Selection of Reference Genes for Expression Normalization by RT-qPCR in Dracocephalum moldavica L.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. RNA Isolation and cDNA Preparation
2.3. Reference Genes Selected and Primer Design
2.4. RT-qPCR Analysis
2.5. Gene Expression Stability Analysis
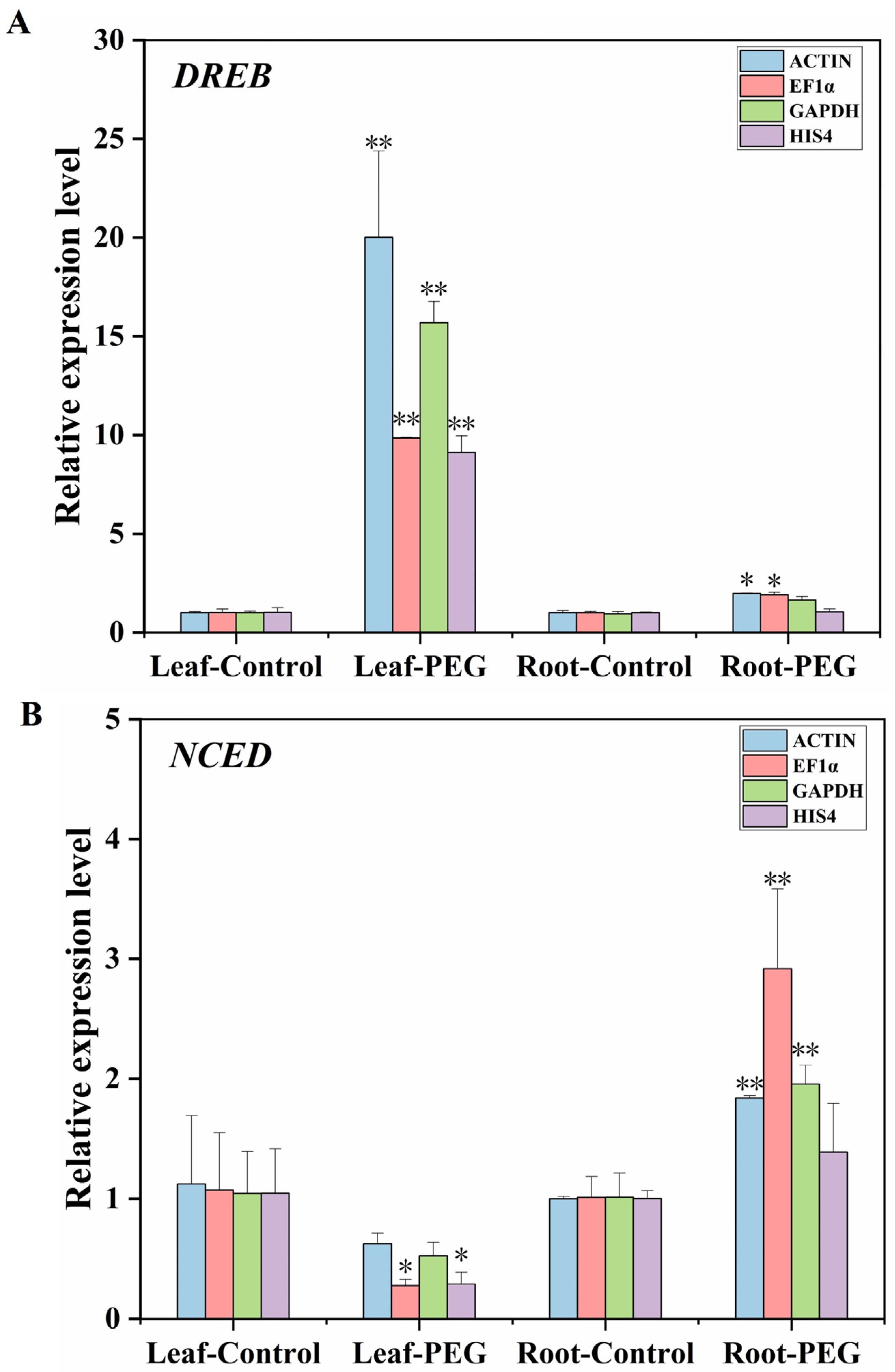
2.6. Validation of Candidate Reference Genes
3. Results
3.1. Expression Profiles of 12 Candidate Reference Genes
3.2. Expression Stability of 12 Candidate Reference Genes
3.2.1. Delta Ct Analysis
3.2.2. GeNorm Analysis
3.2.3. NormFinder Analysis
3.2.4. BestKeeper Analysis
3.3. Comprehensive Stability Analysis of the Reference Genes by RefFinder
3.4. Validation of the Stability of Reference Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhan, M.; Ma, M.; Mo, X.; Zhang, Y.; Li, T.; Yang, Y.; Dong, L. Dracocephalum moldavica L.: An updated comprehensive review of its botany, traditional uses, phytochemistry, pharmacology, and application aspects. Fitoterapia 2024, 172, 105732. [Google Scholar] [CrossRef]
- Khaleghnezhad, V.; Yousefi, A.R.; Tavakoli, A.; Farajmand, B.; Mastinu, A. Concentrations-dependent effect of exogenous abscisic acid on photosynthesis, growth and phenolic content of Dracocephalum moldavica L. under drought stress. Planta 2021, 253, 127. [Google Scholar] [CrossRef]
- Martínez-Vázquez, M.; Estrada-Reyes, R.; Martínez-Laurrabaquio, A.; López-Rubalcava, C.; Heinze, G. Neuropharmacological study of Dracocephalum moldavica L. (Lamiaceae) in mice: Sedative effect and chemical analysis of an aqueous extract. J. Ethnopharmacol. 2012, 141, 908–917. [Google Scholar] [CrossRef]
- Nie, L.; Li, R.; Huang, J.; Wang, L.; Ma, M.; Huang, C.; Wu, T.; Yan, R.; Hu, X. Abietane diterpenoids from Dracocephalum moldavica L. and their anti-inflammatory activities in vitro. Phytochemistry 2021, 184, 112680. [Google Scholar] [CrossRef]
- Jöhrer, K.; Galarza Pérez, M.; Kircher, B.; Çiçek, S.S. Flavones, flavonols, lignans, and caffeic acid derivatives from Dracocephalum moldavica and their In vitro effects on multiple myeloma and acute myeloid leukemia. Int. J. Mol. Sci. 2022, 23, 14219. [Google Scholar] [CrossRef]
