Abstract
The bile acid sodium symporter (BASS) family plays an important role in transporting substances and coordinating plants’ salt tolerance. However, the function of BASS in Brassica rapa has not yet been elucidated. In this study, eight BrBASS genes distributed on five chromosomes were identified that belonged to four subfamilies. Expression profile analysis showed that BrBASS7 was highly expressed in roots, whereas BrBASS4 was highly expressed in flowers. The promoter element analysis also identified several typical homeopathic elements involved in abiotic stress tolerance and stress-related hormonal responses. Notably, under salt stress, the expression of BrBASS2 was significantly upregulated; under osmotic stress, that of BrBASS4 increased and then decreased; and under cold stress, that of BrBASS7 generally declined. The protein–protein interaction analysis revealed that the BrBASS2 homologous gene AtBASS2 interacted with Nhd1 (N-mediated heading date-1) to alleviate salt stress in plants, while the BrBASS4 homologous gene AtBASS3 interacted with BLOS1 (biogenesis of lysosome-related organelles complex 1 subunit 1) via co-regulation with SNX1 (sorting nexin 1) to mitigate an unfavorable growing environment for roots. Further, Bra-miR396 (Bra-microRNA396) targeting BrBASS4 and BrBASS7 played a role in the plant response to osmotic and cold stress conditions, respectively. This research demonstrates that BrBASS2, BrBASS4, and BrBASS7 harbor great potential for regulating abiotic stresses. The findings will help advance the study of the functions of the BrBASS gene family.
1. Introduction
Bile acid sodium symporter (BASS) proteins belong to the sodium-dependent transporter family and are classified as a solute carrier family 10 (SLC10). This family mainly facilitates the transport of substances such as bile acids and steroid hormones within cells []. The BASS gene family is found in many diverse organisms, such as fungi, bacteria, animals, and plants. In mammals, bile acid transporters in the liver and intestine play a crucial role in driving the enterohepatic circulation of bile acids [], which promote the intestinal absorption of lipids and fat-soluble vitamins A, D, E, and K in humans []. Nevertheless, BASS family members from different species retain highly conserved Na-binding sites [,] to effectively participate in Na+ transport within organisms. Ranging in length from 348 to 477 amino acids, BASS proteins consist of 10 transmembrane fragments (TM1–10) and two conserved Na+ binding sites, Na1 and Na2 [,], and have a unique feature: two discontinuous helices that intersect at the center. Specific Na+ binding sites are typically located on these four transmembrane domains: TM3, TM4, TM5, and TM9 [,]. Specific binding of two sodium ions to the Na+ binding site is a prerequisite for structural changes in BASS and substrate binding. This arrangement enables the involvement of BASS gene family proteins in moving sodium ions during substrate transport to help balance ionic contents inside and outside the cell, thereby maintaining normal cellular function [].
In plants, the BASS genes have been studied in several species, namely Triticum aestivum [], Gossypium hirsutum [], Arabidopsis thaliana [], Oryza sativa [], and species of the genera Flaveria and Cleome [], showing them to figure prominently in plant responses to abiotic stressors (especially salt stress) and plant development. For example, overexpression of TaBASS2 in A. thaliana and O. sativa can improve their respective salt tolerance; TaBASS2 enhances the salt tolerance of T. aestivum by inhibiting the expression of ABI4, a node that links ABA (abscisic acid) signaling and plastid retrograde signaling pathways to the salinity response []. Moreover, TaBASS2 overexpression in the Atbass2 mutant also restored the sensitivity of A. thaliana to mevastatin []. In cotton (G. hirsutum), GhBASS1 and GhBASS3 positively regulate that plant’s salt content, while GhBASS2, GhBASS4, and GhBASS5 negatively regulate its salt tolerance []; in this latter respect, GhBASS5 operates by increasing Na+ transport and accumulation at the cellular and tissue levels [,]. Under salt stress conditions, the overexpression of GhBASS5 in A. thaliana can significantly increase the Na+ content of that plant while strongly disrupting plastid function and enhancing salt sensitivity in transgenic A. thaliana. Under salt stress, GhBASS5 overexpression lines significantly express salt-responsive transporter genes regulating K+/Na+ homeostasis [].
In A. thaliana, AtBASS1 is known to transport pantolytic acid in pantothenic acid synthesis, as demonstrated by expressing AtBASS1 in yeast and constructing Atbass1 mutants []. In addition, AtBASS5 in chloroplasts acts as a carrier for 2-ketoacids and is involved in aliphatic glucosinolate (GLS) biosynthesis [,], while the AtBASS6 protein transports glycolic acid in the photorespiratory metabolism of A. thaliana []. In rice (O. sativa), OsSBF1 shows pronounced homology with bile acid transporters in both mammals and bacteria and is possibly involved in the transport of sulfonated brassin-steroids. In studies of Flaviria and Cleome, BASS2 from A. thaliana was highly expressed in both C4 plants and localized to their chloroplast envelope, providing molecular-level evidence for sodium-coupled metabolite transporters in the plastid envelope [].
Brassica plants, including B. Juncea, B. Oleracea and B. napus, and B. rapa, are globally important crops known for their diversity. Chinese cabbage, a B. rapa subspecies, is particularly valued for its economic and nutritional value to humans. However, it is subject to various biotic (e.g., fungi, bacteria, viruses, insects) and abiotic stresses (e.g., low temperature, salt, drought) in its natural environment. This leads to severe growth stagnation, yield loss, and quality reduction of Chinese cabbage crops [,,]. Current research on the BASS gene family has mainly focused on substrate binding and transport mechanisms. However, the function and regulation of BASS genes in Chinese cabbage have not been fully investigated. Accordingly, an analysis of sequence characteristics, gene expression patterns, regulatory mechanisms, and epigenetics of BrBASS genes was carried out in this study. These results are important for future research on abiotic stressors of the BrBASS gene and for the development of stress-tolerant cabbage varieties.
2. Materials and Methods
2.1. Identification and Physicochemical Characterization of BrBASSs
B. rapa and A. thaliana whole genome sequences, gff3 genome annotation data, and BASS amino acid sequences were downloaded from EnsemblPlant (http://plants.ensembl.org/, accessed on 18 February 2024). The BrBASS genes were screened from the B. rapa genome, using the BLASTP program with amino acid sequences of AtBASS serving as input, to predict the BrBASS genes. All B. rapa BASS proteins were predicted by using Expasy technologies (http://www.expasy.org/, accessed on 18 February 2024) and their subcellular distribution was analyzed using the WOLF PSORT website (https://www.genscript.com/wolf-psort.html, accessed on 15 March 2024) [,].
The amino acid sequences of BrBASSs and AtBASSs were aligned using the MUSCLE algorithm in MEGA-X software (v10.0.1) []. A maximum likelihood phylogenetic tree was then constructed using MEGA X (with 1000 bootstrap replicates). The resulting phylogenetic tree was modified for a clearer presentation, using iTOL (https://itol.embl.de/, accessed on 17 March 2024) []. Synteny was analyzed with the ‘Advanced Circles and Table Row Extract’ or ‘Filter’ programs of TBtools software (v1.120) [].
2.2. Gene Structure and Conserved Domain Analysis
Each BrBASS was visualized using the Visualize Gene Structure program of TBtools (v1.120), and their conserved domains were analyzed using NCBI’s CD-Search tools (v3.18) (http://www.ncbi.nlm.nih.gov/cdd, accessed on 19 February 2024). The conserved motifs of the identified BrBASSs were analyzed with the online tool MEME (v5.5.1) (http://meme-suite.org/tools/meme, accessed on 19 February 2024), for which the predicted number of motifs was set to 10.
