Novel Mutations in AKT1 Gene in Prostate Cancer Patients in Jordan
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction
2.3. Primer Design and PCR Amplification
2.4. DNA Sequencing
2.5. Data Analysis
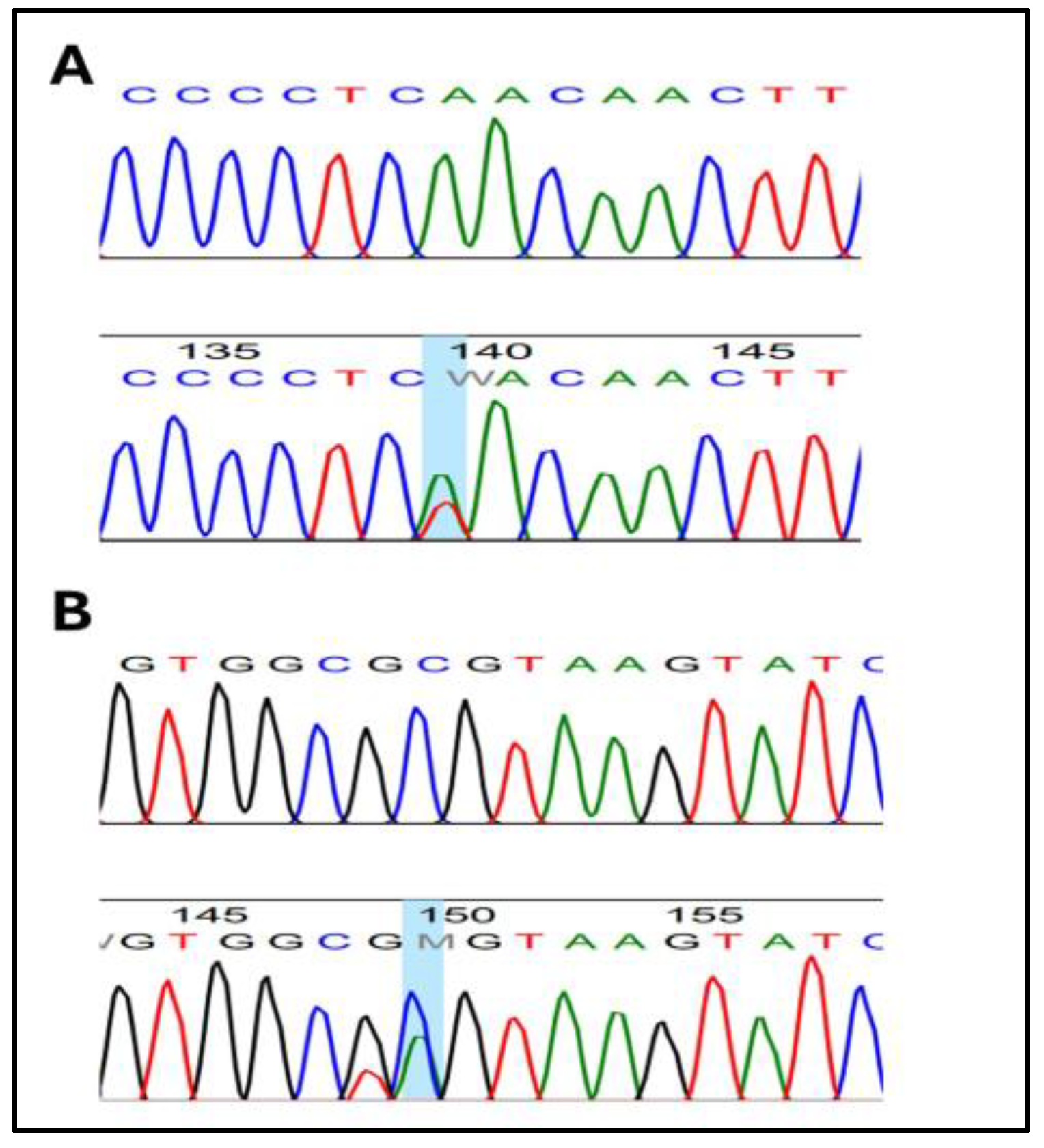
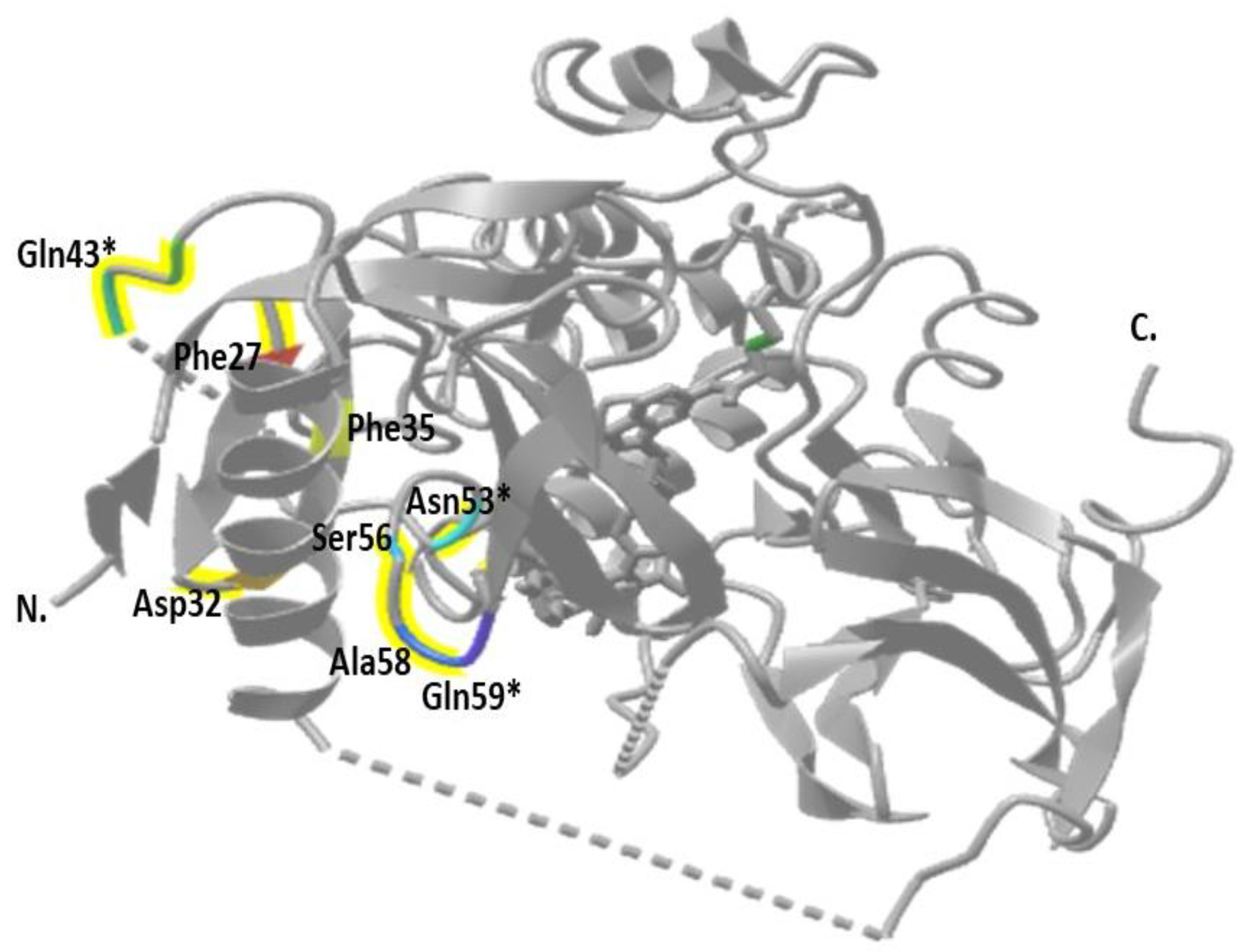
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rizzo, A.; Santoni, M.; Mollica, V.; Fiorentino, M.; Brandi, G.; Massari, F. Microbiota and prostate cancer. Semin. Cancer Biol. 2022, 86 Pt 3, 1058–1065. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Fleshner, K.; Carlsson, S.V.; Roobol, M.J. The effect of the USPSTF PSA screening recommendation on prostate cancer incidence patterns in the USA. Nat. Rev. Urol. 2017, 14, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Hilal, L.; Shahait, M.; Mukherji, D.; Charafeddine, M.; Farhat, Z.; Temraz, S.; Khauli, R.; Shamseddine, A. Prostate Cancer in the Arab World: A View from the Inside. Clin. Genitourin. Cancer 2015, 13, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Daher, M.; Telvizian, T.; Dagher, C.; Abdul-Sater, Z.; Massih, S.A.; Chediak, A.E.; Charafeddine, M.; Shahait, M.; Alameddine, R.; Temraz, S.; et al. High rates of advanced prostate cancer in the Middle East: Analysis from a tertiary care center. Urol. Ann. 2021, 13, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Alqudah, M.A.Y.; Al-Samman, R.; Matalgah, O.; Abu Farhah, R. Early Detection of Prostate Cancer: Self-Reported Knowledge and Attitude of Physicians in Jordan. Inquiry 2022, 59, 469580221095822. [Google Scholar] [CrossRef] [PubMed]
- Grozescu, T.; Popa, F. Prostate cancer between prognosis and adequate/proper therapy. J. Med. Life 2017, 10, 5–12. [Google Scholar]
- Al-Abdin, O.Z.; Al-Beeshi, I.Z. Prostate cancer in the Arab population. An overview. Saudi Med. J. 2018, 39, 453–458. [Google Scholar] [CrossRef]
- Testa, U.; Castelli, G.; Pelosi, E. Cellular and Molecular Mechanisms Underlying Prostate Cancer Development: Therapeutic Implications. Medicines 2019, 6, 82. [Google Scholar] [CrossRef]
- Alves, C.P.; Dey-Guha, I.; Kabraji, S.; Yeh, A.C.; Talele, N.P.; Solé, X.; Chowdhury, J.; Mino-Kenudson, M.; Loda, M.; Sgroi, D. AKT1low quiescent cancer cells promote solid tumor growth. Mol. Cancer Ther. 2018, 17, 254–263. [Google Scholar] [CrossRef]
