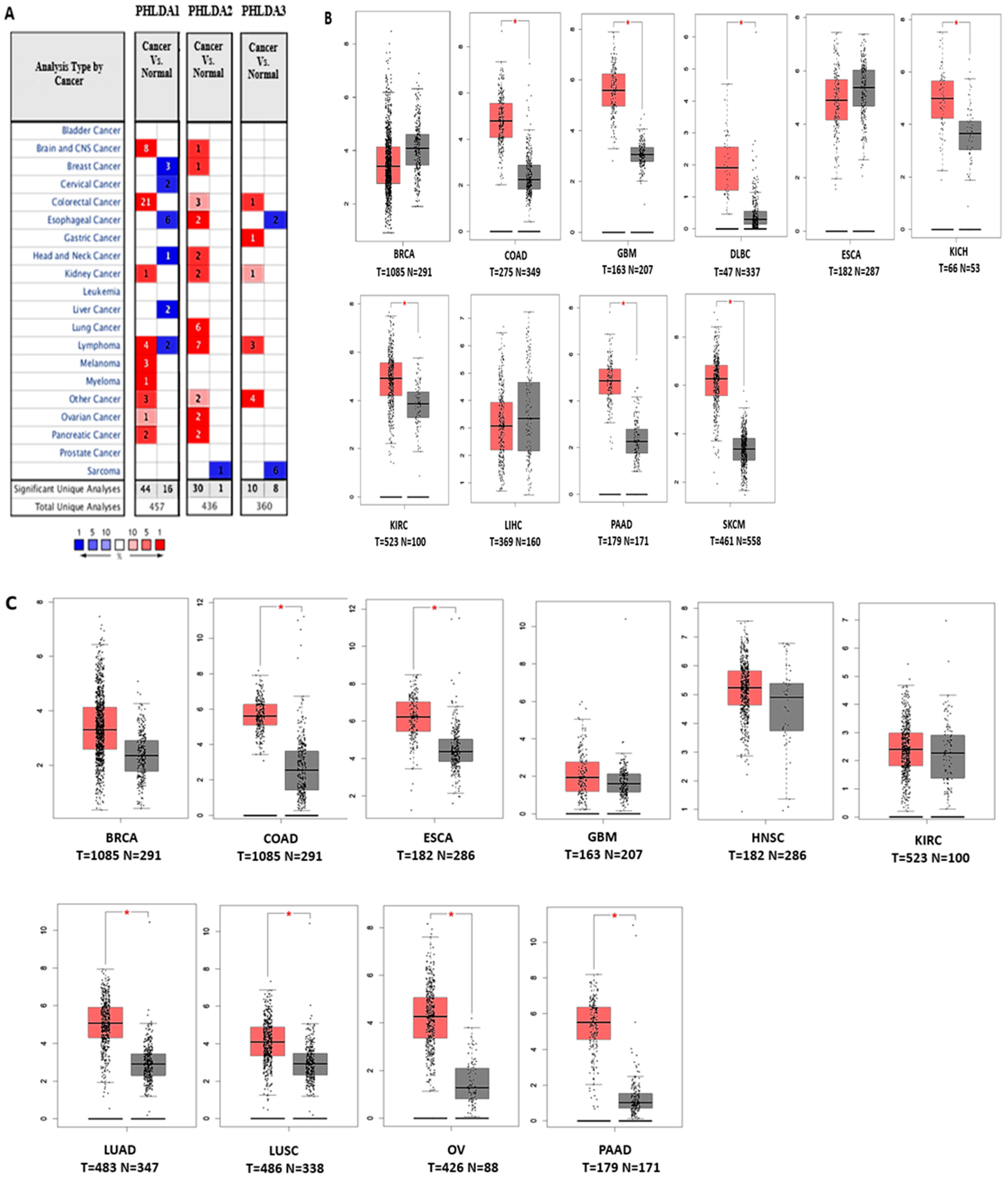
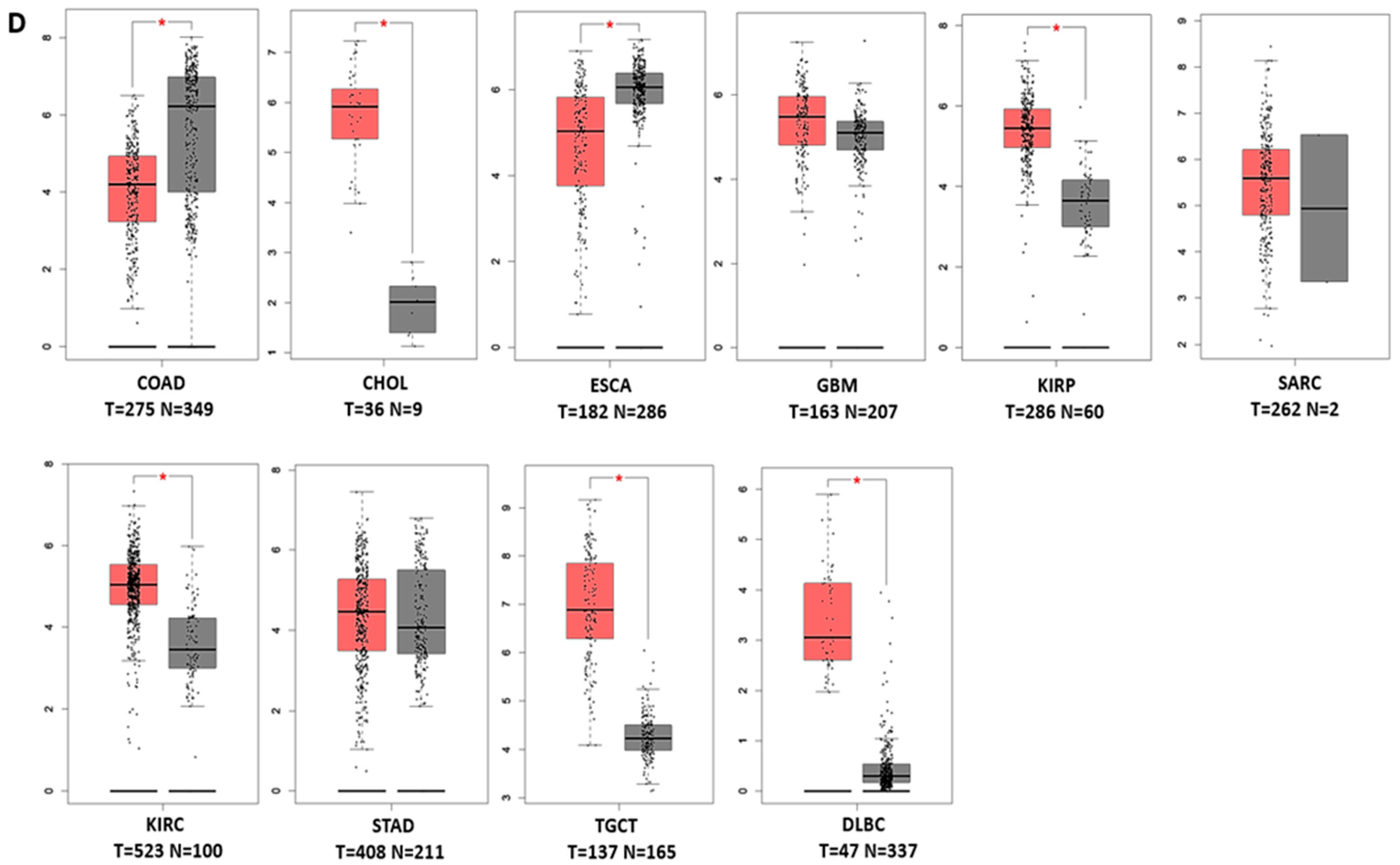
3.1. Transcript Expression Analysis of PHLDA in Various Cancers
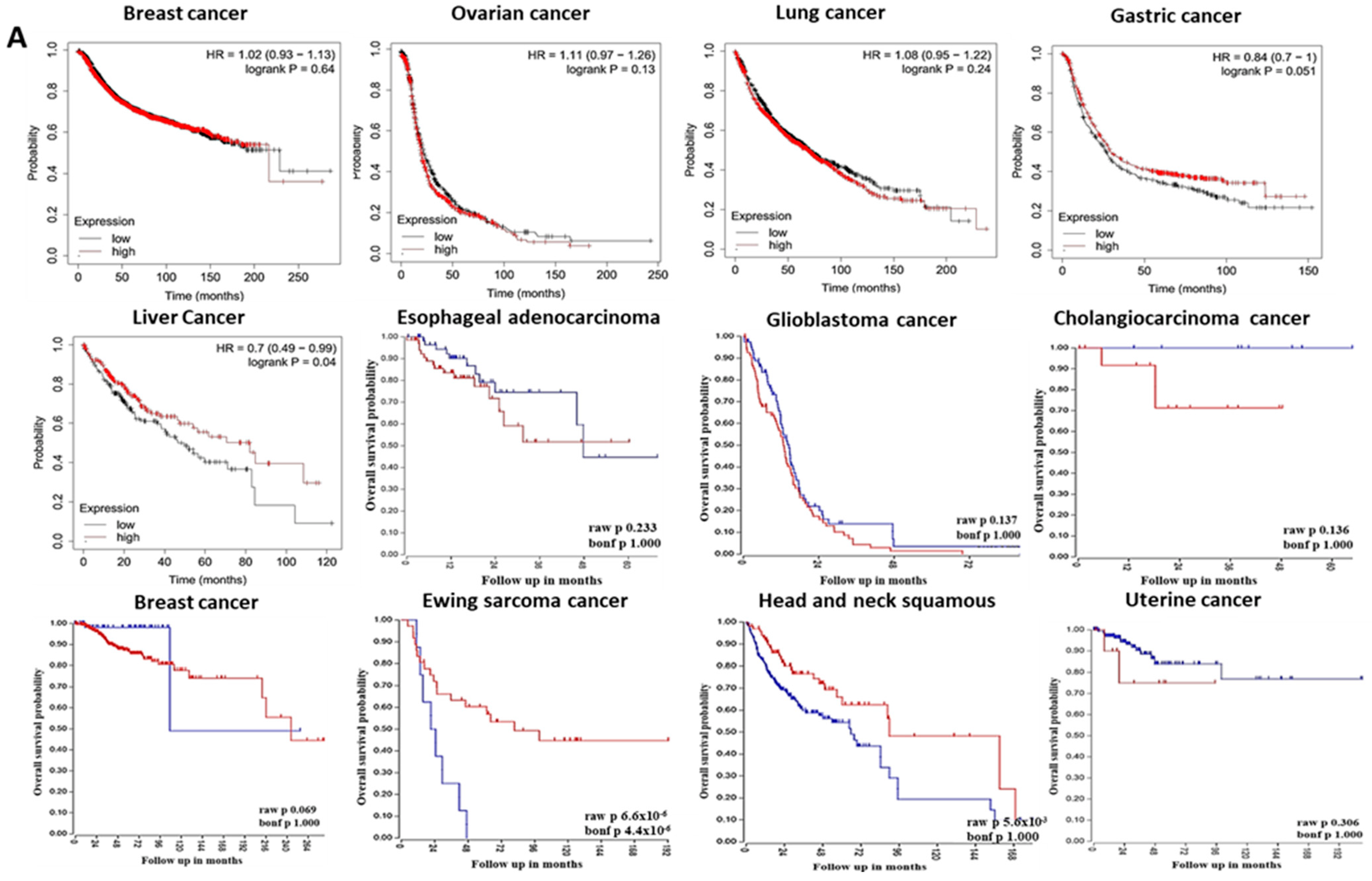
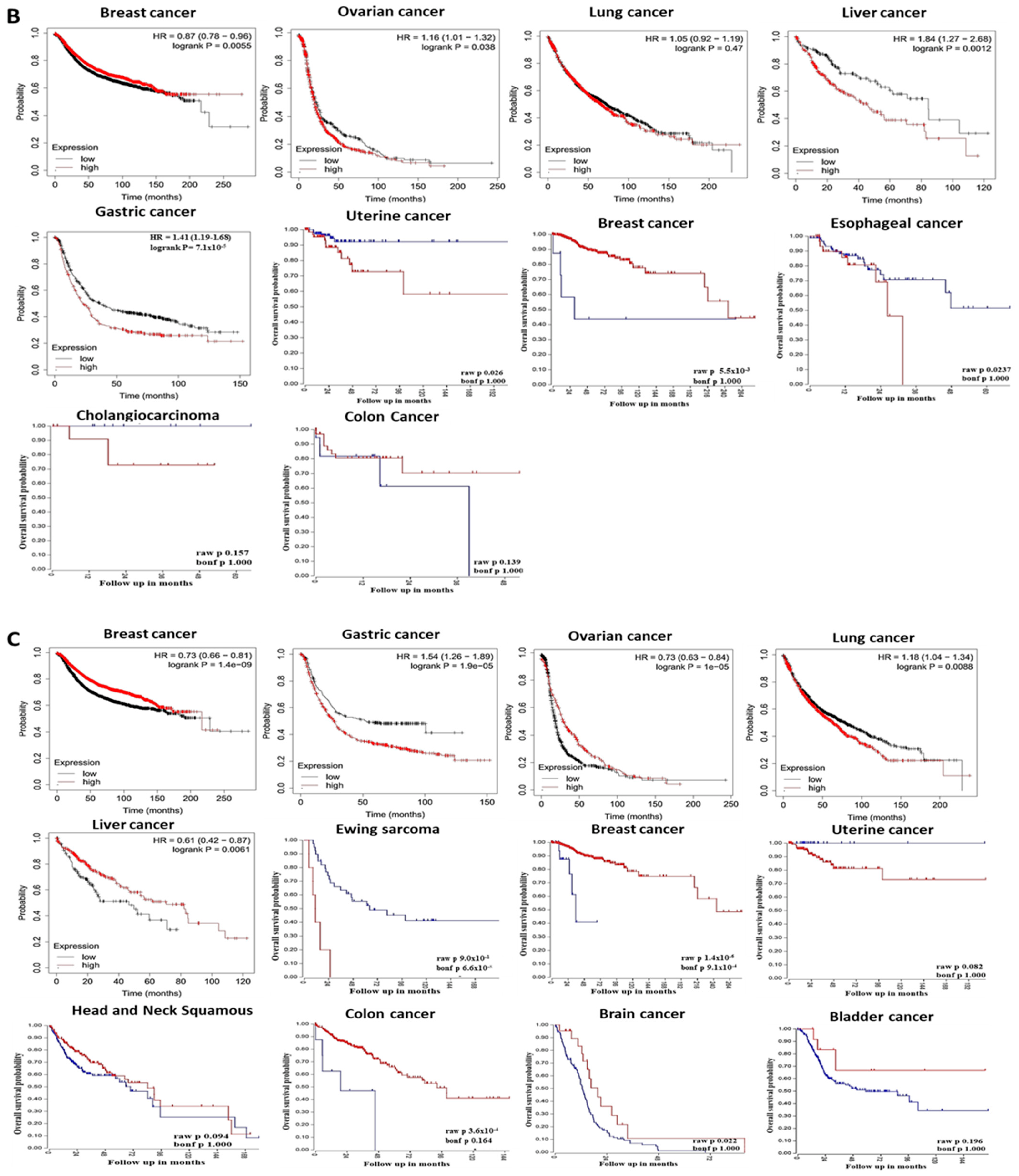
3.2. The Prognostic Value of Pleckstrin Family Member A
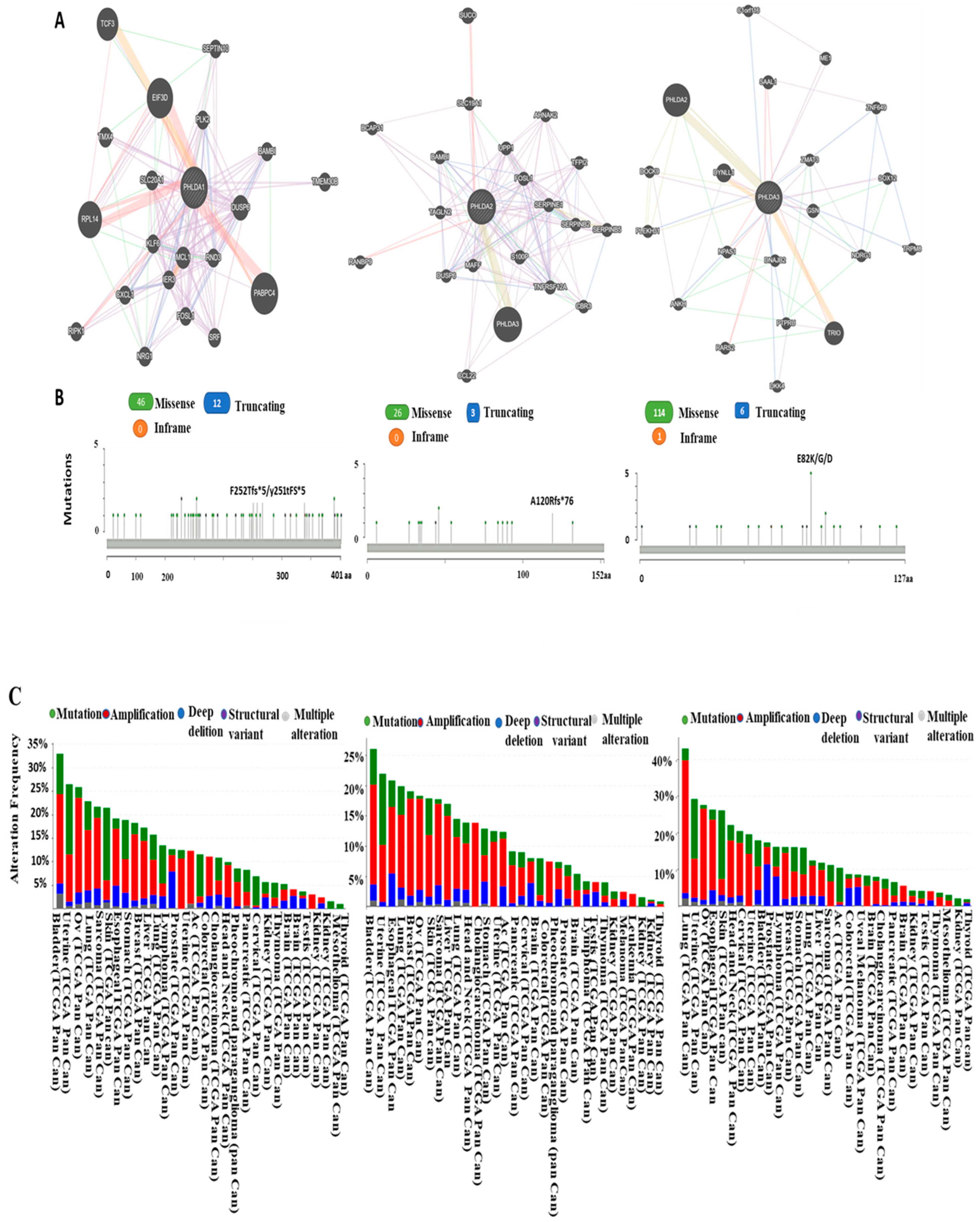
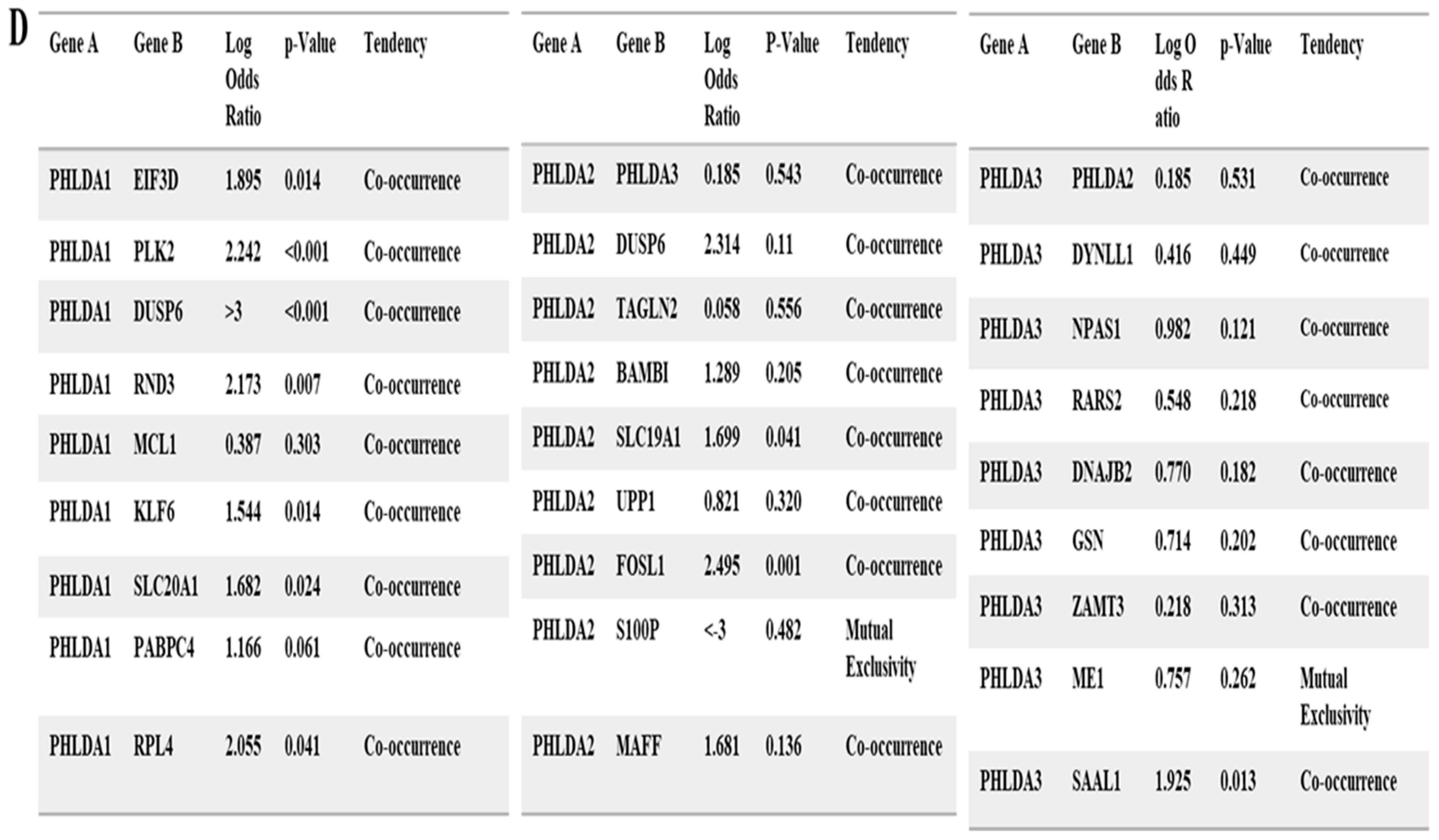
3.3. Predicting Protein–Protein Interaction of Pleckstrin Family Member A
3.4. Analysis of PHLDA Copy Number Alterations and Mutations across Cancers
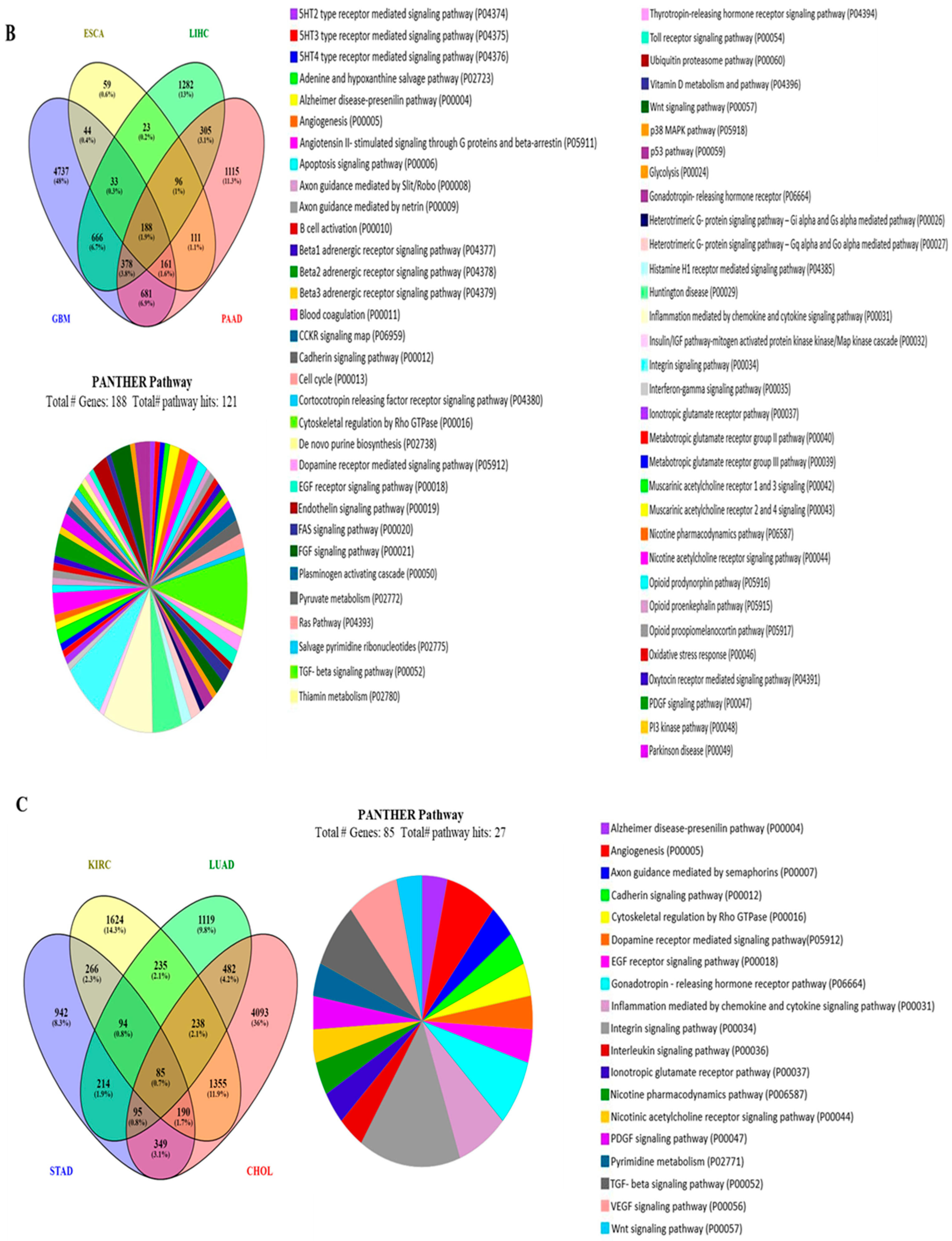
3.5. The Functional Gene Ontology and Pathways of the Genes Associated with the PHLDA Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brader, G.; Compant, S.; Mitter, B.; Trognitz, F.; Sessitsch, A. Metabolic potential of endophytic bacteria. Curr. Opin. Biotechnol. 2014, 27, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, D.R.; Patel, R.; Kirsch, D.G.; Lewis, C.A.; Vander Heiden, M.G.; Locasale, J.W. Metabolomics in cancer research and emerging applications in clinical oncology. CA Cancer J. Clin. 2021, 71, 333–358. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Chandel, N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016, 2, e1600200. [Google Scholar] [CrossRef] [PubMed]
- Baldavira, C.M.; Machado-Rugolo, J.; Prieto, T.G.; Bastos, D.R.; Balancin, M.; Ab’Saber, A.M.; Yaegashi, L.B.; Souza, P.C.; Farhat, C.; Takagaki, T.Y.; et al. The expression patterns and prognostic significance of pleckstrin homology-like domain family A (PHLDA) in lung cancer and malignant mesothelioma. J. Thorac. Dis. 2021, 13, 689–707. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.; Liu, Y.; Gao, Z.; Huang, W. Recent advances in drug delivery systems for targeting cancer stem cells. Acta Pharm. Sin. B 2021, 11, 55–70. [Google Scholar] [CrossRef] [PubMed]