2.3. Analysis of Promoter Cis-Elements of BrBASSs
The upstream 2000-bp sequence of each BrBASS was extracted with TBtools (v1.120), and the eight genes were then analyzed via the PlantCARE online website (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 19 February 2024) []. The findings from this website’s analysis were summarized in MS Excel 2019 to present the results.
2.4. Tissue-Specific Expression of BrBASS Genes and Analysis of MicroRNAs Targeting the Genes of BrBASSs
Transcriptome sequences from various B. rapa tissues were obtained from the NCBI GEO (record number GSE43245) []. These sequences were normalized using Transcripts Per Million (TPM). The HeatMap plug-in of TBtools (v1.120) was utilized to create expression maps, and we collected the CDS sequences of BrBASS genes. MiRNAs targeting BrBASS genes were identified using the psRNATarget online database (accessed on 19 February 2024; https://www.zhaolab.org/psRNATarget/). These data were then visualized in MS Excel 2019 tables [].
2.5. Plant Material and Stress Treatments
Stress treatments were applied to B. rapa plants that have stable self-incompatibility. These plants were grown in an incubator using Murashige and Skoog (MS) Modified Medium (Qingdao Hope Bio-technology, Qingdao, China), which included vitamins, sucrose, and agar. When their true leaves had expanded, the seedlings were transplanted into substrate-filled seedling pots and cultured in a plant incubator (16-h light/8-h dark photoperiod at 25 °C; light intensity of 2000 l×; relative humidity of 67%). Seedlings having six leaves and a similar growth status were then chosen to receive the stress treatments. In a hydroponic system, these seedlings were inserted into a mixture of 150 mM NaCl to simulate salt stress or 15% PEG 6000 (0.15 g/mL) to simulate drought conditions. Plants were exposed to 4 °C for the cold stress treatment. Unstressed seedlings from the same batch and of similar size served as the control (CK). Each stress treatment was applied for durations of 4, 6, and 12 h. Each treatment included three biological replicates; all samples were cryopreserved at –80 °C for subsequent RNA extractions.
2.6. Total RNA Extraction and RT-qPCR
The FastPure® Cell/Tissue Total RNA Isolation Kit V2 (Vazyme Biotech Co., Ltd., Nanjing, China) was used to isolate total RNA from each replicate plant. TransScript Uni all-in-one first-strand cDNA synthesis SuperMix TransGen (AU341-02, Beijing, China) was used for the qPCR; for the RT-qPCR analysis, samples were subjected to reverse transcription. The RT-qPCR primer sequences were designed using the qPrimerDB-qPCR primer database (https://biodb.swu.edu.cn/qprimerdb/, accessed on 24 June 2024). Each RT-qPCR run was conducted on a CFXopus 96 qPCR machine (California, USA) by using the ChamQ SYBR qPCR premix (Nanjing, China) with BrGAPDH serving as the reference gene and applying the 2−ΔΔCt method. Table S5 lists the specific primer sequences used in the experiments that were conducted in technical triplicates.
2.7. Statistical Analysis
The determination of significant differences between means was made using two-tailed t-tests, implemented in IBM SPSS 25 (significance level, α = 0.05).
2.8. BASS Protein Secondary Structure and Tertiary Structure Analysis
For the protein secondary structure analysis, we utilized the predict protein tool (https://predictprotein.org/, accessed on 19 February 2024). The results were analyzed and plotted using MS Excel 2019. To predict the tertiary structure of each BrBASS protein, we used SWISS-MODEL (v1.0) (https://swissmodel.expasy.org/, accessed on 19 February 2024).
2.9. Predicted Protein–Protein Interactions of BrBASSs and Phosphorylation Site Analysis
For the protein interaction network analysis, we employed the protein interaction network prediction website (https://cn.string-db.org/, accessed on 17 March 2024) to obtain B. rapa protein network interaction maps (minimum interaction score required = 0.150; other parameters used at their default settings). To examine the BrBASS protein interaction network, the STRING online server (https://string-db.org/cgi, accessed on 17 March 2024) was used under its default parameters []. The interaction network was constructed in Cytoscape v3.9.1 []. BrBASS gene phosphorylation sites were analyzed using the online site Netphos 2.0, with their visual representations prepared in MS Excel 2019.
3. Results
3.1. Identification and Physicochemical Characterization of BrBASS Family Genes
To analyze the basic features of the BrBASS gene family, we screened the cabbage genome for six amino acid sequences of AtBASSs through HMMER and Blast. Eight members of the BrBASS gene family were finally obtained and renamed BrBASS1–BrBASS8 (Table 1). Relevant bioinformatics analyses showed that the amino acid lengths of BrBASSs ranged from 212 aa (BrBASS8) to 440 aa (BrBASS4), with a molecular weight size of 22,989.89 Da (BrBASS8) to 46,419.66 Da (BrBASS4). Their isoelectric points spanned 8.95 (BrBASS5) to 9.87 (BrBASS3), indicating that all eight members of the BrBASS family were basic proteins. Their subcellular localization showed them distributed in various cytoplasmic compartments within the cell, including the chloroplast, plastid, endoplasmic, reticulum, mitochondrion, cytosol, vascular, cytosol, etc. Still, all eight BrBASS genes’ proteins were located in the chloroplast, suggesting they may regulate vital cellular activity in a specific manner.

Table 1.
Physicochemical properties of the proteins encoded by BrBASS genes.
3.2. Phylogenetic Relationships and Synteny Analysis
The phylogenetic analysis of BrBASSs, along with the BASSs in A. thaliana, was carried out using the maximum likelihood method (Figure 1A). This analysis revealed that the 14 BASSs clustered into four major groups (I, II, III, IV) based on their evolutionary relationships. According to their evolutionary relationships, Group I was the largest, comprising seven members: four B. rapa genes and three A. thaliana genes. Group II consisted of just two members: one from B. rapa and one from A. thaliana. Group III also had two BASS members: one each from B. rapa and A. thaliana. Group IV contained three members: two B. rapa genes and one A. thaliana gene.
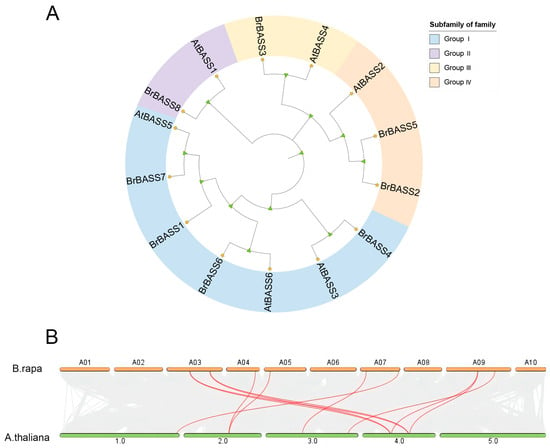
Figure 1.
(A) Evolutionary tree of the BASS family in B. rapa and A. thaliana. The triangles represent leaf nodes, and the five-pointed stars represent branch nodes. (B) Collinearity analysis between B. rapa and A. thaliana. Orange boxes represent the 10 chromosomes of B. rapa, and green boxes represent the five chromosomes of A. thaliana. Red lines indicate homologous gene pairs.