- He, C.; Lv, J.; Khan, G.J.; Duan, H.; Wang, W.; Zhai, K.; Zou, G.; Aisa, H.A. Total flavonoid extract from Dracocephalum moldavica L. improves pulmonary fibrosis by reducing inflammation and inhibiting the hedgehog signaling pathway. Phytother. Res. 2023, 37, 2745–2758. [Google Scholar] [CrossRef]
- Maimaiti, A.; Tao, Y.; Minmin, W.; Weiwei, M.; Wenhui, S.; Aikemu, A.; Maimaitiyiming, D. Improvement of total flavonoids from Dracocephalum moldavica L. in rats with chronic mountain sickness through H-NMR metabonomics. Evid-Based Compl. Alt. 2021, 2021, 6695346. [Google Scholar] [CrossRef]
- Singh, C.; Roy-Chowdhuri, S. Quantitative Real-Time PCR: Recent Advances. In Clinical Applications of PCR; Luthra, R., Singh, R.R., Patel, K.P., Eds.; Springer: New York, NY, USA, 2016; pp. 161–176. [Google Scholar]
- Yu, J.; Su, Y.; Sun, J.; Liu, J.; Li, Z.; Zhang, B. Selection of stable reference genes for gene expression analysis in sweet potato (Ipomoea batatas L.). Mol. Cell. Probes 2020, 53, 101610. [Google Scholar] [CrossRef]
- Li, R.; Cui, K.; Xie, Q.; Xie, S.; Chen, X.; Zhuo, L.; Cao, A.; Shen, H.; Jin, X.; Wang, F.; et al. Selection of the reference genes for quantitative gene expression by RT-qPCR in the desert plant Stipagrostis pennata. Sci. Rep. 2021, 11, 21711. [Google Scholar] [CrossRef]
- Yao, J.; Zhu, G.; Liang, D.; He, B.; Wang, Y.; Cai, Y.; Zhang, Q. Reference gene selection for qPCR analysis in Schima superba under abiotic stress. Genes 2022, 13, 1887. [Google Scholar] [CrossRef]
- Tajti, J.; Pál, M.; Janda, T. Validation of reference genes for studying different abiotic stresses in Oat (Avena sativa L.) by RT-qPCR. Plants 2021, 10, 1272. [Google Scholar] [CrossRef]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 10, 71. [Google Scholar] [CrossRef]
- Lin, Y.; Liu, G.; Rao, Y.; Wang, B.; Tian, R.; Tan, Y.; Peng, T. Identification and validation of reference genes for qRT-PCR analyses under different experimental conditions in Allium wallichii. J. Plant Physiol. 2023, 281, 153925. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, R.; Zhou, Z. Identification and validation of reference genes for gene expression analysis in Schima superba. Genes 2021, 12, 732. [Google Scholar] [CrossRef]