- Dunlap, J.; Le, C.; Shukla, A.; Patterson, J.; Presnell, A.; Heinrich, M.C.; Corless, C.L.; Troxell, M.L. Phosphatidylinositol-3-kinase and AKT1 mutations occur early in breast carcinoma. Breast Cancer Res. Treat. 2010, 120, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Mishima, C.; Kagara, N.; Ikeda, J.-i.; Morii, E.; Miyake, T.; Tanei, T.; Naoi, Y.; Shimoda, M.; Shimazu, K.; Kim, S.J. Mutational analysis of AKT1 and PIK3CA in intraductal papillomas of the breast with special reference to cellular components. Am. J. Pathol. 2018, 188, 1106–1112. [Google Scholar] [CrossRef] [PubMed]
- Troxell, M.L.; Levine, J.; Beadling, C.; Warrick, A.; Dunlap, J.; Presnell, A.; Patterson, J.; Shukla, A.; Olson, N.R.; Heinrich, M.C. High prevalence of PIK3CA/AKT pathway mutations in papillary neoplasms of the breast. Mod. Pathol. 2010, 23, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Cheung, M.; Testa, J.R. Diverse mechanisms of AKT pathway activation in human malignancy. Curr. Cancer Drug Targets 2013, 13, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Virtakoivu, R.; Pellinen, T.; Rantala, J.K.; Perälä, M.; Ivaska, J. Distinct roles of AKT isoforms in regulating β1-integrin activity, migration, and invasion in prostate cancer. Mol. Biol. Cell 2012, 23, 3357–3369. [Google Scholar] [CrossRef]
- Mundi, P.S.; Sachdev, J.; McCourt, C.; Kalinsky, K. AKT in cancer: New molecular insights and advances in drug development. Br. J. Clin. Pharmacol. 2016, 82, 943–956. [Google Scholar] [CrossRef]
- Chandarlapaty, S.; Sawai, A.; Scaltriti, M.; Rodrik-Outmezguine, V.; Grbovic-Huezo, O.; Serra, V.; Majumder, P.K.; Baselga, J.; Rosen, N. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell 2011, 19, 58–71. [Google Scholar] [CrossRef]
- Feng, J.; Park, J.; Cron, P.; Hess, D.; Hemmings, B.A. Identification of a PKB/Akt hydrophobic motif Ser-473 kinase as DNA-dependent protein kinase. J. Biol. Chem. 2004, 279, 41189–41196. [Google Scholar] [CrossRef]
- Hollander, M.C.; Maier, C.R.; Hobbs, E.A.; Ashmore, A.R.; Linnoila, R.I.; Dennis, P.A. Akt1 deletion prevents lung tumorigenesis by mutant K-ras. Oncogene 2011, 30, 1812–1821. [Google Scholar] [CrossRef]