3.3. Gene Structure and Conserved Domain Analysis
We used NCBI-CDD to predict the conserved domains of BrBASSs (Figure 2A). This showed that the BrBASSs encoded four protein domains, namely YfeH, SBF, the YfeH superfamily, and the SBF superfamily. Notably, BrBASS3 and BrBASS8, respectively, contained the SBF and YfeH superfamilies, suggesting that members of these superfamilies may have special functions. Only BrBASS3 and BrBASS6 contained the SBF domain, and the other six family members all harbored the YfeH domain.
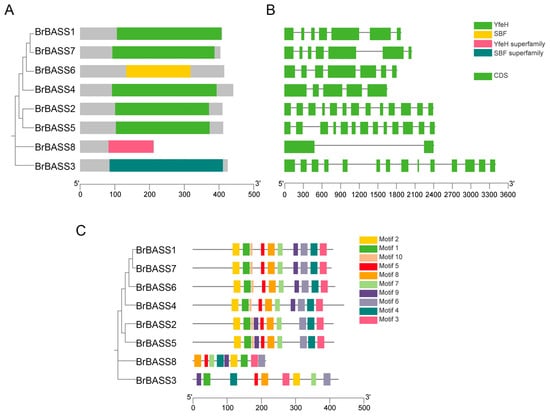
Figure 2.
Gene structure and conserved domain analysis of BrBASSs. (A) Analysis of conserved domains of BrBASS genes. (B) Analysis of their gene structure. The introns and exons are shown as black lines and green boxes, respectively. (C) Analysis of the conserved motifs of BrBASS genes.
Genetic structure analysis revealed 2 to 14 exons in the BrBASS genes (Figure 2B). The protein-conserved motifs of BrBASS genes were analyzed via the MEME website (Figure 2C), with a total of 10 motifs identified. These results showed that most of the motifs had a similar composition, and all contained conserved Motif 1, likely the core motif of the BrBASS family. Except for BrBASS3 and BrBASS8, which had nine motifs, the other genes had 10 motifs. In the same clade, conserved motifs are similar in number, type, and arrangement; hence, functional differences among the BrBASS genes could be due to a different distribution of conserved motifs, so we divided them into four different groups. These findings were consistent with our gene structure analysis and confirmed the subfamily division of the BrBASS genes.
3.4. Analysis of Promoter Cis-Elements of BrBASSs
To better understand their gene expression patterns, we used PlantCARE to analyze the 2000-bp sequence upstream of each BrBASS gene (Figure 3). Concerning those related to growth and development, all eight BrBASS genes contained light-responsive elements but were devoid of endosperm expression and wound-response elements. Among phytohormone-responsive elements, all the genes contained abscisic acid elements; 50% of them contained methyl jasmonate (MeJA)-responsive elements; and 37.5% of them contained gibberellin-responsive and salicylic acid-responsive elements. Among the stress response-related elements, all the genes contained anaerobic induction elements; 62.5% of them contained low-temperature elements; and 50% of them contained drought-inducibility elements or defense and stress-responsive elements.
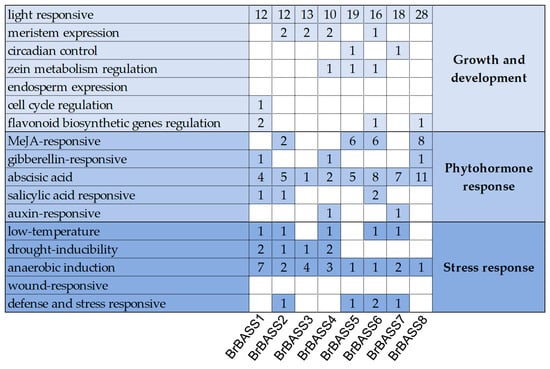
Figure 3.
Analysis of cis-elements in the promoter region of the eight BASS genes in B. rapa. Different shades of blue indicate the presence of cis-elements involved in different biological processes; the number in each cell indicates the number of cis-acting elements for that gene. White cells indicate the absence of the cis-acting element.
3.5. Analysis of Tissue-Specific Expression of BrBASSs
To understand the tissue-specific expression patterns of BrBASS genes, we examined the transcriptome data obtained from various tissues (Figure 4, Table S2). This revealed that BrBASS2 and BrBASS4 were expressed at particularly high levels in flowers, which might be related to their developmental process and potential role in sexual reproduction. Crucially, BrBASS1 and BrBASS7 were distinguished by high expression levels in the roots, which indicated their potential functionality in responding to abiotic stressors (such as salt and osmotic stresses). In addition, BrBASS2 was significantly overexpressed in leaves, which pointed to its possible regulatory role under low- and high-temperature stress conditions.
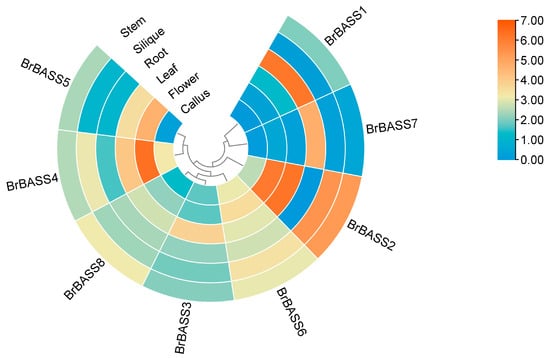
Figure 4.
Tissue-specific expression levels of the eight BrBASS genes. TPM was used to normalize the transcriptome sequences in different B. rapa tissues (from the NCBI GEO, accession number: GSE43245). The heatmap shows the level of BrBASS genes’ expression across six plant tissues. The red-to-blue color gradient indicates higher to lower expression levels.
3.6. Analysis of Expression Patterns in Response to Abiotic Stress
Chinese cabbage is very susceptible to osmotic stress, cold stress, and salt stress during its growth. So, we selected seven genes for RT-qPCR detection under conditions of osmotic stress, cold stress, and salt stress to distinguish which BrBASS genes were associated with each form of abiotic stress. Both BrBASS4 and BrBASS8 always exhibited downregulated expression under either osmotic, low temperature, or salt stress. However, as Figure 5 shows, under osmotic stress, the relative expression levels of BrBASS2, BrBASS3, and BrBASS6 decreased significantly, while those of BrBASS4, BrBASS7, and BrBASS8 increased at first but then decreased, peaking at 4 h and 6 h, respectively.
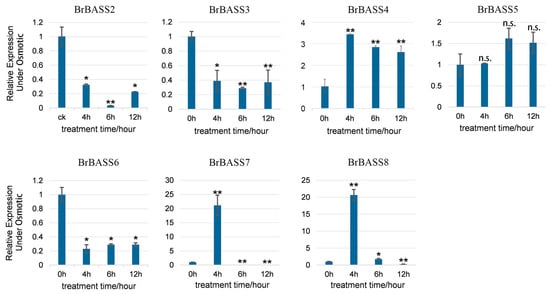
Figure 5.
Expression profiles of seven BrBASS genes under osmotic stress from the RT-qPCR analysis. The above experiments were performed using 0 h as the control, with the treatment time set to 4, 6, and 12 h. Each group had three biological replicates, and the error bars represent the standard error of the mean. Asterisks indicate statistically significant differences (two-tailed t-test; * p < 0.05; ** p < 0.01; n.s., no significance).
In addition to these genes, we also analyzed the expression of BrBASS1 under the same stress conditions. The results showed that the expression level of BrBASS1 remained consistently low and did not exhibit significant changes across all the time points studied. This suggests that BrBASS1 may not play a significant regulatory role under osmotic, cold, or salt stress conditions. Furthermore, its homologous gene in A. thaliana, BASS5, has not been extensively studied under stress conditions. Therefore, BrBASS1 was not included in the detailed analysis and discussion of stress-responsive genes.