- Borges, A.F.; Fonseca, C.; Ferreira, R.B.; Lourenço, A.M.; Monteiro, S. Reference gene validation for quantitative RT-PCR during biotic and abiotic stresses in Vitis vinifera. PLoS ONE 2014, 9, e111399. [Google Scholar] [CrossRef]
- Xu, M.; Zhang, B.; Su, X.; Zhang, S.; Huang, M. Reference gene selection for quantitative real-time polymerase chain reaction in Populus. Anal. Biochem. 2011, 408, 337–339. [Google Scholar] [CrossRef]
- Wang, G.; Wang, L.; Ma, F.; You, Y.; Wang, Y.; Yang, D. Integration of earthworms and arbuscular mycorrhizal fungi into phytoremediation of cadmium-contaminated soil by Solanum nigrum L. J. Hazard. Mater. 2020, 389, 121873. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, L.; Xue, J.; Yang, J.; Hu, H.; Cui, J.; Xu, J. Selection and verification of appropriate reference genes for expression normalization in Cryptomeria fortunei under abiotic stress and hormone treatments. Genes 2021, 12, 791. [Google Scholar] [CrossRef]
- Liu, X.; Guan, H.; Song, M.; Fu, Y.; Han, X.; Lei, M.; Ren, J.; Guo, B.; He, W.; Wei, Y. Reference gene selection for qRT-PCR assays in Stellera chamaejasme subjected to abiotic stresses and hormone treatments based on transcriptome datasets. PeerJ 2018, 6, e4535. [Google Scholar] [CrossRef]
- Rodríguez-Parra, A.; Picazo-Aragonés, J.; Balao, F. Evaluation of reference genes in the polyploid complex Dianthus broteri (Caryophyllaceae) using qPCR. Plants 2022, 11, 518. [Google Scholar] [CrossRef]
- Imai, T.; Ubi, B.E.; Saito, T.; Moriguchi, T. Evaluation of reference genes for accurate normalization of gene expression for real time-quantitative PCR in Pyrus pyrifolia using different tissue samples and seasonal conditions. PLoS ONE 2014, 9, e86492. [Google Scholar] [CrossRef]
- Chi, X.; Hu, R.; Yang, Q.; Zhang, X.; Pan, L.; Chen, N.; Chen, M.; Yang, Z.; Wang, T.; He, Y.; et al. Validation of reference genes for gene expression studies in peanut by quantitative real-time RT-PCR. Mol. Genet. Genomics 2012, 287, 167–176. [Google Scholar] [CrossRef]
