A Novel Dhillonvirus Phage against Escherichia coli Bearing a Unique Gene of Intergeneric Origin
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Sample Collections
2.3. Phage Isolation and Enrichment
2.4. Bacteriophage Plaque Assay
2.5. Phage DNA Extraction
2.6. Genome Sequencing
2.7. Phage Genome Assembly and Characterization
2.8. Transmission Electron Microscopy (TEM) Methodology
2.9. One-Step Growth Curve and Adsorption Assay
2.10. Thermal and pH Stability
2.11. Host Range Determination
2.12. Phylogenetic Analysis
3. Results
3.1. Genome Characterization
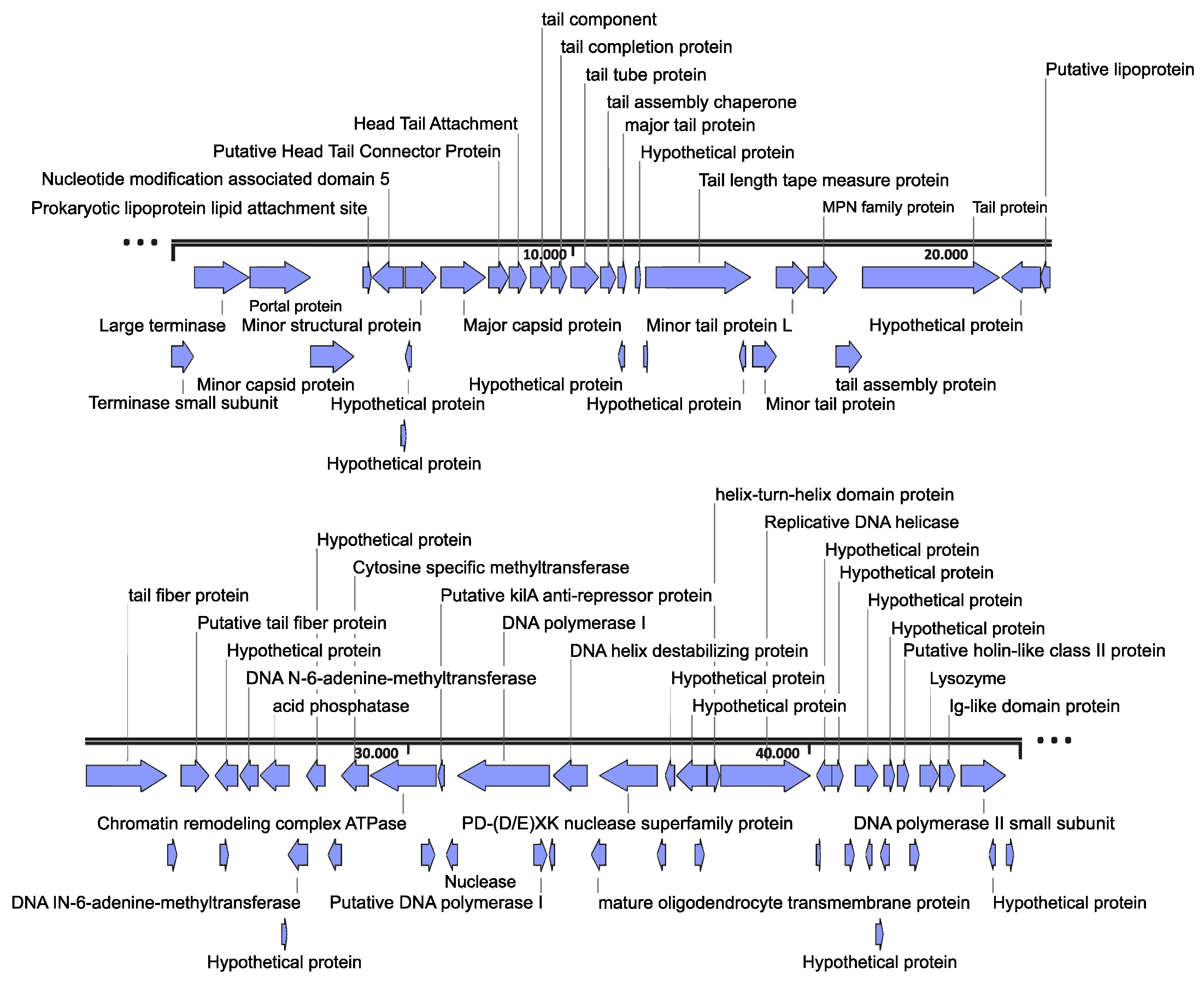
3.2. Genome Organization
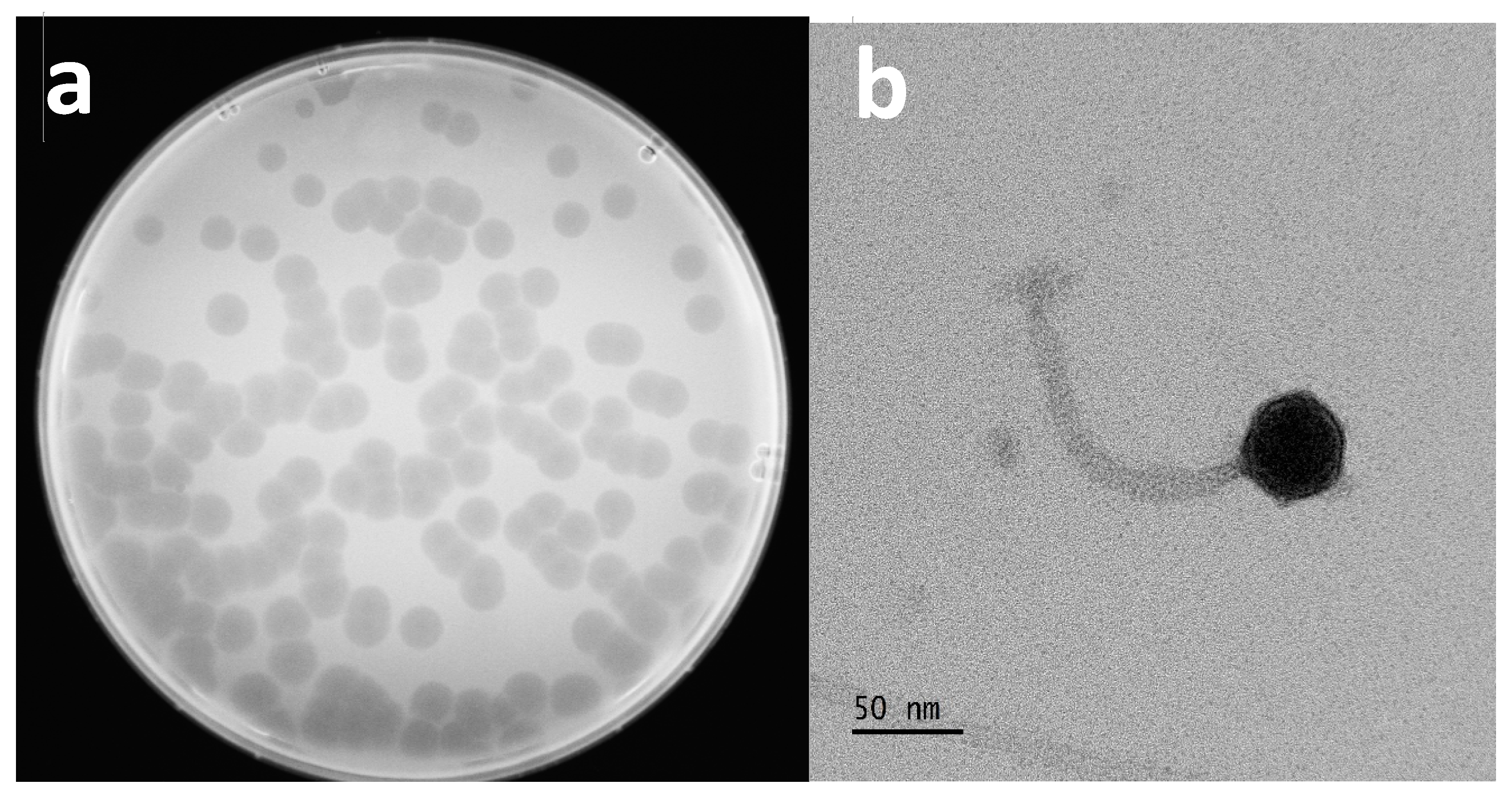
3.3. Bacteriophage Plaque Formation and Morphology
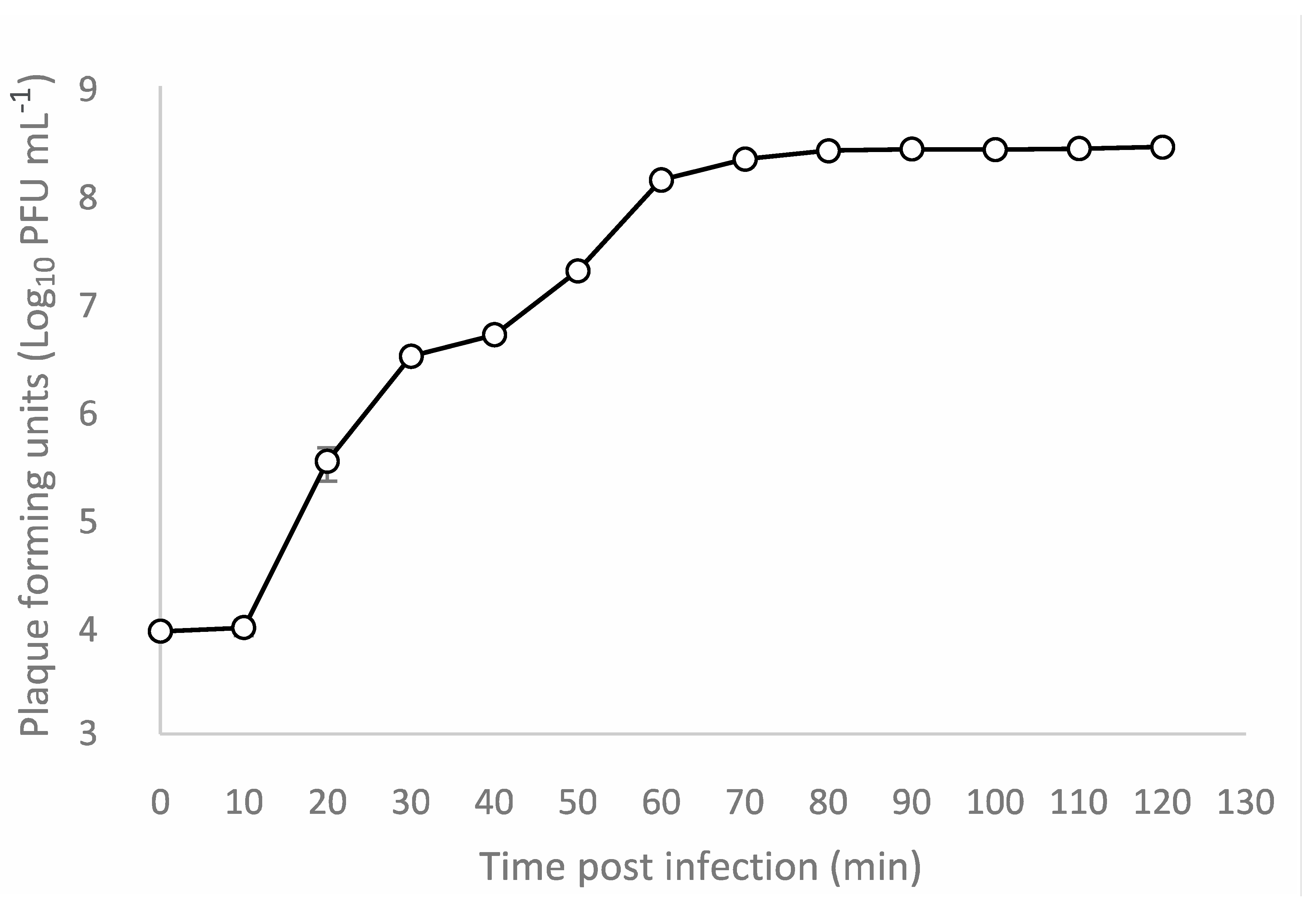
3.4. One-Step Growth Curve and Adsorption Assay
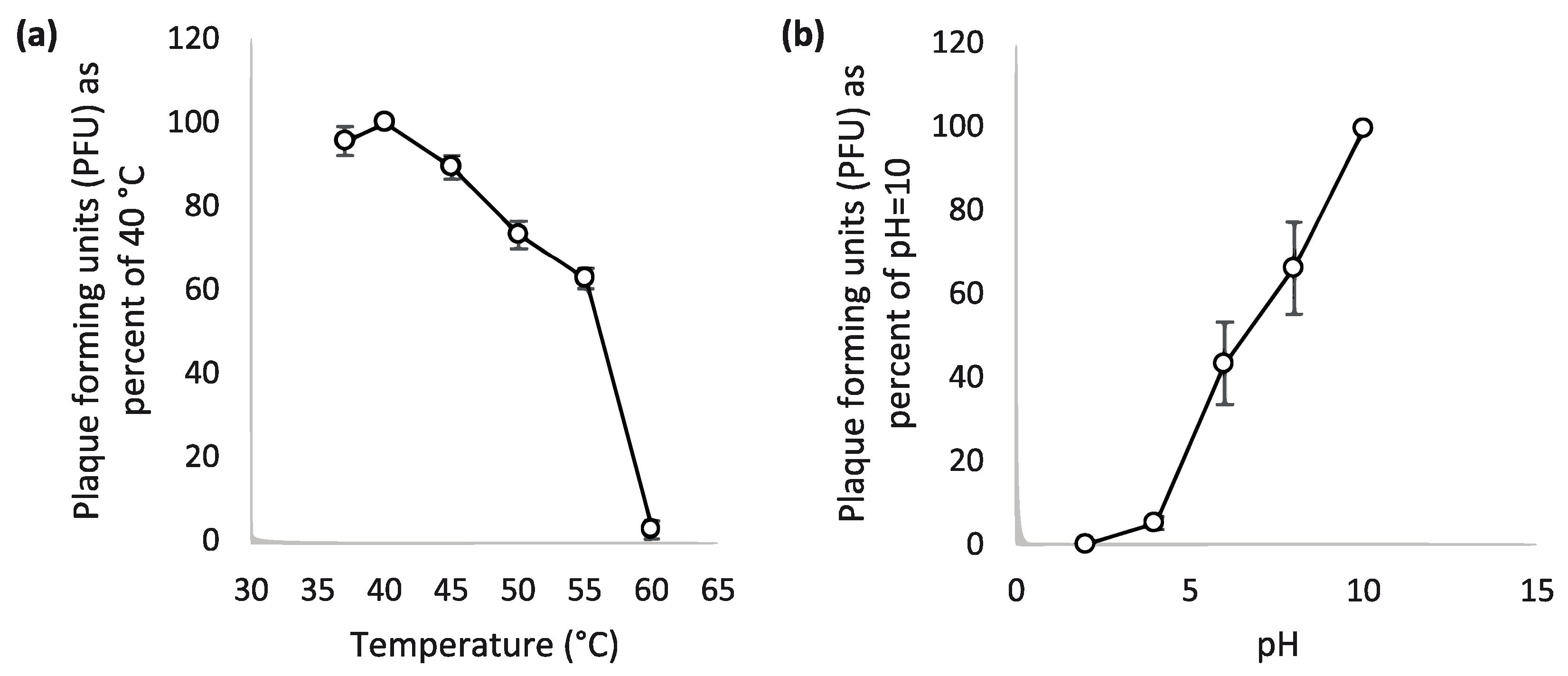
3.5. Thermal and PH Stability of the Phage
3.6. Host Range Determination
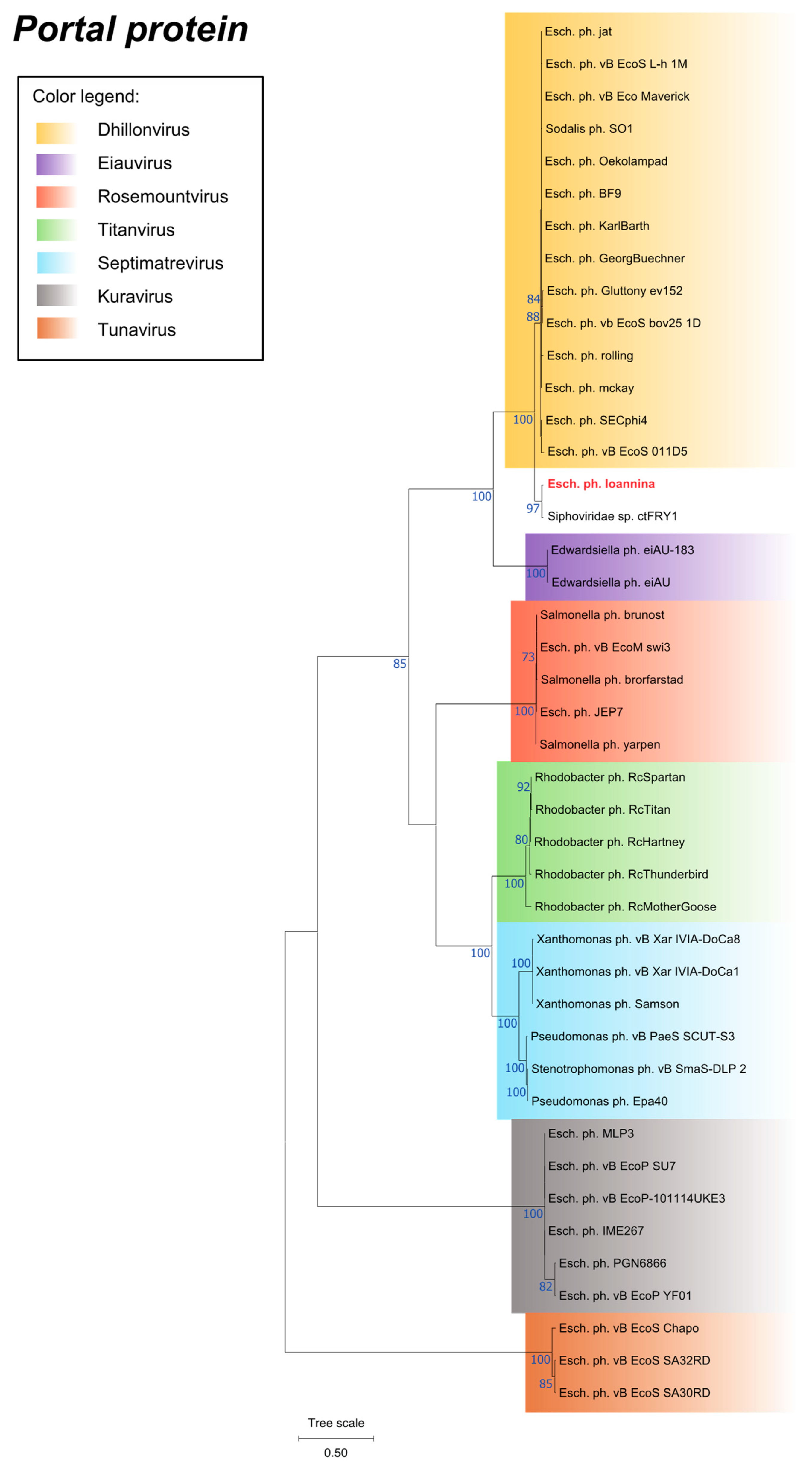
3.7. Phylogenetic Analysis
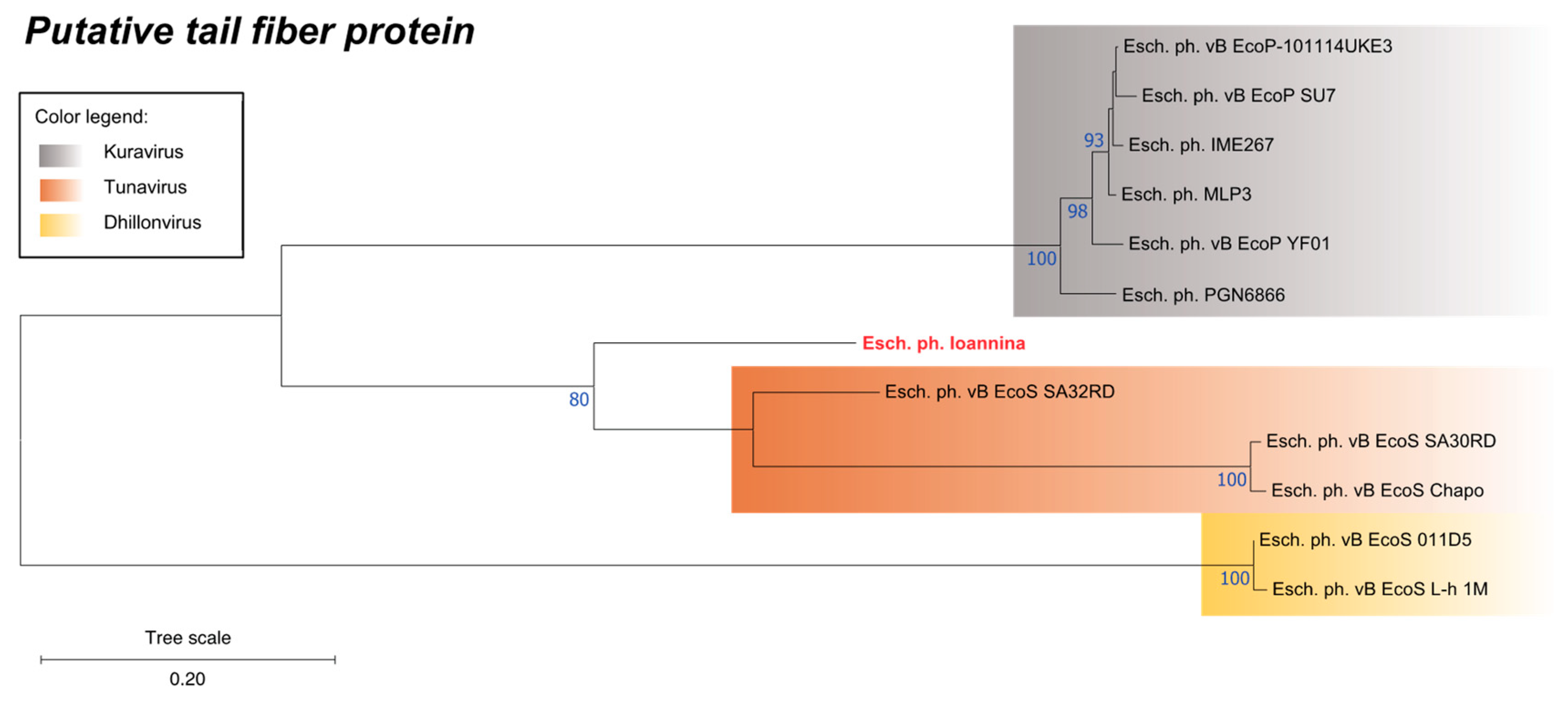
3.8. Phylogenetic Analysis of a Unique Putative Tail Fiber Protein
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Principi, N.; Silvestri, E.; Esposito, S. Advantages and Limitations of Bacteriophages for the Treatment of Bacterial Infections. Front. Pharmacol. 2019, 10, 457104. [Google Scholar] [CrossRef]
- Keen, E.C. A Century of Phage Research: Bacteriophages and the Shaping of Modern Biology. Bioessays 2015, 37, 6–9. [Google Scholar] [CrossRef]
- Naureen, Z.; Dautaj, A.; Anpilogov, K.; Camilleri, G.; Dhuli, K.; Tanzi, B.; Maltese, P.E.; Cristofoli, F.; De Antoni, L.; Beccari, T.; et al. Bacteriophages Presence in Nature and Their Role in the Natural Selection of Bacterial Populations. Acta Biomed. 2020, 91, e2020024. [Google Scholar] [CrossRef]
- Endersen, L.; Coffey, A. The Use of Bacteriophages for Food Safety. Curr. Opin. Food Sci. 2020, 36, 1–8. [Google Scholar] [CrossRef]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage Applications for Food Production and Processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef]
- Harada, L.K.; Silva, E.C.; Campos, W.F.; Del Fiol, F.S.; Vila, M.; Dąbrowska, K.; Krylov, V.N.; Balcão, V.M. Biotechnological Applications of Bacteriophages: State of the Art. Microbiol. Res. 2018, 212–213, 38–58. [Google Scholar] [CrossRef]
- Düzgüneş, N.; Sessevmez, M.; Yildirim, M. Bacteriophage Therapy of Bacterial Infections: The Rediscovered Frontier. Pharmaceuticals 2021, 14, 34. [Google Scholar] [CrossRef]
- Ferriol-González, C.; Domingo-Calap, P. Phages for Biofilm Removal. Antibiotics 2020, 9, 268. [Google Scholar] [CrossRef]