Under cold stress (Figure 6), except for BrBASS3, whose relative expression significantly exceeded that of the control (CK) by three times, all the genes (BrBASS2, BrBASS4–BrBASS8) showed declines in their expression, with BrBASS2, BrBASS7, and BrBASS8 decreasing the most at 4 h and 6 h. The relative expression of BrBASS5 was significantly downregulated as well.
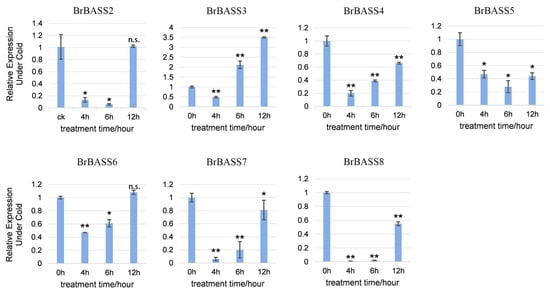
Figure 6.
Expression profiles of seven BrBASS genes under cold stress from the RT-qPCR analysis. The above experiments were performed using 0 h as the control, with the treatment time set to 4, 6, and 12 h. Each group had three biological replicates, and the error bars represent the standard error of the mean. Asterisks indicate statistically significant differences (two-tailed t-test; * p < 0.05; ** p < 0.01; n.s., no significance).
Under salt stress (Figure 7), except for the drastically increased relative expression levels of BrBASS2 and BrBASS7, which were basically reduced to 0, the other five genes’ relative expression levels showed differing degrees of downregulation, this being most pronounced at 4 h. With a longer treatment time, their relative expression increased but was still lower than that of CK. It is worth noting that BrBASS2 was significantly downregulated at 4 h, and its relative expression level showed an opposite trend with prolonged treatment time, peaking at 12 h, which was six times that of CK.
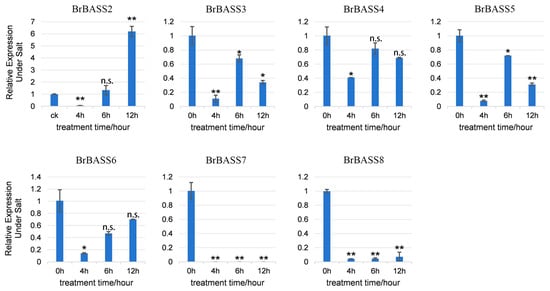
Figure 7.
Expression profiles of seven BrBASS genes under salt stress from the RT-qPCR analysis. The above experiments were performed using 0 h as the control, with the treatment time set to 4, 6, and 12 h. Each group had three biological replicates and the error bars represent the standard error of the mean. Asterisks indicate statistically significant differences (two-tailed t-test; * p < 0.05; ** p < 0.01; n.s., no significance).
3.7. Protein Secondary Structure and Tertiary Structure Prediction of BrBASSs
The structure of proteins is linked to their biological functions, so studying the structure of BASS proteins can provide valuable insights into their functions. Here, we analyzed the predicted protein secondary structures of BrBASSs and found that all members had helixes, extended strands, and loops. The protein secondary structures of BrBASSs are mainly helixes, whereas extended strands are least abundant (Figure 8A). According to the SWISS-MODEL analysis, members of the same subgroup displayed homologous protein tertiary structures (Figure 8B), indicating they have maintained homologous structures during evolution. BrBASS proteins consist of multiple transmembrane fragments (TM helixes) and two conserved sodium-binding sites, Na1 and Na2 (Figure 8B and Figure S1), located on the transmembrane structural domains [,,,]. The two discontinuous helices intersect in the center: a unique structural feature that enables BrBASS proteins to bind sodium ions during substrate transport and to enact structural changes by specifically binding to the sodium-binding site. This binding of sodium ions is a prerequisite for the binding and transport of BASS proteins to substrates, and, through the movement of sodium ions, this helps to balance the ionic content inside vs. outside the cell and to maintain normal cellular functions []. This structure allows BrBASS proteins to play a pivotal role in transmembrane transport dynamics by regulating the movement of sodium ions to facilitate the efficient transport of substrates, thus figuring prominently in the growth and responses of plants to environmental stress. Altogether, this provides basic information that could be helpful for subsequent studies of the functions of BrBASS proteins.
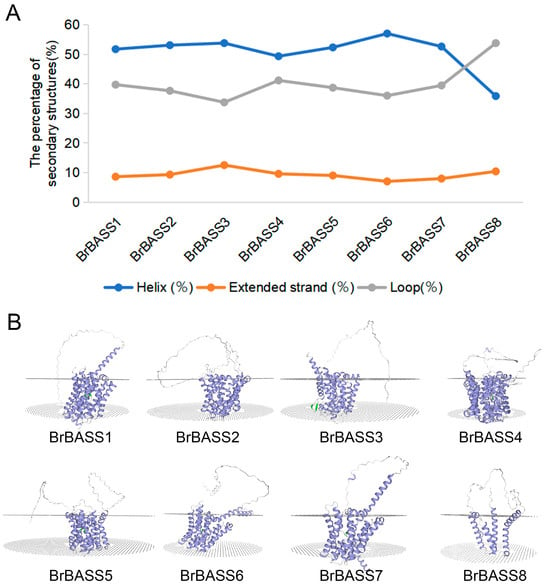
Figure 8.
Protein secondary and tertiary structure prediction of BrBASSs. (A) Predicted secondary structure of BrBASS proteins. Different colors indicate different secondary structures. (B) Predicted tertiary structures of BrBASS proteins. The light blue band represents the alpha helix in the BASS protein structure; the silver-white band indicates irregular coiling; and the green portion indicates the non-helical part of the discontinuous helix. The portion between the dotted planes is the transmembrane region of the protein under prediction.
3.8. Predicted Protein–Protein Interactions
Proteins are essential for the execution of myriad cellular and tissue functions and are implicated in diverse life activities []. PPI networks consist of protein interactions involved in various life processes, including biological signaling, gene expression regulation, regulation of the cell cycle, and intricate coordination of energy and material metabolism [,,,]. It is possible to gain a deeper understanding of the complex biological functions of proteins and biomolecular interactions by identifying connections between unknown functional proteins and PPI interaction networks [,]. The integrated resources and algorithms in the STRING database were utilized in our construction of predicted PPI network maps for detecting strong interactions between BASS members in A. thaliana, namely between BLOS1 and ANTR1 (sodium-dependent phosphate transport protein 1) (Figure 9A, Table S3). BrBASS1 and BrBASS7 were orthologous to AtBASS5, while BrBASS2 was orthologous to AtBASS2.
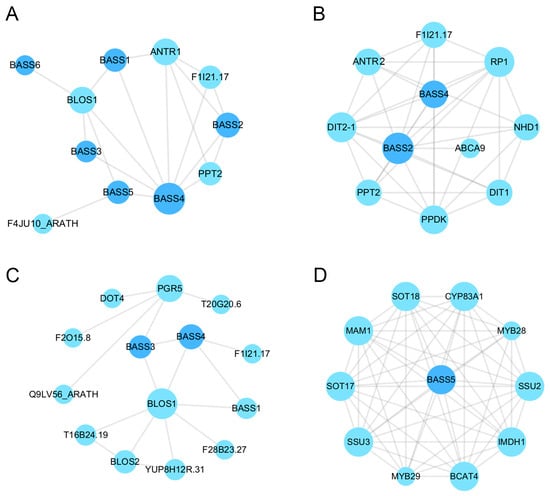
Figure 9.