- Taylor, S.S.; Kornev, A.P. Protein kinases: Evolution of dynamic regulatory proteins. Trends Biochem. Sci. 2011, 36, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Satoh, N.; Nakamura, M.; Suzuki, M.; Suzuki, A.; Seki, G.; Horita, S. Roles of Akt and SGK1 in the Regulation of Renal Tubular Transport. BioMed Res. Int. 2015, 2015, 971697. [Google Scholar] [CrossRef]
- Hemmings, B.A. Akt signaling: Linking membrane events to life and death decisions. Science 1997, 275, 628–630. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Hu, S.; Wang, C.; Song, J.; Chen, C.; Fan, Y.; Ben-David, Y.; Pan, W. Fangchinoline derivatives induce cell cycle arrest and apoptosis in human leukemia cell lines via suppression of the PI3K/AKT and MAPK signaling pathway. Eur. J. Med. Chem. 2020, 186, 111898. [Google Scholar] [CrossRef] [PubMed]
- Carpten, J.D.; Faber, A.L.; Horn, C.; Donoho, G.P.; Briggs, S.L.; Robbins, C.M.; Hostetter, G.; Boguslawski, S.; Moses, T.Y.; Savage, S. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature 2007, 448, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Bellacosa, A.; Etro, D.; Neri, L.M. Mutations of the PIK3CA gene in ovarian and breast cancer. Women’s Oncol. Rev. 2005, 5, 223–225. [Google Scholar] [CrossRef]
- Sjöblom, T.; Jones, S.; Wood, L.D.; Parsons, D.W.; Lin, J.; Barber, T.D.; Mandelker, D.; Leary, R.J.; Ptak, J.; Silliman, N.; et al. The consensus coding sequences of human breast and colorectal cancers. Science 2006, 314, 268–274. [Google Scholar] [CrossRef]
- Sun, M.; Wang, G.; Paciga, J.E.; Feldman, R.I.; Yuan, Z.Q.; Ma, X.L.; Shelley, S.A.; Jove, R.; Tsichlis, P.N.; Nicosia, S.V.; et al. AKT1/PKBalpha kinase is frequently elevated in human cancers and its constitutive activation is required for oncogenic transformation in NIH3T3 cells. Am. J. Pathol. 2001, 159, 431–437. [Google Scholar] [CrossRef]
- Ahmed, M.; Eeles, R. Germline genetic profiling in prostate cancer: Latest developments and potential clinical applications. Future Sci. OA 2016, 2, FSO87. [Google Scholar] [CrossRef]