- Wang, B.; Duan, H.; Chong, P.; Su, S.; Shan, L.; Yi, D.; Wang, L.; Li, Y. Systematic selection and validation of suitable reference genes for quantitative real-time PCR normalization studies of gene expression in Nitraria tangutorum. Sci. Rep. 2020, 10, 15891. [Google Scholar] [CrossRef]
- Saddhe, A.A.; Malvankar, M.R.; Kumar, K. Selection of reference genes for quantitative real-time PCR analysis in halophytic plant Rhizophora apiculata. Peerj 2018, 6, e5226. [Google Scholar] [CrossRef]
- Bisht, A.; Bhalla, S.; Kumar, A.; Kaur, J.; Garg, N. Gene expression analysis for selection and validation of suitable housekeeping gene(s) in cadmium exposed pigeonpea plants inoculated with arbuscular mycorrhizae. Plant Physiol. Biochem. 2021, 162, 592–602. [Google Scholar] [CrossRef]
- Liu, H.; Liu, J.; Chen, P.; Zhang, X.; Wang, K.; Lu, J.; Li, Y. Selection and validation of optimal RT-qPCR reference genes for the normalization of gene expression under different experimental conditions in Lindera megaphylla. Plants 2023, 12, 2185. [Google Scholar] [CrossRef]
- Gao, Z.; Wei, J.; Yang, Y.; Zhang, Z.; Zhao, W. Selection and validation of reference genes for studying stress-related agarwood formation of Aquilaria sinensis. Plant Cell Rep. 2012, 31, 1759–1768. [Google Scholar] [CrossRef]
- Janská, A.; Hodek, J.; Svoboda, P.; Zámečník, J.; Prášil, I.T.; Vlasáková, E.; Milella, L.; Ovesná, J. The choice of reference gene set for assessing gene expression in barley (Hordeum vulgare L.) under low temperature and drought stress. Mol. Genet. Genom. 2013, 288, 639–649. [Google Scholar] [CrossRef]
- Cao, J.; Wang, L.; Lan, H. Validation of reference genes for quantitative RT-PCR normalization in Suaeda aralocaspica, an annual halophyte with heteromorphism and C4 pathway without Kranz anatomy. PeerJ 2016, 4, e1697. [Google Scholar] [CrossRef]
- Park, S.-C.; Kim, Y.-H.; Ji, C.Y.; Park, S.; Jeong, J.c.; Lee, H.-S.; Kwak, S.-S. Stable internal reference genes for the normalization of real-time PCR in different sweet potato cultivars subjected to abiotic stress conditions. PLoS ONE 2012, 7, e51502. [Google Scholar] [CrossRef]