Predictive PPI interaction networks for BASSs in A. thaliana. (A) Predictive analysis of BASS interaction networks in A. thaliana. (B) Predictive analysis of BASS2 in A. thaliana. (C) Predictive analysis of the interaction network of BASS3 in A. thaliana. (D) BASS5 interaction network prediction in A. thaliana. The minimum engagement score requirement is 0.150, and all other parameters were set to default. Nodes represent proteins, and edges represent protein-protein interactions.
BLOS1 possibly regulates PIN1 (PIN-formed 1) and PIN2 (PIN-formed 2) homeostasis through its interaction with SNX1 in auxin-mediated root growth and development of A. thaliana [,,].
The analysis of the protein-protein interaction (PPI) network uncovered a notable resemblance in the anticipated PPI networks of BASS2, 3, and 5 in A. thaliana and the proteins encoded by the BrBASS2 and BrBASS3 homologs (AtBASS2 and AtBASS3, respectively), which were found to interact with the abovementioned Nhd1 and BOLS1 in A. thaliana, respectively (Figure 9B,C). Nhd1 functions as a chloroplast sodium exporter, and its transport activity plays an important role in protecting vital chloroplast reactions such as photosynthesis from toxicity due to high sodium levels []. Furthermore, MYB (myeloblastosis) transcription factors are involved, including MYB28 and MYB29, which interact with the proteins encoded by homologs of BrBASS5 (Figure 9D). Brassica plants respond to abiotic stresses via proteins encoded by BrBASSs, which interact with proteins encoded by different gene families.
3.9. Phosphorylation Site Analysis
Protein phosphorylation modifications play a key role in signal transduction in the development and environmental adaptation of plants []. Protein phosphorylation mainly occurs on serine, threonine, and tyrosine residues []. To identify these processes—better known as protein phosphorylation—in BrBASSs, we conducted a phosphorylation site analysis (Figure 10). All proteins encoded by the eight members of the BrBASS gene family contained phosphorylation sites. We uncovered 330 phosphorylation sites, of which 66.7% were serine residues, 29.3% were threonine residues, with just 3.93% accounted for by tyrosine residues. Further examination revealed that all of the BASS genes in B. rapa, except BrBASS6, contained phospho-serine, phospho-tryptophan, and phospho-tyrosine sites. It should be noted that BrBASS4 harbored the highest number of phosphate sites.
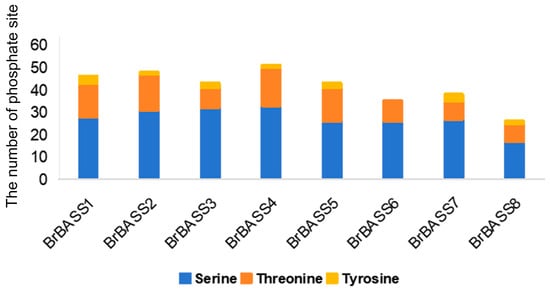
Figure 10.
Distribution of predicted phosphorylation sites in the amino acid sequences of BrBASSs.
3.10. Prediction of microRNAs Targeting BrBASSs
MicroRNAs (miRNAs) regulate their target genes post-transcriptionally in a negative manner [,]. Since they can have important functions in regulating gene expression, we investigated the miRNAs associated with BrBASSs genes (Table 2 and Table S4). These results revealed a total of 11 miRNA types regulating the eight BrBASS genes. BrBASS4, BrBASS5, and BrBASS8 were targeted by differing miRNAs, and BrBASS6 was the target of two different miRNAs. Notably, BrBASS3 and BrBASS7 were both targeted by three different types of miRNAs.

Table 2.
Details of BrBASSs and targeted miRNAs.
4. Discussion
Bile acid sodium symporter (BASS) family proteins are a class of Na+/solute isotropic transporters. Several studies have revealed that the BASS gene family is instrumental in sodium transport activities in mammals and for mediating how plants adapt to abiotic stresses, especially in coordinating their salt tolerance traits [,,], but its function has not been systematically studied in B. rapa. In the present study, eight BrBASS genes were identified and obtained, with the aim of elucidating their role in abiotic stress response by analyzing not only the structure and expression pattern of this family but also their physicochemical properties, gene structure, and cis-acting elements, etc. This study deepens our understanding of the functioning of BrBASSs.
Collinearity analysis can help to infer local events based on incongruity with the surrounding environment, often resulting in gene conversion []. When applied here to BrBASSs and AtBASSs, this analysis uncovered as many as seven groups of collinear genes between both, involving six BrBASSs (75% of the entire BrBASS gene family) and five AtBASSs (83.3%). This indicates that the evolutionary relationship between the BrBASS gene family and the AtBASS gene family is quite close. Accordingly, the functional study of AtBASS genes can provide a key reference for the functional research of BrBASS genes.
In B. rapa, the subcellular localization of its BrBASS gene family shows that all eight genes are distributed on chloroplasts. As a plastid, chloroplasts are not only the primary site for the excessive production of reactive oxygen species (ROS) in response to abiotic stresses [,], but also an important site for the production of antioxidant enzymes (e.g., superoxide dismutase (SOD), catalase (CAT), and peroxidase (POD)) to mitigate ROS toxicity [,]. In plants, ROS metabolism is a crucial component of their responsive adaptation to osmotic, cold, and salt stresses [,,]. Many studies have suggested that chloroplasts may be important in plant responses to abiotic stress conditions [,,,,,]. Under cold stress, chloroplasts can sense low-temperature signals via photoreceptors and membranes and promote photosynthesis by regulating enzyme activity and hormonal balance, thereby enhancing plant tolerance to cold stress []. SAL1-PAP chloroplasts in plants retrograde in response to nuclear stress, while the closure of stomata in leaves is responsive to osmotic stress []. Hence, we speculate the BrBASS gene family plays a key role in the abiotic stress dynamics of B. rapa plants.
BrBASS4 may contribute to coping with osmotic stress. The RT-qPCR results showed that this gene’s expression was higher at 4 h and peaked at 6 h under the osmotic treatment. But at 12 h, the expression level of BrBASS4 was significantly lower than that of CK. Analysis of its promoter cis-elements showed that BrBASS4 contained drought-responsive elements as well as gibberellin-responsive, abscisic acid, and auxin-responsive elements associated with hormone regulation in plants. Gibberellin (GA) is a key phytohormone that enables plants to cope with osmotic stress via signal transduction pathways and active oxygen clearance. The biosynthesis and degradation of GA in the early stage of osmotic stress alters the stomatal opening of plants, which in turn affects the rate of transpiration; hence, it has been postulated that GA regulates plants’ osmotic tolerance by reducing leaf transpiration []. Under osmotic conditions, plants synthesize ABA to regulate the plasma membrane ion channels of guard cells, leading to the long-term outflow of negative ions and K+, which in turn causes the guard cells to shrink and the stomata to close, enabling plants to better cope with osmotic stress []. The ABA response element (ABRE) enhances plant tolerance by regulating gene expression under drought and osmotic stress conditions []. Furthermore, the key node of the DELLA protein may jointly regulate osmotic stress response in plants between GA signaling and ABA signaling []. It has been shown that the auxin-responsive factor (ARF) can bind directly to the promoter of auxin-responsive genes, thereby augmenting tomato’s response to osmotic stress [].