- Haslam, R.J.; Koide, H.B.; Hemmings, B.A. Pleckstrin domain homology. Nature 1993, 363, 309–310. [Google Scholar] [CrossRef]
- Frank, D.; Mendelsohn, C.L.; Ciccone, E.; Svensson, K.; Ohlsson, R.; Tycko, B. A novel pleckstrin homology-related gene family defined by Ipl/Tssc3, TDAG51, and Tih1: Tissue-specific expression, chromosomal location, and parental imprinting. Mamm. Genome. 1999, 10, 1150–1159. [Google Scholar] [CrossRef] [PubMed]
- Nagai, M.A. Pleckstrin homology-like domain, family A, member 1 (PHLDA1) and cancer. Biomed. Rep. 2016, 4, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Fearon, A.E.; Carter, E.P.; Clayton, N.S.; Wilkes, E.H.; Baker, A.-M.; Kapitonova, E.; Bakhouche, B.A.; Tanner, Y.; Wang, J.; Gadaleta, E.; et al. PHLDA1 Mediates Drug Resistance in Receptor Tyrosine Kinase-Driven Cancer. Cell Rep. 2018, 22, 2469–2481. [Google Scholar] [CrossRef]
- Li, G.; Wang, X.; Hibshoosh, H.; Jin, C.; Halmos, B. Modulation of ErbB2 blockade in ErbB2-positive cancers: The role of ErbB2 Mutations and PHLDA1. PLoS ONE 2014, 9, e106349. [Google Scholar] [CrossRef]
- Duan, Y.; Du, Y.; Gu, Z.; Zheng, X.; Wang, C. Prognostic Value, Immune Signature, and Molecular Mechanisms of the PHLDA Family in Pancreatic Adenocarcinoma. Int. J. Mol. Sci. 2022, 23, 10316. [Google Scholar] [CrossRef] [PubMed]
- Ertas, Y.N.; Abedi Dorcheh, K.; Akbari, A.; Jabbari, E. Nanoparticles for Targeted Drug Delivery to Cancer Stem Cells: A Review of Recent Advances. Nanomaterials 2021, 11, 1755. [Google Scholar] [CrossRef]
- Purba, T.S.; Haslam, I.S.; Poblet, E.; Jiménez, F.; Gandarillas, A.; Izeta, A.; Paus, R. Human epithelial hair follicle stem cells and their progeny: Current state of knowledge, the widening gap in translational research and future challenges. Bioessays 2014, 36, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Latil, M.; Nassar, D.; Beck, B.; Boumahdi, S.; Wang, L.; Brisebarre, A.; Dubois, C.; Nkusi, E.; Lenglez, S.; Checinska, A.; et al. Cell-Type-Specific Chromatin States Differentially Prime Squamous Cell Carcinoma Tumor-Initiating Cells for Epithelial to Mesenchymal Transition. Cell Stem Cell 2017, 20, 191–204.e5. [Google Scholar] [CrossRef]