- Lv, Y.; Li, Y.; Liu, X.; Xu, K. Identification of ginger (Zingiber officinale Roscoe) reference genes for gene expression analysis. Front. Genet. 2020, 11, 586098. [Google Scholar] [CrossRef]
- Chandna, R.; Augustine, R.; Bisht, N.C. Evaluation of candidate reference genes for gene expression normalization in Brassica juncea using real time quantitative RT-PCR. PLoS ONE 2012, 7, e36918. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.0031. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genom. 2023, 23, 125. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Mei, F.; Chen, B.; Du, L.; Li, S.; Zhu, D.; Chen, N.; Zhang, Y.; Li, F.; Wang, Z.; Cheng, X.; et al. A gain-of-function allele of a DREB transcription factor gene ameliorates drought tolerance in wheat. The Plant Cell 2022, 34, 4472–4494. [Google Scholar] [CrossRef]
- Lee, S.U.; Mun, B.G.; Bae, E.K.; Kim, J.Y.; Kim, H.H.; Shahid, M.; Choi, Y.I.; Hussain, A.; Yun, B.W. Drought stress-mediated transcriptome profile reveals NCED as a key player modulating drought tolerance in Populus davidiana. Front. Plant Sci. 2021, 12, 755539. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhang, Y.; Xue, J.; Zhu, L.; Hu, H.; Yang, J.; Cui, J.; Xu, J. Selection and optimization of reference genes for MicroRNA expression normalization by qRT-PCR in Chinese cedar (Cryptomeria fortunei) under multiple stresses. Int. J. Mol. Sci. 2021, 22, 7246. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, C.; Yu, X.; Yang, J.; Feng, C. Screening of reference genes for differentially expressed genes in Pyrus betulaefolia plant under salt stress by qRT-PCR. Acta Hortic. Sin. 2022, 49, 1557–1570. [Google Scholar]
- Zhang, S.; Zeng, Y.; Yi, X.; Zhang, Y. Selection of suitable reference genes for quantitative RT-PCR normalization in the halophyte Halostachys caspica under salt and drought stress. Sci. Rep. 2016, 6, 30363. [Google Scholar] [CrossRef]
- Fu, J.; Wang, Y.; Huang, H.; Zhang, C.; Dai, S. Reference gene selection for RT-qPCR analysis of Chrysanthemum lavandulifolium during its flowering stages. Mol. Breed. 2013, 31, 205–215. [Google Scholar] [CrossRef]