Since BrBASS4 contains the most serine and threonine residues, we speculate that the biological function of this gene may be determined by the phosphorylation of serine residues. This would play a key role in cell signal transduction [], thus providing evidence for the osmotic stress response of the BrBASS4 gene. According to the PPI results, AtBASS3, the homologous gene of BrBASS4, interacted with both BLOS1 and PGR5 (proton gradient regulation 5). In Arabidopsis, there is evidence showing that BLOS1 figures prominently in the response to osmotic stress. BLOS1 is capable of interacting directly with SNX1 and regulates the homeostasis of PIN1 and PIN2 by mediating vacuolar degradation transport []. This regulation affects the distribution of auxin in roots, which in turn modulates the process of cell division and expansion [], allowing them to grow more actively under favorable soil conditions and to better tolerate arid conditions []. In addition, prediction of the target gene showed that Bra-miR396 targeted BrBASS4. Research using A. thaliana has found that miR396 is responsive to osmotic and salinity [], and that overexpression of miR396 in tobacco confers osmotic tolerance []. Thus, we speculate that BrBASS4 plays an important role in coping with osmotic stress.
BrBASS7 harbors great potential for regulating cold stress. The RT-qPCR showed that this gene’s relative expression fell considerably under low temperature stress. Analysis of the promoter elements showed that BrBASS7 contained a low-temperature response element, an abscisic acid, and an auxin-responsive hormone response element. When plants are exposed to cold conditions, stomatal opening in their leaves is inhibited, resulting in the accumulation of ABA during stomatal closure that in turn enables plants to cope with cold stress []. When A. thaliana is subjected to cold stress, the auxin transport rate is affected, allowing this plant to endure cold stress [].
The target gene prediction analysis showed that Bra-miR396, Bra-miR9565, and Bra-miR5725 all targeted BrBASS7. A recent study of Jerusalem artichoke found that htu-miR396 was upregulated under cryogenic stress []. Analysis of the PPI network has revealed that AtBASS5, a homologous gene of BrBASS7, interacts with MYB28 and MYB29 proteins; MYB28 is a positive regulator of aliphatic methionine-derived glucosinolates (GLS), and MYB29 is involved in hormone-mediated pathways and signal transduction, playing a role in GLS biosynthesis and promoting it in conjunction with proteins such as MYB28/HAG1 [,]. Other research has confirmed that the synthesis of GLS in Brassica plants was also affected by ambient temperature, in that low temperatures promoted the production of GLS in response to external factors []. However, since BASS5 was a candidate transporter gene involved in GLS synthesis in A. thaliana, it was posited that BASS5 may promote GLS biosynthesis via its regulation at low temperatures []. In other words, BASS5 could also be related to cold stress.
Salt stress could be alleviated by BrBASS2. The economic productivity of vegetable crops is greatly limited by salinity stress, being one of the most important environmental constraints []. The RT-qPCR results show that the relative expression of BrBASS2 is upregulated under conditions of salt stress. The promoter cis-acting element of BrBASS2 contains abscisic acid-responsive elements, MeJA-responsive elements, and salicylic acid-responsive elements. In cotton, ABA was continuously upregulated under salt stress, which implied its potential pertinence for mitigating that type of stress, and ABA’s accumulation underpinned the salt tolerance mechanism []. MeJA plays an important role in resisting salt stress, osmotic stress, cold stress, heavy metal stress, and the toxicity of other elements. After applying MeJA to stressed plants, they grow better, accumulate active compounds, and begin to undergo changes in their physiological and biochemical properties as well as their endogenous hormone levels []. The application of salicylic acid can also improve the salt tolerance ability of plants []. Here, the protein–protein interaction results showed that AtBASS2, the homologous gene of BrBASS2, interacted with Nhd1. In O. sativa, Nhd1 has been found to alleviate the toxic effects of salt stress on crops by activating nitrogen transporters to regulate root growth and nitrogen uptake [,]. When A. thaliana is subjected to salt stress, the chloroplast Na content of Atnhd1 mutants increases considerably, resulting in the pronounced impairment of photosynthetic performance; moreover, high Na levels may reduce the activity of plastid bile acid/sodium co-transporter family protein 2 (BASS2) []. In studies of T. aestivum, it was found that a pyruvate transporter called TaBASS2 is a homolog of BASS2. By applying a NaCl treatment to A. thaliana and T. aestivum, it was found that TaBASS2-overexpressing plants could alleviate the growth inhibition caused by high salinity and enhance the salt tolerance of both plant species []. Likewise, we deduced that the homologous gene BrBASS2 of BASS2 has a similar function in Brassica plants’ response to salt stress.
5. Conclusions
In summary, we identified eight BrBASS genes in the B. rapa genome. By analyzing their sequence features, cis-elements, protein-protein interactions, predicted microRNA targets, abiotic stress tolerance, and published data, we further determined that BrBASS2 may play a key role in regulating tolerance to salt stress, that BrBASS4 has the greatest potential for regulating the osmotic stress response, and that BrBASS7 has substantial potential for regulating cold stress tolerance.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/cimb46070396/s1.
Author Contributions
Conceptualization, Y.C.; Data curation, Y.C.; Formal analysis, Y.W.; Investigation, M.Z. and L.C.; Project administration, J.H.; Resources, Y.C.; Supervision, Y.C.; Validation, S.H. and B.L.; Visualization, Q.J.; Writing—original draft, Z.J. and R.W.; Writing—review and editing, Y.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China (32002045).
Data Availability Statement
All the data that support the findings of this study are available in the paper and its Supplementary Materials published online.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Alrefai, W.A.; Gill, R.K. Bile acid transporters: Structure, function, regulation and pathophysiological implications. Pharm. Res. 2007, 24, 1803–1823. [Google Scholar] [CrossRef] [PubMed]
- Pols, T.W.H.; Noriega, L.G.; Nomura, M.; Auwerx, J.; Schoonjans, K. The bile acid membrane receptor TGR5 as an emerging target in metabolism and inflammation. J. Hepatol. 2011, 54, 1263–1272. [Google Scholar] [CrossRef] [PubMed]