- Carascal, M.B.; dela Cruz-Papa, D.M.; Remenyi, R.; Cruz, M.C.B.; Destura, R.V. Phage Revolution against Multidrug-Resistant Clinical Pathogens in Southeast Asia. Front. Microbiol. 2022, 13, 820572. [Google Scholar] [CrossRef]
- Aranaga, C.; Pantoja, L.D.; Martínez, E.A.; Falco, A. Phage Therapy in the Era of Multidrug Resistance in Bacteria: A Systematic Review. Int. J. Mol. Sci. 2022, 23, 4577. [Google Scholar] [CrossRef]
- Manrique, P.; Bolduc, B.; Walk, S.T.; van der Oost, J.; de Vos, W.M.; Young, M.J. Healthy Human Gut Phageome. Proc. Natl. Acad. Sci. USA 2016, 113, 10400–10405. [Google Scholar] [CrossRef]
- Tiamani, K.; Luo, S.; Schulz, S.; Xue, J.; Costa, R.; Khan Mirzaei, M.; Deng, L. The Role of Virome in the Gastrointestinal Tract and Beyond. FEMS Microbiol. Rev. 2022, 46, fuac027. [Google Scholar] [CrossRef]
- Federici, S.; Nobs, S.P.; Elinav, E. Phages and Their Potential to Modulate the Microbiome and Immunity. Cell Mol. Immunol. 2021, 18, 889–904. [Google Scholar] [CrossRef]
- Liu, B.; Furevi, A.; Perepelov, A.V.; Guo, X.; Cao, H.; Wang, Q.; Reeves, P.R.; Knirel, Y.A.; Wang, L.; Widmalm, G. Structure and Genetics of Escherichia Coli O Antigens. FEMS Microbiol. Rev. 2020, 44, 655–683. [Google Scholar] [CrossRef]
- van der Hooft, J.J.J.; Goldstone, R.J.; Harris, S.; Burgess, K.E.V.; Smith, D.G.E. Substantial Extracellular Metabolic Differences Found between Phylogenetically Closely Related Probiotic and Pathogenic Strains of Escherichia Coli. Front. Microbiol. 2019, 10, 252. [Google Scholar] [CrossRef]
- Pang, W.; Wang, H.; Shi, L.; Sun, Y.; Wang, X.; Wang, M.; Li, J.; Wang, H.; Shi, G. Immunomodulatory Effects of Escherichia Coli ATCC 25922 on Allergic Airway Inflammation in a Mouse Model. PLoS ONE 2013, 8, e59174. [Google Scholar] [CrossRef]
- Minogue, T.D.; Daligault, H.A.; Davenport, K.W.; Bishop-Lilly, K.A.; Broomall, S.M.; Bruce, D.C.; Chain, P.S.; Chertkov, O.; Coyne, S.R.; Freitas, T.; et al. Complete Genome Assembly of Escherichia Coli ATCC 25922, a Serotype O6 Reference Strain. Genome Announc. 2014, 2, e00969-14. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a Full-Length Transcriptome without a Genome from RNA-Seq Data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Huang, X.; Madan, A. CAP3: A DNA Sequence Assembly Program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. Methods Mol. Biol. 2019, 1962, 1–14. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A Genome Comparison Visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Pertics, B.Z.; Kovács, T.; Schneider, G. Characterization of a Lytic Bacteriophage and Demonstration of Its Combined Lytic Effect with a K2 Depolymerase on the Hypervirulent Klebsiella Pneumoniae Strain 52145. Microorganisms 2023, 11, 669. [Google Scholar] [CrossRef]