- Dave, B.; Mittal, V.; Tan, N.M.; Chang, J.C. Epithelial-mesenchymal transition, cancer stem cells and treatment resistance. Breast Cancer Res. 2012, 14, 202. [Google Scholar] [CrossRef]
- Sakthianandeswaren, A.; Christie, M.; D’Andreti, C.; Tsui, C.; Jorissen, R.N.; Li, S.; Fleming, N.I.; Gibbs, P.; Lipton, L.; Malaterre, J.; et al. PHLDA1 expression marks the putative epithelial stem cells and contributes to intestinal tumorigenesis. Cancer Res. 2011, 71, 3709–3719. [Google Scholar] [CrossRef]
- Bonatto, N.; Carlini, M.J.; de Bessa Garcia, S.A.; Nagai, M.A. PHLDA1 (pleckstrin homology-like domain, family A, member 1) knockdown promotes migration and invasion of MCF10A breast epithelial cells. Cell Adh Migr. 2018, 12, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Hossain, G.S.; Lynn, E.G.; Maclean, K.N.; Zhou, J.; Dickhout, J.G.; Lhoták, S.; Trigatti, B.; Capone, J.; Rho, J.; Tang, D.; et al. Deficiency of TDAG51 protects against atherosclerosis by modulating apoptosis, cholesterol efflux, and peroxiredoxin-1 expression. J. Am. Heart Assoc. 2013, 2, e000134. [Google Scholar] [CrossRef]
- Zimnicka, A.M.; Sharma, T.; Regan, M.; Merrill, B.J.; Frasor, J. Knockout of the PHLDA1 gene in breast cancer cells reveals multiple roles for PHLDA1 in cancer phenotypes. FASEB J. 2017, 31 (Suppl. S1), 178.8. [Google Scholar] [CrossRef]
- Neef, R.; Kuske, M.A.; Pröls, E.; Johnson, J.P. Identification of the human PHLDA1/TDAG51 gene: Down-regulation in metastatic melanoma contributes to apoptosis resistance and growth deregulation. Cancer Res. 2002, 62, 5920–5929. [Google Scholar]