- Al Bashir, S.; Alzoubi, A.; Alfaqih, M.A.; Kheirallah, K.; Smairat, A.; Haddad, H.; Al-Dwairy, A.; Fawwaz, B.A.; Alzoubi, M.; Trpkov, K. PTEN loss in a prostate cancer cohort from Jordan. Appl. Immunohistochem. Mol. Morphol. 2020, 28, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Al Zoubi, M.S.; Otoum, R.; Alorjani, M.S.; Al Bashir, S.; Al Trad, B.; Abualrja, M.I.; Al-Khatib, S.M.; Al-Batayneh, K. TP53, SPOP and PIK3CA genes status in prostate cancer. Asian Pac. J. Cancer Prev. APJCP 2020, 21, 3365. [Google Scholar] [CrossRef] [PubMed]
- Alorjani, M.; Aburub, M.; Al-Trad, B.; Al Hamad, M.; AbuAlarja, M.; Al Bashir, S.; Al-Batayneh, K.; Al Zoubi, M. The prevalence of CHEK1 and CHEK2 mutations in prostate cancer: A retrospective cohort study. Med. Arch. 2023, 77, 8. [Google Scholar] [CrossRef]
- Cuzick, J.; Thorat, M.A.; Andriole, G.; Brawley, O.W.; Brown, P.H.; Culig, Z.; Eeles, R.A.; Ford, L.G.; Hamdy, F.C.; Holmberg, L.; et al. Prevention and early detection of prostate cancer. Lancet Oncol. 2014, 15, e484–e492. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.; Alkhushaym, N.; Fallatah, S.; Althagafi, A.; Aljadeed, R.; Alsowaida, Y.; Jeter, J.; Martin, J.R.; Babiker, H.M.; McBride, A. The association of BRCA1 and BRCA2 mutations with prostate cancer risk, frequency, and mortality: A meta-analysis. Prostate 2019, 79, 880–895. [Google Scholar] [CrossRef] [PubMed]
- Salim, A.-Z.M.; Khalid, A.-B.; Bahaa, A.-T.; Mohammed, A.; Samir, A.-B.; Mohammad, A.-H.; Riyad, M.; Ismail, M. Polymorphisms of 5′-UTR of rad51 gene in prostate cancer. Экoлoгическая генетика 2018, 16, 24–29. [Google Scholar]
- Adjakly, M.; Ngollo, M.; Dagdemir, A.; Judes, G.; Pajon, A.; Karsli-Ceppioglu, S.; Penault-Llorca, F.; Boiteux, J.P.; Bignon, Y.J.; Guy, L.; et al. Prostate cancer: The main risk and protective factors-Epigenetic modifications. In Annales D’endocrinologie; Elsevier Masson: Paris, France, 2015; Volume 76, pp. 25–41. [Google Scholar] [CrossRef]
- Weiss, R.E.; Refetoff, S. Genetic Diagnosis of Endocrine Disorders; Academic Press: Cambridge, MA, USA, 2015. [Google Scholar]
- Powis, G.; Meuillet, E.J.; Indarte, M.; Booher, G.; Kirkpatrick, L. Pleckstrin Homology [PH] domain, structure, mechanism, and contribution to human disease. Biomed. Pharmacother. 2023, 165, 115024. [Google Scholar] [CrossRef]