- Dioli, C.; Pappa, O.; Siatravani, E.; Bratakou, S.; Tatsiopoulos, A.; Giakkoupi, P.; Miriagou, V.; Beloukas, A. Molecular Characterization and Prevalence of Antimicrobial-Resistant Escherichia Coli Isolates Derived from Clinical Specimens and Environmental Habitats. Microorganisms 2023, 11, 1399. [Google Scholar] [CrossRef]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and Simple Determination of the Escherichia Coli Phylogenetic Group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [PubMed]
- Kutter, E. Phage Host Range and Efficiency of Plating. In Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions; Clokie, M.R.J., Kropinski, A.M., Eds.; Methods in Molecular BiologyTM; Humana Press: Totowa, NJ, USA, 2009; pp. 141–149. ISBN 978-1-60327-164-6. [Google Scholar]
- Yazdi, M.; Bouzari, M.; Ghaemi, E.A.; Shahin, K. Isolation, Characterization and Genomic Analysis of a Novel Bacteriophage VB_EcoS-Golestan Infecting Multidrug-Resistant Escherichia Coli Isolated from Urinary Tract Infection. Sci. Rep. 2020, 10, 7690. [Google Scholar] [CrossRef]
- Mardiana, M.; Teh, S.-H.; Lin, L.-C.; Lin, N.-T. Isolation and Characterization of a Novel Siphoviridae Phage, vB_AbaS_TCUP2199, Infecting Multidrug-Resistant Acinetobacter Baumannii. Viruses 2022, 14, 1240. [Google Scholar] [CrossRef] [PubMed]
- Tabassum, R.; Shafique, M.; Khawaja, K.A.; Alvi, I.A.; Rehman, Y.; Sheik, C.S.; Abbas, Z.; Rehman, S. ur Complete Genome Analysis of a Siphoviridae Phage TSK1 Showing Biofilm Removal Potential against Klebsiella Pneumoniae. Sci. Rep. 2018, 8, 17904. [Google Scholar] [CrossRef] [PubMed]
- Swanson, M.M.; Reavy, B.; Makarova, K.S.; Cock, P.J.; Hopkins, D.W.; Torrance, L.; Koonin, E.V.; Taliansky, M. Novel Bacteriophages Containing a Genome of Another Bacteriophage within Their Genomes. PLoS ONE 2012, 7, e40683. [Google Scholar] [CrossRef] [PubMed]
- Kans, J. Entrez Direct: E-Utilities on the Unix Command Line; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2023. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Felsenstein, J. Maximum Likelihood and Minimum-Steps Methods for Estimating Evolutionary Trees from Data on Discrete Characters. Syst. Biol. 1973, 22, 240–249. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A Multiple Sequence Alignment Method with Reduced Time and Space Complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Moraru, C.; Varsani, A.; Kropinski, A.M. VIRIDIC—A Novel Tool to Calculate the Intergenomic Similarities of Prokaryote-Infecting Viruses. Viruses 2020, 12, 1268. [Google Scholar] [CrossRef]
- Baral, B. Phages against Killer Superbugs: An Enticing Strategy against Antibiotics-Resistant Pathogens. Front. Pharmacol. 2023, 14, 1036051. [Google Scholar] [CrossRef] [PubMed]