- Ren, L.; Mendoza, A.; Zhu, J.; Briggs, J.W.; Halsey, C.; Hong, E.S.; Burkett, S.S.; Morrow, J.J.; Lizardo, M.M.; Osborne, T.; et al. Characterization of the metastatic phenotype of a panel of established osteosarcoma cells. Oncotarget 2015, 6, 29469–29481. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.P.; Reeves, C.; Schmitt, A.; Su, K.; Connors, T.D.; Hu, R.J.; Brandenburg, S.; Lee, M.J.; Miller, G.; Feinberg, A.P. Somatic mutation of TSSC5, a novel imprinted gene from human chromosome 11p15.5. Cancer Res. 1998, 58, 4155–4159. [Google Scholar] [PubMed]
- Ma, Z.; Lou, S.; Jiang, Z. PHLDA2 regulates EMT and autophagy in colorectal cancer via the PI3K/AKT signaling pathway. Aging 2020, 12, 7985–8000. [Google Scholar] [CrossRef]
- Idichi, T.; Seki, N.; Kurahara, H.; Fukuhisa, H.; Toda, H.; Shimonosono, M.; Okato, A.; Arai, T.; Kita, Y.; Mataki, Y.; et al. Molecular pathogenesis of pancreatic ductal adenocarcinoma: Impact of passenger strand of pre-miR-148a on gene regulation. Cancer Sci. 2018, 109, 2013–2026. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Song, X.; Liu, Z.; Li, Q.; Huang, M.; Su, B.; Mao, Y.; Wang, Y.; Mo, W.; Chen, H. Upregulation of miR-214 Induced Radioresistance of Osteosarcoma by Targeting PHLDA2 via PI3K/Akt Signaling. Front. Oncol. 2019, 9, 298. [Google Scholar] [CrossRef] [PubMed]
- Chou, W.-H.; Liu, K.-L.; Shih, Y.-L.; Chuang, Y.-Y.; Chou, J.; Lu, H.-F.; Jair, H.-W.; Lee, M.-Z.; Au, M.-K.; Chung, J.-G. Ouabain Induces Apoptotic Cell Death Through Caspase- and Mitochondria-dependent Pathways in Human Osteosarcoma U-2 OS Cells. Anticancer Res. 2018, 38, 169–178. [Google Scholar] [PubMed]
- Chen, Y.; Ohki, R. p53-PHLDA3-Akt Network: The Key Regulators of Neuroendocrine Tumorigenesis. Int. J. Mol. Sci. 2020, 21, 4098. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.G.; Kang, Y.J.; Kim, H.S.; Moon, A.; Kim, S.G. Phlda3, a urine-detectable protein, causes p53 accumulation in renal tubular cells injured by cisplatin. Cell Biol. Toxicol. 2015, 31, 121–130. [Google Scholar] [CrossRef]