- Gutierrez, L.; Mauriat, M.; Guénin, S.; Pelloux, J.; Lefebvre, J.-F.; Louvet, R.; Rusterucci, C.; Moritz, T.; Guerineau, F.; Bellini, C.; et al. The lack of a systematic validation of reference genes: A serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J. 2008, 6, 609–618. [Google Scholar] [CrossRef]
- Walling, J.G.; Zalapa, L.A.; Vinje, M.A. Evaluation and selection of internal reference genes from two- and six-row U.S. malting barley varieties throughout micromalting for use in RT-qPCR. PLoS ONE 2018, 13, e0196966. [Google Scholar] [CrossRef]
- Chakraborty, S.; Dutta, S.; Biswas, P.; Das, M. Identification of candidate reference genes in tropical bamboos stable across species, tissues, and developmental stages. Biol. Plant. 2019, 63, 253–261. [Google Scholar] [CrossRef]
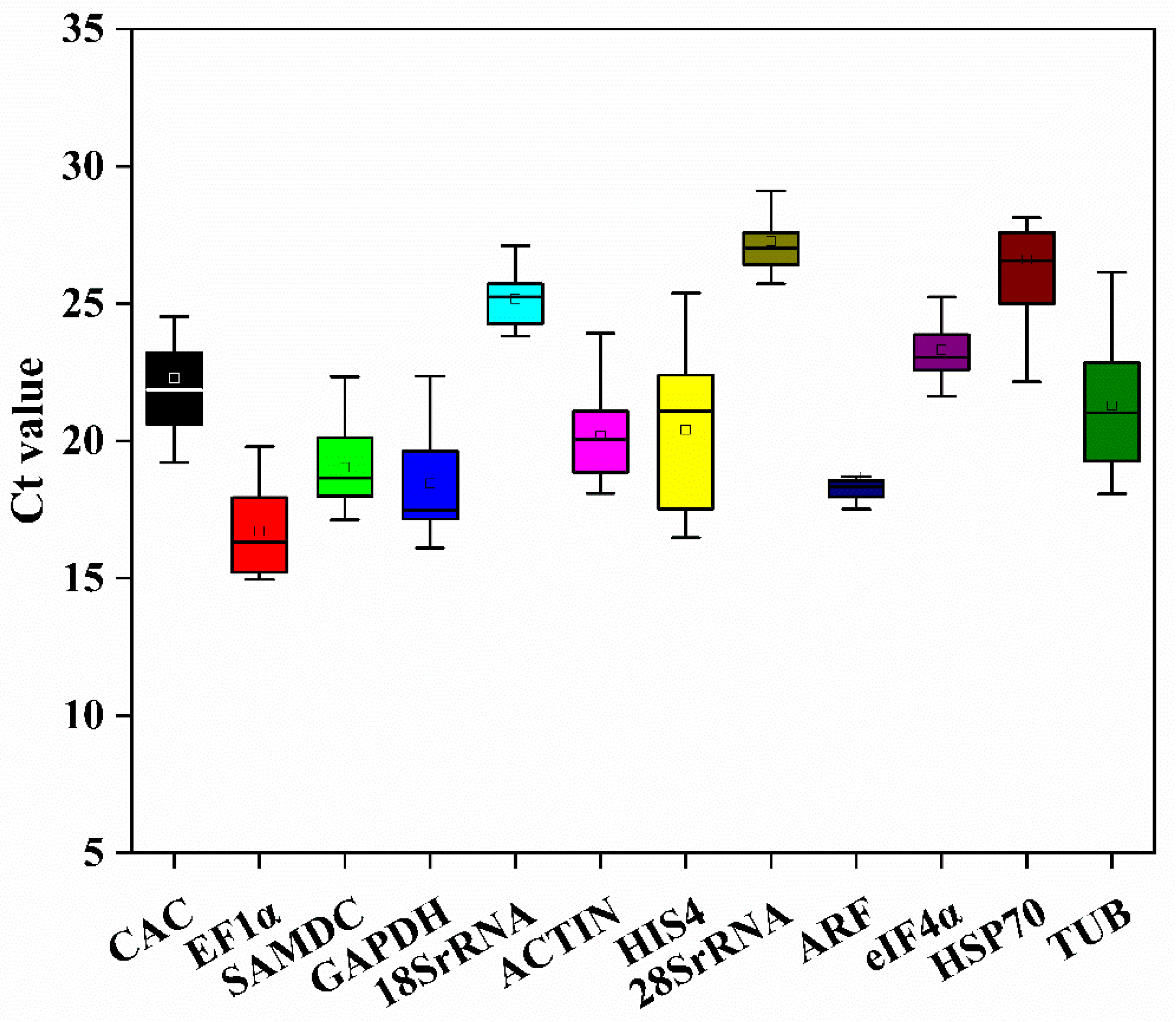
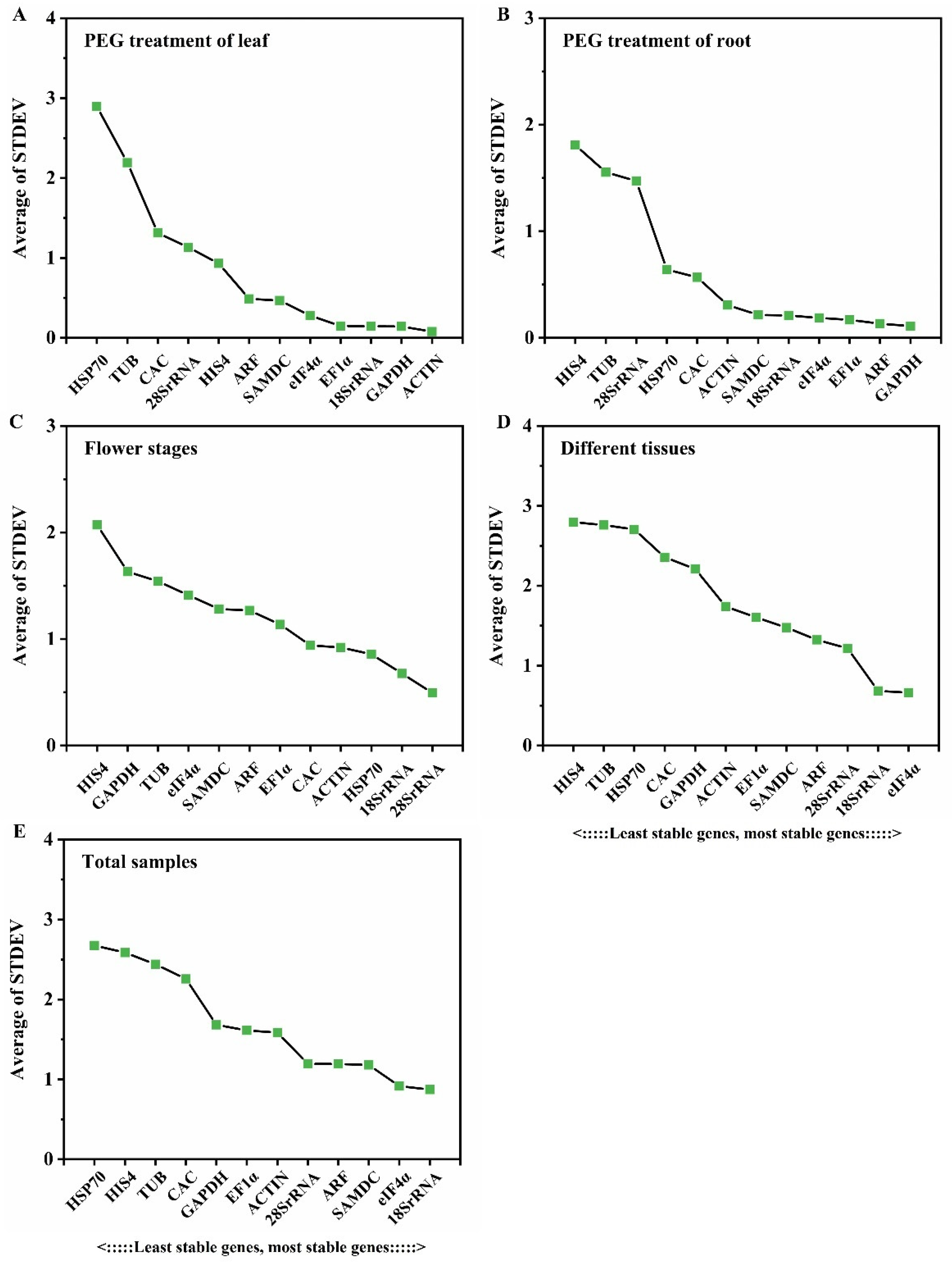
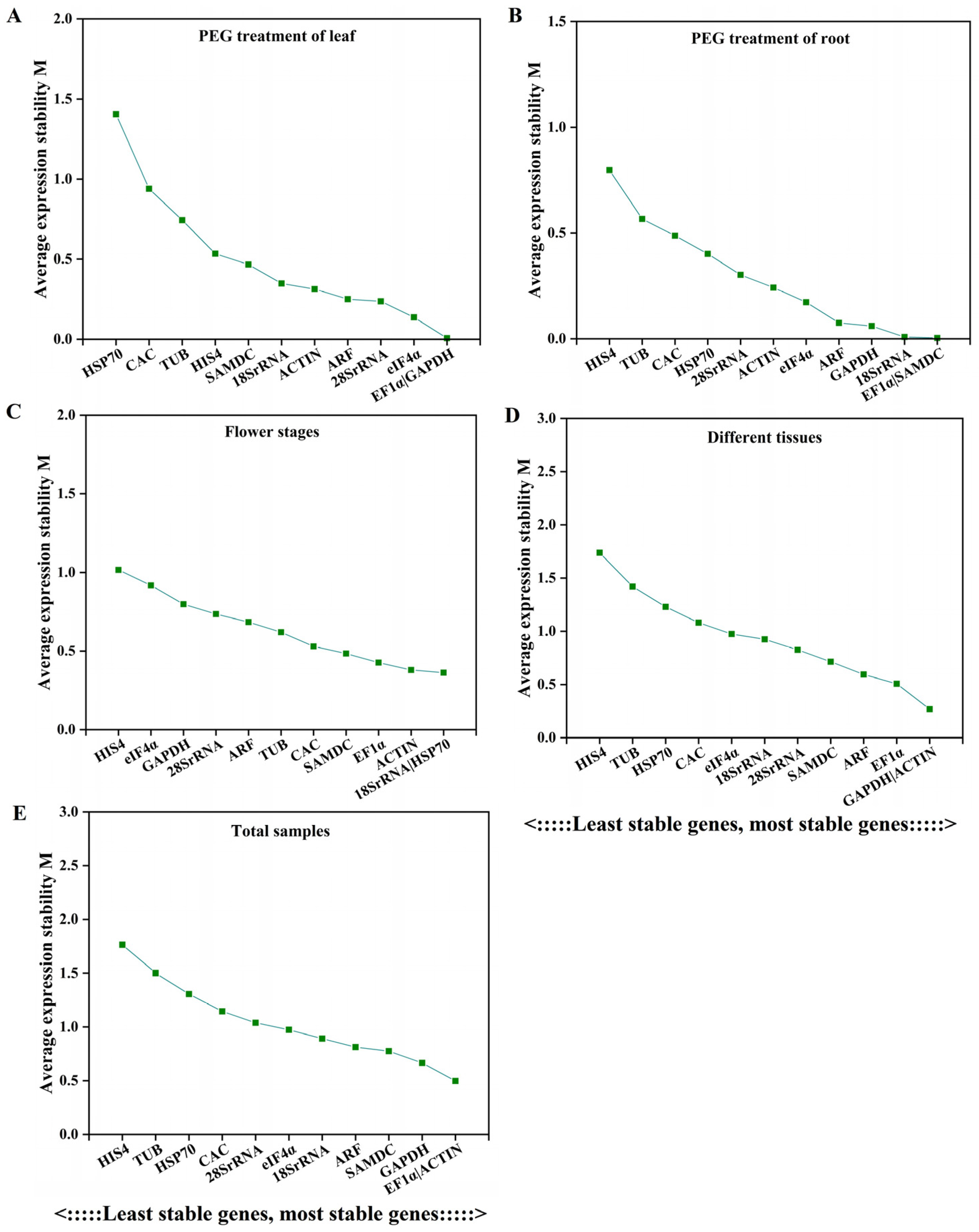
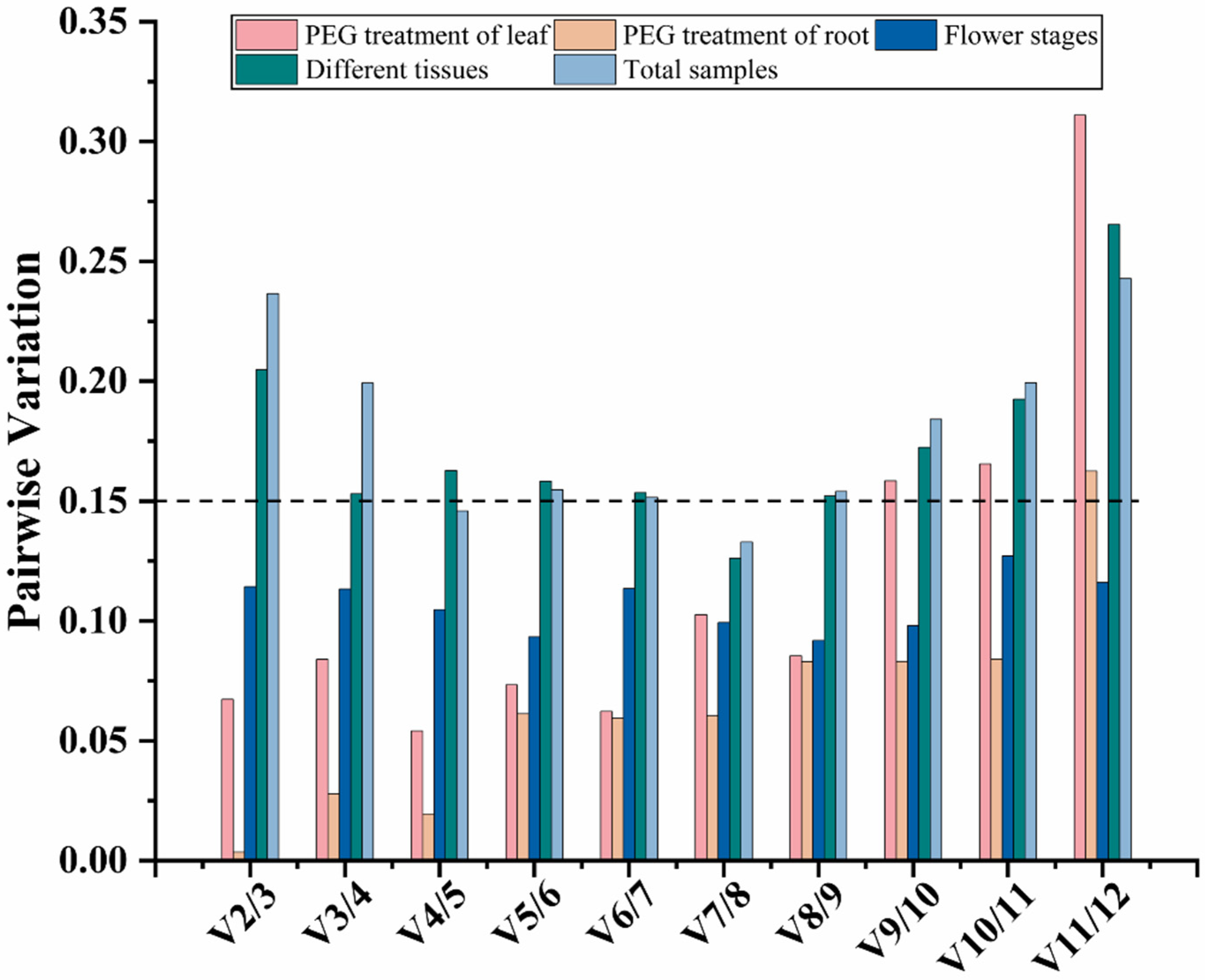

| Gene Name | Gene ID | Expression (FPKM) | Gene Annotation | |||
|---|---|---|---|---|---|---|
| Leaf | Root | |||||
| Control | PEG | Control | PEG | |||
| 18S rRNA | c100609 | 10.37 | 9.46 | 10.77 | 8.07 | 18S ribosomal RNA |
| ACTIN | c110032 | 669.04 | 393.55 | 519.34 | 512.14 | Actin |
| HIS4 | c103664 | 118.31 | 172.25 | 181.46 | 153.77 | Histone 4 |
| HSP70 | c112390 | 267.91 | 77.51 | 44.82 | 39.42 | Heat shock protein 70 |
| 28S rRNA | c117403 | 4.97 | 3.89 | 5.58 | 4.14 | 28S ribosomal RNA |
| ARF | c111206 | 613.04 | 488.67 | 608.69 | 599.65 | ADP-ribosylation factor |
| CAC | c100772 | 37.13 | 73.32 | 61.93 | 64.44 | Clathrin adaptor complex |
| EF1α | c118069 | 2997.05 | 1986.06 | 2565.10 | 2184.92 | Elongation factor 1-alpha |
| SAMDC | c102943 | 457.44 | 236.29 | 374.79 | 324.16 | S-adenosylmethionine decarboxylase |
| GAPDH | c118460 | 449.81 | 274.44 | 485.34 | 452.65 | Glyceraldehyde 3-phosphate dehydrogenase |
| eIF4α | c118665 | 21.25 | 7.11 | 15.18 | 13.63 | ATP-dependent RNA helicase eukaryotic initiation factor 4-α |
| TUB | c117718 | 85.25 | 133.54 | 55 | 63.32 | tubulin alpha-3 |
| Ranking | PEG6000 Treatment of Leaf | PEG6000 Treatment of Root | Flower Stages | Different Tissues | Total Samples |
|---|---|---|---|---|---|