- Ezzatpour, S.; Mondragon Portocarrero, A.d.C.; Cardelle-Cobas, A.; Lamas, A.; López-Santamarina, A.; Miranda, J.M.; Aguilar, H.C. The Human Gut Virome and Its Relationship with Nontransmissible Chronic Diseases. Nutrients 2023, 15, 977. [Google Scholar] [CrossRef]
- Aggarwala, V.; Liang, G.; Bushman, F.D. Viral Communities of the Human Gut: Metagenomic Analysis of Composition and Dynamics. Mob. DNA 2017, 8, 12. [Google Scholar] [CrossRef]
- Li, N.; Ma, W.-T.; Pang, M.; Fan, Q.-L.; Hua, J.-L. The Commensal Microbiota and Viral Infection: A Comprehensive Review. Front. Immunol. 2019, 10, 1551. [Google Scholar] [CrossRef] [PubMed]
- Hibstu, Z.; Belew, H.; Akelew, Y.; Mengist, H.M. Phage Therapy: A Different Approach to Fight Bacterial Infections. Biologics 2022, 16, 173–186. [Google Scholar] [CrossRef]
- Khan Mirzaei, M.; Nilsson, A.S. Isolation of Phages for Phage Therapy: A Comparison of Spot Tests and Efficiency of Plating Analyses for Determination of Host Range and Efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef]
- Ye, M.; Sun, M.; Huang, D.; Zhang, Z.; Zhang, H.; Zhang, S.; Hu, F.; Jiang, X.; Jiao, W. A Review of Bacteriophage Therapy for Pathogenic Bacteria Inactivation in the Soil Environment. Environ. Int. 2019, 129, 488–496. [Google Scholar] [CrossRef]
- Abdulhussein, A.A.; Abdulsattar, B.O. Isolation and Characterization of Two Novel Phages as a Possible Therapeutic Alternative against Multi-Drug Resistant E. Coli. Gene Rep. 2022, 28, 101644. [Google Scholar] [CrossRef]
- Kulshrestha, M.; Tiwari, M.; Tiwari, V. Bacteriophage Therapy against ESKAPE Bacterial Pathogens: Current Status, Strategies, Challenges, and Future Scope. Microb. Pathog. 2024, 186, 106467. [Google Scholar] [CrossRef]
- Mani, I. Chapter Seven—Phage and Phage Cocktails Formulations. In Progress in Molecular Biology and Translational Science; Singh, V., Ed.; Phage Therapy—Part A; Academic Press: Cambridge, MA, USA, 2023; Volume 200, pp. 159–169. [Google Scholar]
- Osman, A.-H.; Kotey, F.C.N.; Odoom, A.; Darkwah, S.; Yeboah, R.K.; Dayie, N.T.K.D.; Donkor, E.S. The Potential of Bacteriophage-Antibiotic Combination Therapy in Treating Infections with Multidrug-Resistant Bacteria. Antibiotics 2023, 12, 1329. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Yuan, Y. Characterization of a Newly Isolated Phage Infecting Pathogenic Escherichia Coli and Analysis of Its Mosaic Structural Genes. Sci. Rep. 2018, 8, 8086. [Google Scholar] [CrossRef]
- Abdelrahman, F.; Rezk, N.; Fayez, M.S.; Abdelmoteleb, M.; Atteya, R.; Elhadidy, M.; El-Shibiny, A. Isolation, Characterization, and Genomic Analysis of Three Novel E. Coli Bacteriophages That Effectively Infect E. Coli O18. Microorganisms 2022, 10, 589. [Google Scholar] [CrossRef] [PubMed]