- Brugge, J.; Hung, M.-C.; Mills, G.B. A new mutational AKTivation in the PI3K pathway. Cancer Cell 2007, 12, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Askham, J.; Platt, F.; Chambers, P.; Snowden, H.; Taylor, C.; Knowles, M. AKT1 mutations in bladder cancer: Identification of a novel oncogenic mutation that can co-operate with E17K. Oncogene 2010, 29, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Bleeker, F.; Felicioni, L.; Buttitta, F.; Lamba, S.; Cardone, L.; Rodolfo, M.; Scarpa, A.; Leenstra, S.; Frattini, M.; Barbareschi, M. AKT1E17K in human solid tumours. Oncogene 2008, 27, 5648–5650. [Google Scholar] [CrossRef]
- Boormans, J.; Korsten, H.; Ziel-Van Der Made, A.; Van Leenders, G.; Verhagen, P.; Trapman, J. E17K substitution in AKT1 in prostate cancer. Br. J. Cancer 2010, 102, 1491–1494. [Google Scholar] [CrossRef] [PubMed]
- Do, H.; Salemi, R.; Murone, C.; Mitchell, P.L.; Dobrovic, A. Rarity of AKT1 and AKT3 E17K mutations in squamous cell carcinoma of lung. Cell Cycle 2010, 9, 4411–4412. [Google Scholar] [CrossRef]
- Do, H.; Solomon, B.; Mitchell, P.L.; Fox, S.B.; Dobrovic, A. Detection of the transforming AKT1 mutation E17K in non-small cell lung cancer by high resolution melting. BMC Res. Notes 2008, 1, 14. [Google Scholar] [CrossRef] [PubMed]
- Malanga, D.; Scrima, M.; De Marco, C.; Fabiani, F.; De Rosa, N.; De Gisi, S.; Malara, N.; Savino, R.; Rocco, G.; Chiappetta, G. Activating E17K mutation in the gene encoding the protein kinase AKT in a subset of squamous cell carcinoma of the lung. Cell Cycle 2008, 7, 665–669. [Google Scholar] [CrossRef]
- Smyth, L.M.; Zhou, Q.; Nguyen, B.; Yu, C.; Lepisto, E.M.; Arnedos, M.; Hasset, M.J.; Lenoue-Newton, M.L.; Blauvelt, N.; Dogan, S. Characteristics and outcome of AKT1 E17K-mutant breast cancer defined through AACR project GENIE, a clinicogenomic registry. Cancer Discov. 2020, 10, 526–535. [Google Scholar] [CrossRef]