| 1 | SAMDC 0.018 | GAPDH 0.005 | HSP70 0.092 | ARF 0.246 | ARF 0.256 |
| 2 | HIS4 0.018 | ARF 0.005 | CAC 0.139 | 28S rRNA 0.263 | SAMDC 0.397 |
| 3 | 18S rRNA 0.024 | eIF4α 0.038 | SAMDC 0.273 | ACTIN 0.399 | ACTIN 0.478 |
| 4 | ACTIN 0.024 | ACTIN 0.043 | ARF 0.373 | SAMDC 0.558 | GAPDH 0.505 |
| 5 | GAPDH 0.126 | EF1α 0.163 | ACTIN 0.388 | GAPDH 0.571 | 28S rRNA 0.628 |
| 6 | EF1α 0.138 | SAMDC 0.167 | TUB 0.404 | EF1α 0.633 | EF1α 0.640 |
| 7 | eIF4α 0.374 | 18S rRNA 0.176 | 18S rRNA 0.433 | 18S rRNA 0.830 | 18S rRNA 0.695 |
| 8 | 28S rRNA 0.550 | 28S rRNA 0.208 | EF1α 0.453 | CAC 0.840 | eIF4α 0.738 |
| 9 | ARF 0.571 | HSP70 0.478 | 28S rRNA 0.591 | eIF4α 0.869 | CAC 0.931 |
| 10 | TUB 0.926 | CAC 0.607 | GAPDH 0.770 | HSP70 1.136 | HSP70 1.294 |
| 11 | CAC 1.465 | TUB 0.746 | eIF4α 0.905 | TUB 1.297 | TUB 1.371 |
| 12 | HSP70 2.588 | HIS4 1.348 | HIS4 0.954 | HIS4 2.196 | HIS4 1.957 |
| Ranking | Samples | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PEG Treatment of Leaf | PEG Treatment of Root | Different Flower Stages | Different Tissues | Total Samples | |||||||||||
| Gene Name | SD | CV [%] | Gene Name | SD | CV [%] | Gene Name | SD | CV [%] | Gene Name | SD | CV [%] | Gene Name | SD | CV [%] | |
| 1 | ACTIN | 0.06 | 0.34 | EF1α | 0.06 | 0.41 | 28S rRNA | 0.38 | 1.42 | 18S rRNA | 0.59 | 2.35 | 18S rRNA | 0.76 | 3.01 |
| 2 | GAPDH | 0.11 | 0.61 | ACTIN | 0.14 | 0.76 | 18S rRNA | 0.61 | 2.33 | eIF4α | 0.60 | 2.58 | eIF4α | 0.77 | 3.32 |
| 3 | 18S rRNA | 0.11 | 0.47 | 18S rRNA | 0.14 | 0.59 | HSP70 | 0.73 | 2.69 | ARF | 0.93 | 4.96 | ARF | 0.87 | 4.64 |
| 4 | EF1α | 0.12 | 0.78 | CAC | 0.15 | 0.78 | ACTIN | 0.77 | 3.62 | 28S rRNA | 1.02 | 3.71 | 28S rRNA | 0.93 | 3.41 |
| 5 | eIF4α | 0.25 | 1.09 | GAPDH | 0.18 | 1.04 | CAC | 0.81 | 3.46 | SAMDC | 1.27 | 6.61 | SAMDC | 1.09 | 5.71 |
| 6 | ARF | 0.41 | 2.25 | ARF | 0.24 | 1.31 | EF1α | 0.99 | 1.03 | ACTIN | 1.27 | 6.29 | ACTIN | 1.26 | 6.25 |
| 7 | SAMDC | 0.43 | 2.33 | eIF4α | 0.27 | 1.17 | SAMDC | 1.03 | 5.37 | EF1α | 1.31 | 7.84 | EF1α | 1.37 | 8.18 |
| 8 | 28S rRNA | 0.93 | 3.31 | SAMDC | 0.30 | 1.61 | ARF | 1.11 | 5.83 | CAC | 1.70 | 7.51 | GAPDH | 1.52 | 8.23 |