- Liu, J.; Liu, X.; Hui, X.; Cai, L.; Li, X.; Yang, Y.; Shu, S.; Wang, F.; Xia, H.; Li, S. Novel Role for Pleckstrin Homology-Like Domain Family A, Member 3 in the Regulation of Pathological Cardiac Hypertrophy. J. Am. Heart Assoc. 2019, 8, e011830. [Google Scholar] [CrossRef]
- Kawase, T.; Ohki, R.; Shibata, T.; Tsutsumi, S.; Kamimura, N.; Inazawa, J.; Ohta, T.; Ichikawa, H.; Aburatani, H.; Tashiro, F.; et al. PH domain-only protein PHLDA3 is a p53-regulated repressor of Akt. Cell 2009, 136, 535–550. [Google Scholar] [CrossRef]
- Indarte, M.; Puentes, R.; Maruggi, M.; Ihle, N.T.; Grandjean, G.; Scott, M.; Ahmed, Z.; Meuillet, E.J.; Zhang, S.; Lemos, R.; et al. An Inhibitor of the Pleckstrin Homology Domain of CNK1 Selectively Blocks the Growth of Mutant KRAS Cells and Tumors. Cancer Res. 2019, 79, 3100–3111. [Google Scholar] [CrossRef] [PubMed]
- Fuselier, T.T.; Lu, H. PHLD Class Proteins: A Family of New Players in the p53 Network. Int. J. Mol. Sci. 2020, 21, 3543. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.R.; Yu, J.; Shanker, K.; Deshpande, N.; Varambally, R.; Ghosh, D.; Barrette, T.; Pander, A.; Chinnaiyan, A.M. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia 2004, 6, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.E.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, H.; Kitada, K.; Nakai, K.; Sarai, A. PrognoScan: A new database for meta-analysis of the prognostic value of genes. BMC Med. Genom. 2009, 2, 18. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Lanczky, A.; Nagy, A.; Bottai, G.; Munkacsy, G.; Szabo, A.; Santarpia, L.; Gyorffy, B. miRpower: A web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016, 160, 439–446. [Google Scholar] [CrossRef]