- Hu, N.J.; Iwata, S.; Cameron, A.D.; Drew, D. Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature 2011, 478, 408–411. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Levin, E.J.; Pan, Y.; McCoy, J.G.; Sharma, R.; Kloss, B.; Bruni, R.; Quick, M.; Zhou, M. Structural basis of the alternating-access mechanism in a bile acid transporter. Nature 2014, 505, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Claro da Silva, T.; Polli, J.E.; Swaan, P.W. The solute carrier family 10 (SLC10): Beyond bile acid transport. Mol. Asp. Med. 2013, 34, 252–269. [Google Scholar] [CrossRef] [PubMed]
- Myo, T.; Wei, F.; Zhang, H.; Hao, J.; Zhang, B.; Liu, Z.; Cao, G.; Tian, B.; Shi, G. Genome-wide identification of the BASS gene family in four Gossypium species and functional characterization of GhBASSs against salt stress. Sci. Rep. 2021, 11, 11342. [Google Scholar] [CrossRef] [PubMed]
- Becker, P.; Naughton, F.; Brotherton, D.; Pacheco-Gomez, R.; Beckstein, O.; Cameron, A.D. Mechanism of substrate binding and transport in BASS transporters. eLife 2023, 12, RP89167. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ai, X.; Wang, M.; Xiao, L.; Xia, G. A putative pyruvate transporter TaBASS2 positively regulates salinity tolerance in wheat via modulation of ABI4 expression. BMC Plant Biol. 2016, 16, 109. [Google Scholar] [CrossRef]
- Myo, T.; Tian, B.; Zhang, Q.; Niu, S.; Liu, Z.; Shi, Y.; Cao, G.; Ling, H.; Wei, F.; Shi, G. Ectopic overexpression of a cotton plastidial Na+ transporter GhBASS5 impairs salt tolerance in Arabidopsis via increasing Na+ loading and accumulation. Planta 2020, 252, 41. [Google Scholar] [CrossRef]
- Sawada, Y.; Toyooka, K.; Kuwahara, A.; Sakata, A.; Nagano, M.; Saito, K.; Hirai, M.Y. Arabidopsis Bile Acid:Sodium Symporter Family Protein 5 is Involved in Methionine-Derived Glucosinolate Biosynthesis. Plant Cell Physiol. 2009, 50, 1579–1586. [Google Scholar] [CrossRef]
- Rzewuski, G.; Sauter, M. The novel rice (Oryza sativa L.) gene OsSbf1 encodes a putative member of the Na+/bile acid symporter family. J. Exp. Bot. 2002, 53, 1991–1993. [Google Scholar] [CrossRef]
- Furumoto, T.; Yamaguchi, T.; Ohshima-Ichie, Y.; Nakamura, M.; Tsuchida-Iwata, Y.; Shimamura, M.; Ohnishi, J.; Hata, S.; Gowik, U.; Westhoff, P.; et al. A plastidial sodium-dependent pyruvate transporter. Nature 2011, 476, 472–475. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Suzuki, N.; Miller, G.; Tognetti, V.B.; Vandepoele, K.; Gollery, M.; Shulaev, V.; Van Breusegem, F. ROS signaling: The new wave? Trends Plant Sci. 2011, 16, 300–309. [Google Scholar] [CrossRef]
- Huang, L.; Pyc, M.; Alseekh, S.; McCarty, D.R.; de Crécy-Lagard, V.; Gregory, J.F., 3rd; Henry, C.S.; Fernie, A.R.; Mullen, R.T.; Hanson, A.D. A plastidial pantoate transporter with a potential role in pantothenate synthesis. Biochem. J. 2018, 475, 813–825. [Google Scholar] [CrossRef]
- Gigolashvili, T.; Yatusevich, R.; Rollwitz, I.; Humphry, M.; Gershenzon, J.; Flügge, U.-I. The Plastidic Bile Acid Transporter 5 Is Required for the Biosynthesis of Methionine-Derived Glucosinolates in Arabidopsis thaliana. Plant Cell 2009, 21, 1813–1829. [Google Scholar] [CrossRef]
- South, P.F.; Walker, B.J.; Cavanagh, A.P.; Rolland, V.; Badger, M.; Ort, D.R. Bile Acid Sodium Symporter BASS6 Can Transport Glycolate and Is Involved in Photorespiratory Metabolism in Arabidopsis thaliana. Plant Cell 2017, 29, 808–823. [Google Scholar] [CrossRef] [PubMed]
- Pavlović, I.; Mlinarić, S.; Tarkowská, D.; Oklestkova, J.; Novák, O.; Lepeduš, H.; Bok, V.V.; Brkanac, S.R.; Strnad, M.; Salopek-Sondi, B. Early Brassica Crops Responses to Salinity Stress: A Comparative Analysis Between Chinese Cabbage, White Cabbage, and Kale. Front. Plant Sci. 2019, 10, 450. [Google Scholar] [CrossRef]
- Ahmed, N.U.; Park, J.-I.; Jung, H.-J.; Seo, M.-S.; Kumar, T.S.; Lee, I.-H.; Nou, I.-S. Identification and characterization of stress resistance related genes of Brassicarapa. Biotechnol. Lett. 2012, 34, 979–987. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Xiong, L. General mechanisms of drought response and their application in drought resistance improvement in plants. Cell. Mol. Life Sci. 2015, 72, 673–689. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Chen, C.C.H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Rombauts, S.; Déhais, P.; Van Montagu, M.; Rouzé, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef] [PubMed]
- Tong, C.; Wang, X.; Yu, J.; Wu, J.; Li, W.; Huang, J.; Dong, C.; Hua, W.; Liu, S. Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genom. 2013, 14, 689. [Google Scholar] [CrossRef]
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, W155–W159. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Zhang, L.; Guo, Y.; Zhang, Y.; Li, Y.; Pei, Y.; Zhang, M. Regulation of PIN-FORMED Protein Degradation. Int. J. Mol. Sci. 2023, 24, 843. [Google Scholar] [CrossRef]
- Liu, C.; Liu, L.; Zhou, C.; Zhuang, J.; Wang, L.; Sun, Y.; Sun, C. Protein-protein interaction networks and different clustering analysis in Burkitt’s lymphoma. Hematology 2018, 23, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Bader, G.D.; Hogue, C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Movahedi, A.; Liu, G.; Li, Y.; Liu, S.; Yu, C.; Chen, Y.; Zhong, F.; Zhang, J. Comprehensive Analysis of Carotenoid Cleavage Dioxygenases Gene Family and Its Expression in Response to Abiotic Stress in Poplar. Int. J. Mol. Sci. 2022, 23, 1418. [Google Scholar] [CrossRef] [PubMed]
- Alberts, B. The cell as a collection of protein machines: Preparing the next generation of molecular biologists. Cell 1998, 92, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, Y.; Zhu, J.; Ma, W.; Li, Z.; Bi, Z.; Sun, C.; Bai, J.; Zhang, J.; Liu, Y. Genome-Wide Identification and Analysis of the NF-Y Gene Family in Potato (Solanum tuberosum L.). Front. Genet. 2021, 12, 739989. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Guo, X.; Liu, J.; Zhou, F.; Liu, W.; Wu, J.; Zhang, H.; Cao, H.; Su, H.; Wen, R. Genome-Wide Identification, Characterization, and Expression Analysis of the NAC Transcription Factor in Chenopodium quinoa. Genes 2019, 10, 500. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Kamiya, T.; Fujiwara, T. Differential Roles of PIN1 and PIN2 in Root Meristem Maintenance Under Low-B Conditions in Arabidopsis thaliana. Plant Cell Physiol. 2015, 56, 1205–1214. [Google Scholar] [CrossRef] [PubMed]
- Blilou, I.; Xu, J.; Wildwater, M.; Willemsen, V.; Paponov, I.; Friml, J.; Heidstra, R.; Aida, M.; Palme, K.; Scheres, B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 2005, 433, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Kunz, H.H.; Schroeder, J.I.; Kemp, G.; Young, H.S.; Neuhaus, H.E. Decreased capacity for sodium export out of Arabidopsis chloroplasts impairs salt tolerance, photosynthesis and plant performance. Plant J. 2014, 78, 646–658. [Google Scholar] [CrossRef]
- Zhang, W.J.; Zhou, Y.; Zhang, Y.; Su, Y.H.; Xu, T. Protein phosphorylation: A molecular switch in plant signaling. Cell Rep. 2023, 42, 112729. [Google Scholar] [CrossRef]