- Strickland, M.R.; Gill, C.M.; Nayyar, N.; D’Andrea, M.R.; Thiede, C.; Juratli, T.A.; Schackert, G.; Borger, D.R.; Santagata, S.; Frosch, M.P. Targeted sequencing of SMO and AKT1 in anterior skull base meningiomas. J. Neurosurg. 2017, 127, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Yesilöz, Ü.; Kirches, E.; Hartmann, C.; Scholz, J.; Kropf, S.; Sahm, F.; Nakamura, M.; Mawrin, C. Frequent AKT1 E17K mutations in skull base meningiomas are associated with mTOR and ERK1/2 activation and reduced time to tumor recurrence. Neuro-Oncology 2017, 19, 1088–1096. [Google Scholar] [CrossRef]
- Youngblood, M.W.; Miyagishima, D.F.; Jin, L.; Gupte, T.; Li, C.; Duran, D.; Montejo, J.D.; Zhao, A.; Sheth, A.; Tyrtova, E. Associations of meningioma molecular subgroup and tumor recurrence. Neuro-Oncology 2021, 23, 783–794. [Google Scholar] [CrossRef] [PubMed]
- Boetto, J.; Bielle, F.; Sanson, M.; Peyre, M.; Kalamarides, M. SMO mutation status defines a distinct and frequent molecular subgroup in olfactory groove meningiomas. Neuro-Oncology 2017, 19, 345–351. [Google Scholar]
- De Marchi, F.; Haley, L.; Fryer, H.; Ibrahim, J.; Beierl, K.; Zheng, G.; Gocke, C.D.; Eshleman, J.R.; Belchis, D.; Illei, P. Clinical validation of coexisting activating mutations within EGFR, mitogen-activated protein kinase, and phosphatidylinositol 3-kinase pathways in lung cancers. Arch. Pathol. Lab. Med. 2019, 143, 174–182. [Google Scholar] [CrossRef] [PubMed]
- De Marco, C.; Malanga, D.; Rinaldo, N.; De Vita, F.; Scrima, M.; Lovisa, S.; Fabris, L.; Carriero, M.V.; Franco, R.; Rizzuto, A. Mutant AKT1-E17K is oncogenic in lung epithelial cells. Oncotarget 2015, 6, 39634. [Google Scholar] [CrossRef]
- Kim, M.; Jeong, E.; Yoo, N.; Lee, S. Mutational analysis of oncogenic AKT E17K mutation in common solid cancers and acute leukaemias. Br. J. Cancer 2008, 98, 1533–1535. [Google Scholar] [CrossRef]
- Rekhtman, N.; Paik, P.K.; Arcila, M.E.; Tafe, L.J.; Oxnard, G.R.; Moreira, A.L.; Travis, W.D.; Zakowski, M.F.; Kris, M.G.; Ladanyi, M. Clarifying the Spectrum of Driver Oncogene Mutations in Biomarker-Verified Squamous Carcinoma of Lung: Lack of EGFR/KRA S and Presence of PIK3CA/AKT1 Mutations. Clin. Cancer Res. 2012, 18, 1167–1176. [Google Scholar] [CrossRef] [PubMed]