- Koster, J.; Molenaar, J.J.; Versteeg, R. Abstract A2-45: R2: Accessible web-based genomics analysis and visualization platform for biomedical researchers. Cancer Res. 2015, 75, A2-45. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Chiu, S.T.; Hsieh, F.J.; Chen, S.W.; Chen, C.L.; Shu, H.F.; Li, H. Clinicopathologic correlation of up-regulated genes identified using cDNA microarray and real-time reverse transcription-PCR in human colorectal cancer. Cancer Epidemiol. Biomark. Prev. 2005, 14, 437–443. [Google Scholar] [CrossRef]
- Kastrati, I.; Canestrari, E.; Frasor, J. PHLDA1 expression is controlled by an estrogen receptor-NFκB-miR-181 regulatory loop and is essential for formation of ER+ mammospheres. Oncogene 2015, 34, 2309–2316. [Google Scholar] [CrossRef]
- Yoo, N.J.; Kim, Y.R.; Lee, S.H. Expressional and mutational analysis of PHLDA3 gene in common human cancers. Pathology 2011, 43, 510–511. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.K.; Islam, S.M.R.; Kwak, K.S.; Rahman, M.S.; Cho, S.G. PROM1 and PROM2 expression differentially modulates clinical prognosis of cancer: A multiomics analysis. Cancer Gene Ther. 2020, 27, 147–167. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Yang, R.; Song, J.; Wang, X.; Dong, W. Calpain2 Upregulation Regulates EMT-Mediated Pancreatic Cancer Metastasis via the Wnt/β-Catenin Signaling Pathway. Front. Med. 2022, 9, 783592. [Google Scholar] [CrossRef] [PubMed]
- Powis, G.; Meuillet, E.J.; Indarte, M.; Booher, G.; Kirkpatrick, L. Pleckstrin Homology [PH] domain, structure, mechanism, and contribution to human disease. Biomed. Pharmacother. 2023, 165, 115024. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.O.; Chang, K.-H.; de Pablo, Y.; Ghosh, S.; Mehta, R.; Badve, S.; Shah, K. PHLDA1 is a crucial negative regulator and effector of Aurora A kinase in breast cancer. J. Cell Sci. 2011, 124 Pt 16, 2711–2722. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Wang, Y.; Li, Z.