- Hon, K.; Liu, S.; Camens, S.; Bouras, G.S.; Psaltis, A.J.; Wormald, P.-J.; Vreugde, S. APTC-EC-2A: A Lytic Phage Targeting Multidrug Resistant E. Coli Planktonic Cells and Biofilms. Microorganisms 2022, 10, 102. [Google Scholar] [CrossRef]
- Fan, C.; Tie, D.; Sun, Y.; Jiang, J.; Huang, H.; Gong, Y.; Zhao, C. Characterization and Genomic Analysis of Escherichia Coli O157:H7 Bacteriophage FEC14, a New Member of Genus Kuttervirus. Curr. Microbiol. 2021, 78, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Olsen, N.S.; Forero-Junco, L.; Kot, W.; Hansen, L.H. Exploring the Remarkable Diversity of Culturable Escherichia Coli Phages in the Danish Wastewater Environment. Viruses 2020, 12, 986. [Google Scholar] [CrossRef]
- Sváb, D.; Falgenhauer, L.; Chakraborty, T.; Tóth, I. Complete Genome Sequences of Novel Bovine T4, Rv5-Like, and Dhillonviruses Effective against Escherichia Coli O157. Microbiol. Resour. Announc. 2021, 10, e01261-20. [Google Scholar] [CrossRef]
- Skaradzińska, A.; Śliwka, P.; Kuźmińska-Bajor, M.; Skaradziński, G.; Rząsa, A.; Friese, A.; Roschanski, N.; Murugaiyan, J.; Roesler, U.H. The Efficacy of Isolated Bacteriophages from Pig Farms against ESBL/AmpC-Producing Escherichia Coli from Pig and Turkey Farms. Front. Microbiol. 2017, 8, 530. [Google Scholar] [CrossRef] [PubMed]
- Sultan-Alolama, M.I.; Amin, A.; El-Tarabily, K.A.; Vijayan, R. Characterization and Genomic Analysis of Escherichia Coli O157:H7 Phage UAE_MI-01 Isolated from Birds. Int. J. Mol. Sci. 2022, 23, 14846. [Google Scholar] [CrossRef]
- Sellvam, D.; Lau, N.S.; Arip, Y.M. Genome Organization of Escherichia Phage YD-2008.s: A New Entry to Siphoviridae Family. Trop. Life Sci. Res. 2018, 29, 37–50. [Google Scholar] [CrossRef]
- Li, S.; Lu, S.; Huang, H.; Tan, L.; Ni, Q.; Shang, W.; Yang, Y.; Hu, Z.; Zhu, J.; Li, M.; et al. Comparative Analysis and Characterization of Enterobacteria Phage SSL-2009a and ‘HK578likevirus’ Bacteriophages. Virus Res. 2019, 259, 77–84. [Google Scholar] [CrossRef]
- Chen, L.; Liu, Q.; Fan, J.; Yan, T.; Zhang, H.; Yang, J.; Deng, D.; Liu, C.; Wei, T.; Ma, Y. Characterization and Genomic Analysis of ValSw3-3, a New Siphoviridae Bacteriophage Infecting Vibrio Alginolyticus. J. Virol. 2020, 94, e00066-20. [Google Scholar] [CrossRef]
- Qadri, I.; Harakeh, S.; Demeke Teklemariam, A.; Al Amri, T.; Al-Hindi, R. Isolation and Identification of a Wastewater Siphoviridae Bacteriophage Targeting Multidrug-Resistant Klebsiella Pneumoniae. Jundishapur J. Microbiol. 2021, 14, e118910. [Google Scholar] [CrossRef]
- Mondal, P.; Mallick, B.; Dutta, M.; Dutta, S. Isolation, Characterization, and Application of a Novel Polyvalent Lytic Phage STWB21 against Typhoidal and Nontyphoidal Salmonella Spp. Front. Microbiol. 2022, 13, 980025. [Google Scholar] [CrossRef]