- Li, P.; Liu, J. Protein Phosphorylation in Plant Cell Signaling. In Plant Phosphoproteomics: Methods and Protocols; Wu, X.N., Ed.; Springer: New York, NY, USA, 2021; pp. 45–71. [Google Scholar]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Wang, X.; Sun, Y.; Joseph, P.V.; Paterson, A.H. Detection of colinear blocks and synteny and evolutionary analyses based on utilization of MCScanX. Nat. Protoc. 2024, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Hanke, G. ROS production and signalling in chloroplasts: Cornerstones and evolving concepts. Plant J. 2022, 111, 642–661. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Smith, J.A.; Harberd, N.P.; Jiang, C. The regulatory roles of ethylene and reactive oxygen species (ROS) in plant salt stress responses. Plant Mol. Biol. 2016, 91, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Zandalinas, S.I.; Mittler, R.; Balfagón, D.; Arbona, V.; Gómez-Cadenas, A. Plant adaptations to the combination of drought and high temperatures. Physiol. Plant. 2018, 162, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Agurla, S.; Gahir, S.; Munemasa, S.; Murata, Y.; Raghavendra, A.S. Mechanism of Stomatal Closure in Plants Exposed to Drought and Cold Stress. Adv. Exp. Med. Biol. 2018, 1081, 215–232. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, A.; Li, X.; Lu, C. The Role of Chloroplast Gene Expression in Plant Responses to Environmental Stress. Int. J. Mol. Sci. 2020, 21, 6082. [Google Scholar] [CrossRef]
- Littlejohn, G.R.; Breen, S.; Smirnoff, N.; Grant, M. Chloroplast immunity illuminated. New Phytol. 2021, 229, 3088–3107. [Google Scholar] [CrossRef] [PubMed]
- Serrano, I.; Audran, C.; Rivas, S. Chloroplasts at work during plant innate immunity. J. Exp. Bot. 2016, 67, 3845–3854. [Google Scholar] [CrossRef]
- Watson, S.J.; Sowden, R.G.; Jarvis, P. Abiotic stress-induced chloroplast proteome remodelling: A mechanistic overview. J. Exp. Bot. 2018, 69, 2773–2781. [Google Scholar] [CrossRef] [PubMed]
- Hameed, A.; Ahmed, M.Z.; Hussain, T.; Aziz, I.; Ahmad, N.; Gul, B.; Nielsen, B.L. Effects of Salinity Stress on Chloroplast Structure and Function. Cells 2021, 10, 2023. [Google Scholar] [CrossRef]
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 2012, 35, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Gan, P.; Liu, F.; Li, R.; Wang, S.; Luo, J. Chloroplasts—Beyond Energy Capture and Carbon Fixation: Tuning of Photosynthesis in Response to Chilling Stress. Int. J. Mol. Sci. 2019, 20, 5046. [Google Scholar] [CrossRef]
- Estavillo, G.M.; Crisp, P.A.; Pornsiriwong, W.; Wirtz, M.; Collinge, D.; Carrie, C.; Giraud, E.; Whelan, J.; David, P.; Javot, H.; et al. Evidence for a SAL1-PAP Chloroplast Retrograde Pathway That Functions in Drought and High Light Signaling in Arabidopsis. Plant Cell 2011, 23, 3992–4012. [Google Scholar] [CrossRef]
- Iqbal, S.; Wang, X.; Mubeen, I.; Kamran, M.; Kanwal, I.; Díaz, G.A.; Abbas, A.; Parveen, A.; Atiq, M.N.; Alshaya, H.; et al. Phytohormones Trigger Drought Tolerance in Crop Plants: Outlook and Future Perspectives. Front. Plant Sci. 2021, 12, 799318. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, L.; Cheng, C.; Ren, Z.; Xu, S.; Li, X. GAI Functions in the Plant Response to Dehydration Stress in Arabidopsis thaliana. Int. J. Mol. Sci. 2020, 21, 819. [Google Scholar] [CrossRef] [PubMed]
- Bouzroud, S.; Gouiaa, S.; Hu, N.; Bernadac, A.; Mila, I.; Bendaou, N.; Smouni, A.; Bouzayen, M.; Zouine, M. Auxin Response Factors (ARFs) are potential mediators of auxin action in tomato response to biotic and abiotic stress (Solanum lycopersicum). PLoS ONE 2018, 13, e0193517. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Liu, J. Protein Phosphorylation in Plant Cell Signaling. Methods Mol. Biol. 2021, 2358, 45–71. [Google Scholar] [CrossRef] [PubMed]
- Karlova, R.; Boer, D.; Hayes, S.; Testerink, C. Root plasticity under abiotic stress. Plant Physiol. 2021, 187, 1057–1070. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.H.; Tian, X.; Li, Y.J.; Wu, C.A.; Zheng, C.C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 2008, 14, 836–843. [Google Scholar] [CrossRef]
- Fengxi, Y.; Di-qiu, Y. Overexpression of Arabidopsis MiR396 enhances drought tolerance in transgenic tobacco plants. Acta Bot. Yunnanica 2009, 31, 421–426. [Google Scholar]
- Morris, D.A. The effect of temperature on the velocity of exogenous auxin transport in intact chilling-sensitive and chilling-resistant plants. Planta 1979, 146, 603–605. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Yue, Y.; Chen, X.; Long, X.; Zhou, Z. Identification and expression analysis of miR396 and its target genes in Jerusalem artichoke under temperature stress. Gene 2024, 893, 147908. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ivanova, A.; Vandepoele, K.; Radomiljac, J.; Van de Velde, J.; Berkowitz, O.; Willems, P.; Xu, Y.; Ng, S.; Van Aken, O.; et al. The Transcription Factor MYB29 Is a Regulator of ALTERNATIVE OXIDASE1a. Plant Physiol. 2017, 173, 1824–1843. [Google Scholar] [CrossRef]
- Gigolashvili, T.; Yatusevich, R.; Berger, B.; Müller, C.; Flügge, U.I. The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007, 51, 247–261. [Google Scholar] [CrossRef]
- Kissen, R.; Eberl, F.; Winge, P.; Uleberg, E.; Martinussen, I.; Bones, A.M. Effect of growth temperature on glucosinolate profiles in Arabidopsis thaliana accessions. Phytochemistry 2016, 130, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Behera, T.K.; Krishna, R.; Ansari, W.A.; Aamir, M.; Kumar, P.; Kashyap, S.P.; Pandey, S.; Kole, C. Approaches Involved in the Vegetable Crops Salt Stress Tolerance Improvement: Present Status and Way Ahead. Front. Plant Sci. 2021, 12, 787292. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Cui, R.; Wang, D.; Huang, H.; Rui, C.; Malik, W.A.; Wang, J.; Zhang, H.; Xu, N.; Liu, X.; et al. Combined transcriptomic and metabolomic analyses elucidate key salt-responsive biomarkers to regulate salt tolerance in cotton. BMC Plant Biol. 2023, 23, 245. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhang, W.; Zhang, Y.; Zhang, X.; Lang, D.; Zhang, X. The roles of methyl jasmonate to stress in plants. Funct. Plant Biol. 2019, 46, 197–212. [Google Scholar] [CrossRef] [PubMed]
- Abdi, N.; Van Biljon, A.; Steyn, C.; Labuschagne, M.T. Salicylic Acid Improves Growth and Physiological Attributes and Salt Tolerance Differentially in Two Bread Wheat Cultivars. Plants 2022, 11, 1853. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Wang, J.; Wang, H.; Cui, J.; Shi, X.; Song, J.; Li, W.; Zhong, M.; Qiu, Y.; Xu, T. Nitrogen application alleviates salt stress by enhancing osmotic balance, ROS scavenging, and photosynthesis of rapeseed seedlings (Brassica napus). Plant Signal. Behav. 2022, 17, 2081419. [Google Scholar] [CrossRef]
- Li, K.; Zhang, S.; Tang, S.; Zhang, J.; Dong, H.; Yang, S.; Qu, H.; Xuan, W.; Gu, M.; Xu, G. The rice transcription factor Nhd1 regulates root growth and nitrogen uptake by activating nitrogen transporters. Plant Physiol. 2022, 189, 1608–1624. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).