-H.; Fei, L.-R.; Huang, W.-J.; Zheng, Y.-W.; Liu, C.-C.; Yang, M.-Q.; Wang, Z.; Zou, Z.-F.; et al. PHLDA3 promotes lung adenocarcinoma cell proliferation and invasion via activation of the Wnt signaling pathway. Lab. Investig. 2021, 101, 1130–1141. [Google Scholar] [CrossRef] [PubMed]
- Christgen, M.; Noskowicz, M.; Heil, C.; Schipper, E.; Christgen, H.; Geffers, R.; Kreipe, H.; Lehmann, U. IPH-926 lobular breast cancer cells harbor a p53 mutant with temperature-sensitive functional activity and allow for profiling of p53-responsive genes. Lab. Investig. 2012, 92, 1635–1647. [Google Scholar] [CrossRef]
- Saffarzadeh, N.; Ghafouri-Fard, S.; Rezaei, Z.; Aghazadeh, K.; Yazdani, F.; Mohebi, M.; Ahmadi, M.; Farahani, A.S.; Tavakkoly-Bazzaz, J. Expression Analysis of GRHL3 and PHLDA3 in Head and Neck Squamous Cell Carcinoma. Cancer Manag. Res. 2020, 12, 4085–4096. [Google Scholar] [CrossRef]
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Purohit, R. Cancer associated E17K mutation causes rapid conformational drift in AKT1 pleckstrin homology (PH) domain. PLoS ONE 2013, 8, e64364. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, G.; Koul, S.; Ohki, R.; Maurer, M.; Borczuk, A.; Halmos, B. PHLDA2 is a key oncogene-induced negative feedback inhibitor of EGFR/ErbB2 signaling via interference with AKT signaling. Oncotarget 2018, 9, 24914–24926. [Google Scholar] [CrossRef] [PubMed]
- Ohki, R.; Saito, K.; Chen, Y.; Kawase, T.; Hiraoka, N.; Saigawa, R.; Minegishi, M.; Aita, Y.; Yanai, G.; Shimizu, H.; et al. PHLDA3 is a novel tumor suppressor of pancreatic neuroendocrine tumors. Proc. Natl. Acad. Sci. USA 2014, 111, E2404–E2413. [Google Scholar] [CrossRef]
- Lu, H.; Zhou, Q.; He, J.; Jiang, Z.; Peng, C.; Tong, R.; Shi, J. Recent advances in the development of protein-protein interactions modulators: Mechanisms and clinical trials. Signal Transduct. Target. Ther. 2020, 5, 213. [Google Scholar] [CrossRef] [PubMed]