- Chen, X.; Xi, Y.; Zhang, H.; Wang, Z.; Fan, M.; Liu, Y.; Wu, W. Characterization and Adsorption of Lactobacillus Virulent Phage P1. J. Dairy Sci. 2016, 99, 6995–7001. [Google Scholar] [CrossRef]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A Roadmap for Genome-Based Phage Taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef]
- Taslem Mourosi, J.; Awe, A.; Guo, W.; Batra, H.; Ganesh, H.; Wu, X.; Zhu, J. Understanding Bacteriophage Tail Fiber Interaction with Host Surface Receptor: The Key “Blueprint” for Reprogramming Phage Host Range. Int. J. Mol. Sci. 2022, 23, 12146. [Google Scholar] [CrossRef]
- Zhang, J.; Ning, H.; Lin, H.; She, J.; Wang, L.; Jing, Y.; Wang, J. Expansion of the Plaquing Host Range and Improvement of the Absorption Rate of a T5-like Salmonella Phage by Altering the Long Tail Fibers. Appl. Environ. Microbiol. 2022, 88, e00895-22. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhang, L.; Abdelgader, S.A.; Yu, L.; Xu, J.; Yao, H.; Lu, C.; Zhang, W. Alterations in Gp37 Expand the Host Range of a T4-Like Phage. Appl. Environ. Microbiol. 2017, 83, e01576-17. [Google Scholar] [CrossRef]
- Burrowes, B.H.; Molineux, I.J.; Fralick, J.A. Directed in Vitro Evolution of Therapeutic Bacteriophages: The Appelmans Protocol. Viruses 2019, 11, 241. [Google Scholar] [CrossRef] [PubMed]
- Górniak, M.; Zalewska, A.; Jurczak-Kurek, A. Recombination Events in Putative Tail Fibre Gene in Litunavirus Phages Infecting Pseudomonas Aeruginosa and Their Phylogenetic Consequences. Viruses 2022, 14, 2669. [Google Scholar] [CrossRef]
| Sample No. | Bacterial Strains/ Isolates * | Samples Source | Antibiotics Resistance $ | Phylogroup & | Phage Lytic Activity ¶ |
|---|---|---|---|---|---|
| 1 | Escherichia coli 823 | Wastewater treatment plant | S | A | + |
| 2 | Escherichia coli 668 | Wastewater treatment plant | S | B1 | + |
| 3 | Escherichia coli 824 | Wastewater treatment plant | S | B2 | − |
| 4 | Escherichia coli 663 | Wastewater treatment plant | S | D | |
| 5 | Escherichia coli 792 | Wastewater treatment plant | MDR | A | − |
| 6 | Escherichia coli 494 | Wastewater treatment plant | MDR | B1 | − |
| 7 | Escherichia coli 810 | Wastewater treatment plant | MDR | B2 | − |
| 8 | Escherichia coli 638 | Wastewater treatment plant | MDR | D | − |
| 9 | Escherichia coli 640 | Wastewater treatment plant | R | A | − |
| 10 | Escherichia coli 643 | Wastewater treatment plant | R | B1 | − |
| 11 | Escherichia coli 809 | Wastewater treatment plant | R | B2 | − |
| 12 | Escherichia coli 635 | Wastewater treatment plant | R | D | − |
| 13 | Escherichia coli 865 | Hospital wastewater | WT | A | + |