| 9 | CAC | 1.20 | 5.82 | HSP70 | 0.78 | 3.40 | eIF4α | 1.14 | 4.81 | GAPDH | 1.77 | 9.39 | CAC | 1.64 | 7.38 |
| 10 | HIS4 | 1.79 | 7.45 | 28S rRNA | 1.40 | 5.11 | TUB | 1.32 | 6.59 | HSP70 | 1.84 | 6.67 | HSP70 | 1.91 | 7.16 |
| 11 | TUB | 1.82 | 8.64 | TUB | 2.38 | 10.84 | GAPDH | 1.43 | 7.74 | TUB | 2.30 | 10.58 | TUB | 2.13 | 10.01 |
| 12 | HSP70 | 2.63 | 10.58 | HIS4 | 2.47 | 10.54 | HIS4 | 1.82 | 9.72 | HIS4 | 2.30 | 11.17 | HIS4 | 2.45 | 11.87 |
| Ranking | PEG Treatment of Leaf | PEG Treatment of Root | Different Flower Stages | Different Tissues | Total Samples |
|---|---|---|---|---|---|
| 1 | ACTIN | EF1α | HSP70 | ARF | ARF |
| 2 | EF1α | eIF4α | SAMDC | EF1α | ACTIN |
| 3 | 18S rRNA | GAPDH | ACTIN | ACTIN | EF1α |
| 4 | GAPDH | ARF | EF1α | 28S rRNA | 18S rRNA |
| 5 | eIF4α | ACTIN | CAC | 18S rRNA | SAMDC |
| 6 | SAMDC | SAMDC | ARF | eIF4α | eIF4α |
| 7 | ARF | 18S rRNA | 28S rRNA | CAC | 28S rRNA |
| 8 | HIS4 | CAC | 18S rRNA | SAMDC | GAPDH |
| 9 | 28S rRNA | HSP70 | TUB | GAPDH | CAC |
| 10 | CAC | 28S rRNA | eIF4α | HSP70 | HSP70 |
| 11 | TUB | HIS4 | GAPDH | TUB | TUB |
| 12 | HSP70 | TUB | HIS4 | HIS4 | HIS4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Ge, X.; Bai, G.; Chen, C. Selection of Reference Genes for Expression Normalization by RT-qPCR in Dracocephalum moldavica L. Curr. Issues Mol. Biol. 2024, 46, 6284-6299. https://doi.org/10.3390/cimb46060375
Li S, Ge X, Bai G, Chen C. Selection of Reference Genes for Expression Normalization by RT-qPCR in Dracocephalum moldavica L. Current Issues in Molecular Biology. 2024; 46(6):6284-6299. https://doi.org/10.3390/cimb46060375
Chicago/Turabian StyleLi, Shasha, Xiaomin Ge, Guoqing Bai, and Chen Chen. 2024. "Selection of Reference Genes for Expression Normalization by RT-qPCR in Dracocephalum moldavica L." Current Issues in Molecular Biology 46, no. 6: 6284-6299. https://doi.org/10.3390/cimb46060375
APA StyleLi, S., Ge, X., Bai, G., & Chen, C. (2024). Selection of Reference Genes for Expression Normalization by RT-qPCR in Dracocephalum moldavica L. Current Issues in Molecular Biology, 46(6), 6284-6299. https://doi.org/10.3390/cimb46060375