- Korkut, A.; Zaidi, S.; Kanchi, R.S.; Rao, S.; Gough, N.R.; Schultz, A.; Li, X.; Lorenzi, P.L.; Berger, A.C.; Robertson, G.; et al. A Pan-Cancer Analysis Reveals High-Frequency Genetic Alterations in Mediators of Signaling by the TGF-β Superfamily. Cell Syst. 2018, 7, 422–437.e7. [Google Scholar] [CrossRef]
- Kraus, V.B. Biomarkers as drug development tools: Discovery, validation, qualification and use. Nat. Rev. Rheumatol. 2018, 14, 354–362. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iqbal, S.; Karim, M.R.; Mohammad, S.; Mathiyalagan, R.; Morshed, M.N.; Yang, D.-C.; Bae, H.; Rupa, E.J.; Yang, D.U. Multiomics Analysis of the PHLDA Gene Family in Different Cancers and Their Clinical Prognostic Value. Curr. Issues Mol. Biol. 2024, 46, 5488-5510. https://doi.org/10.3390/cimb46060328
Iqbal S, Karim MR, Mohammad S, Mathiyalagan R, Morshed MN, Yang D-C, Bae H, Rupa EJ, Yang DU. Multiomics Analysis of the PHLDA Gene Family in Different Cancers and Their Clinical Prognostic Value. Current Issues in Molecular Biology. 2024; 46(6):5488-5510. https://doi.org/10.3390/cimb46060328
Chicago/Turabian StyleIqbal, Safia, Md. Rezaul Karim, Shahnawaz Mohammad, Ramya Mathiyalagan, Md. Niaj Morshed, Deok-Chun Yang, Hyocheol Bae, Esrat Jahan Rupa, and Dong Uk Yang. 2024. "Multiomics Analysis of the PHLDA Gene Family in Different Cancers and Their Clinical Prognostic Value" Current Issues in Molecular Biology 46, no. 6: 5488-5510. https://doi.org/10.3390/cimb46060328
APA StyleIqbal, S., Karim, M. R., Mohammad, S., Mathiyalagan, R., Morshed, M. N., Yang, D.-C., Bae, H., Rupa, E. J., & Yang, D. U. (2024). Multiomics Analysis of the PHLDA Gene Family in Different Cancers and Their Clinical Prognostic Value. Current Issues in Molecular Biology, 46(6), 5488-5510. https://doi.org/10.3390/cimb46060328