| 14 | Escherichia coli 866 | Hospital wastewater | WT | B1 | − |
| 15 | Escherichia coli 843 | Hospital wastewater | WT | B2 | − |
| 16 | Escherichia coli 580 | Hospital wastewater | WT | D | − |
| 17 | Escherichia coli 426 | Hospital wastewater | MDR | D | + |
| 18 | Escherichia coli 858 | Hospital wastewater | MDR | A | − |
| 19 | Escherichia coli 546 | Hospital wastewater | MDR | B2 | − |
| 20 | Escherichia coli 576 | Hospital wastewater | R | B2 | − |
| 21 | Escherichia coli 545 | Hospital wastewater | R | D | + |
| 22 | Escherichia coli 674 | Hospital wastewater | R | A | − |
| 23 | Escherichia coli 759 | River water | WT | A | − |
| 24 | Escherichia coli 774 | River water | WT | B1 | − |
| 25 | Escherichia coli 624 | River water | WT | B2 | − |
| 26 | Escherichia coli 769 | River water | WT | D | − |
| 27 | Escherichia coli 472 | River water | MDR | A | − |
| 28 | Escherichia coli 607 | River water | MDR | B1 | − |
| 29 | Escherichia coli 737 | River water | MDR | B2 | − |
| 30 | Escherichia coli 408 | River water | MDR | D | − |
| 31 | Escherichia coli 614 | River water | R | A | − |
| 32 | Escherichia coli 372 | River water | R | D | − |
| 33 | Escherichia coli 743 | River water | R | B2 | − |
| 34 | Escherichia coli 784 | River water | R | B1 | − |
| 35 | Escherichia coli 117 | Clinical | MDR | A | + |
| 36 | Escherichia coli 60 | Clinical | MDR | B2 | + |
| 37 | Escherichia coli 203 | Clinical | MDR | D | + |
| 38 | Escherichia coli 325 | Clinical | R | A | + |
| 39 | Escherichia coli 5 | Clinical | R | A | − |
| 40 | Escherichia coli 264 | Clinical | R | B1 | − |
| 41 | Escherichia coli 294 | Clinical | R | B2 | − |
| 42 | Escherichia coli 378 | Clinical | R | B2 | + |
| 43 | Escherichia coli 313 | Clinical | R | D | − |
| 44 | Escherichia coli 324 | Clinical | S | A | + |
| 45 | Escherichia coli 368 | Clinical | S | D | − |
| 46 | Escherichia coli 387 | Clinical | S | B2 | − |
| 47 | Escherichia coli 301 | Clinical | S | B1 | − |
| 48 | Escherichia coli 25922 | Reference strain from Becton Dickinson, France S.A.S | S | − | + |
| 49 | Escherichia coli 35218 | Reference strain from Becton Dickinson, France S.A.S | S | − | − |
| 50 | Escherichia coli 13846 | Reference strain from Becton Dickinson, France S.A.S | S | − | − |
| 51 | Escherichia coli O157:H7 | Clinical | MDR | − | − |
| 52 | Klebsiella pneumoniae 13883 | Reference strain from Becton Dickinson, France S.A.S | S | − | − |
| 53 | Klebsiella pneumoniae 700603 | Reference strain from Becton Dickinson, France S.A.S | S | − | − |
| 54 | Pseudomonas aeruginosa 27853 | Reference strain from Becton Dickinson, France S.A.S | S | − | − |
| 55 | Yersinia enterocolitica 9610 | Reference strain from Becton Dickinson, France S.A.S | S | − | − |
| 56 | Acinetobacter baumannii 17978 | Reference strain from Becton Dickinson, France S.A.S | S | − | − |
| 57 | Acinetobacter baumannii 19668 | Reference strain from Becton Dickinson, France S.A.S | S | − | − |
| Phage Name | BLASTp Coverage (%) | BLASTp Percent Identity (%) | Genus | Family/ Morphotype | NCBI Accession Number |
|---|---|---|---|---|---|
| vB_EcoS_SA32RD | 98 | 72.5 | Tunavirus | Drexlerviridae | UIU27553.1 |
| PGN6866 | 92 | 74.22 | Kuravirus | podoviruses | QKL16987.1 |
| vB_EcoP_YF01 | 92 | 72.89 | Kuravirus | podoviruses | WBF04932.1 |
| IME267 | 92 | 72.44 | Kuravirus | podoviruses | YP_010673185.1 |
| MLP3 | 92 | 72.89 | Kuravirus | podoviruses | UEN68517.1 |
| vB_EcoP-101114UKE3 | 92 | 72.44 | Kuravirus | podoviruses | YP_010673043.1 |
| νB_EcoP_SU7 | 92 | 72 | Kuravirus | podoviruses | YP_010672804.1 |
| vB_EcoS_011D5 | 99 | 38.7 | Dhillonvirus | siphoviruses | QMP82830.1 |
| vB_EcoS_L-h 1M | 99 | 37.79 | Dhillonvirus | siphoviruses | UNY42316.1 |
| vB_EcoS_SA30RD | 72 | 48.02 | Tunavirus | Drexlerviridae | UIU27628.1 |
| vB_EcoS_Chapo | 72 | 47.46 | Tunavirus | Drexlerviridae | QLF82390.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vasileiadis, A.; Bozidis, P.; Konstantinidis, K.; Kesesidis, N.; Potamiti, L.; Kolliopoulou, A.; Beloukas, A.; Panayiotidis, M.I.; Havaki, S.; Gorgoulis, V.G.; et al. A Novel Dhillonvirus Phage against Escherichia coli Bearing a Unique Gene of Intergeneric Origin. Curr. Issues Mol. Biol. 2024, 46, 9312-9329. https://doi.org/10.3390/cimb46090551
Vasileiadis A, Bozidis P, Konstantinidis K, Kesesidis N, Potamiti L, Kolliopoulou A, Beloukas A, Panayiotidis MI, Havaki S, Gorgoulis VG, et al. A Novel Dhillonvirus Phage against Escherichia coli Bearing a Unique Gene of Intergeneric Origin. Current Issues in Molecular Biology. 2024; 46(9):9312-9329. https://doi.org/10.3390/cimb46090551
Chicago/Turabian StyleVasileiadis, Anastasios, Petros Bozidis, Konstantinos Konstantinidis, Nikolaos Kesesidis, Louiza Potamiti, Anna Kolliopoulou, Apostolos Beloukas, Mihalis I. Panayiotidis, Sophia Havaki, Vassilis G. Gorgoulis, and et al. 2024. "A Novel Dhillonvirus Phage against Escherichia coli Bearing a Unique Gene of Intergeneric Origin" Current Issues in Molecular Biology 46, no. 9: 9312-9329. https://doi.org/10.3390/cimb46090551
APA StyleVasileiadis, A., Bozidis, P., Konstantinidis, K., Kesesidis, N., Potamiti, L., Kolliopoulou, A., Beloukas, A., Panayiotidis, M. I., Havaki, S., Gorgoulis, V. G., Gartzonika, K., & Karakasiliotis, I. (2024). A Novel Dhillonvirus Phage against Escherichia coli Bearing a Unique Gene of Intergeneric Origin. Current Issues in Molecular Biology, 46(9), 9312-9329. https://doi.org/10.3390/cimb46090551