- De Marco, C.; Laudanna, C.; Rinaldo, N.; Oliveira, D.M.; Ravo, M.; Weisz, A.; Ceccarelli, M.; Caira, E.; Rizzuto, A.; Zoppoli, P. Specific gene expression signatures induced by the multiple oncogenic alterations that occur within the PTEN/PI3K/AKT pathway in lung cancer. PLoS ONE 2017, 12, e0178865. [Google Scholar] [CrossRef]
- Eom, H.S.; Kim, M.S.; Hur, S.Y.; Yoo, N.J.; Lee, S.H. Absence of oncogenic AKT1 E17K mutation in prostate, esophageal, laryngeal and urothelial carcinomas, hepatoblastomas, gastrointestinal stromal tumors and malignant meningiomas. Acta Oncol. 2009, 48, 1084–1085. [Google Scholar] [CrossRef] [PubMed]
- Herberts, C.; Murtha, A.J.; Fu, S.; Wang, G.; Schönlau, E.; Xue, H.; Lin, D.; Gleave, A.; Yip, S.; Angeles, A. Activating AKT1 and PIK3CA mutations in metastatic castration-resistant prostate cancer. Eur. Urol. 2020, 78, 834–844. [Google Scholar] [CrossRef] [PubMed]
- Kirkegaard, T.; Witton, C.J.; Edwards, J.; Nielsen, K.V.; Jensen, L.B.; Campbell, F.M.; Cooke, T.G.; Bartlett, J.M. Molecular alterations in AKT1, AKT2 and AKT3 detected in breast and prostatic cancer by FISH. Histopathology 2010, 56, 203–211. [Google Scholar] [CrossRef]
- Fu, S.; Murtha, A.; Herberts, C.; Yip, S.; Angeles, A.; Hotte, S.; Alshangiti, A.; Tran, B.; Taavitsainen, S.; Annala, M. Activating mutations in AKT1/PIK3CA are associated with poor clinical outcomes in metastatic prostate cancer (mPC). Ann. Oncol. 2019, 30, v347. [Google Scholar] [CrossRef]
- Sarker, D.; Reid, A.H.; Yap, T.A.; De Bono, J.S. Targeting the PI3K/AKT pathway for the treatment of prostate cancer. Clin. Cancer Res. 2009, 15, 4799–4805. [Google Scholar] [CrossRef]
- Toren, P.; Zoubeidi, A. Targeting the PI3K/Akt pathway in prostate cancer: Challenges and opportunities. Int. J. Oncol. 2014, 45, 1793–1801. [Google Scholar] [CrossRef]

| Exon 3 | Exon 4 | |||||
|---|---|---|---|---|---|---|
| Forward 5′-3′ | GTC AGA GAG CTT AGA GGG AT | GGT CTG ACG GGT AGA GTG TG | ||||
| Reverse 5′-3′ | AGG GCA CAG GCA CTC ACA GA | ACG CAG TGC TTG TTG CTT CG | ||||
| Operation | Temp (°C) | Time (second) | Cycles | Temp (°C) | Time (second) | Cycles |
| Initial denaturation | 95 | 300 | 1 | 95 | 300 | 1 |
| Denaturation | 95 | 30 | 40 | 95 | 30 | 45 |
| Annealing | 60 | 30 | 60 | 30 | ||
| Elongation | 72 | 40 | 72 | 40 | ||
| Final elongation | 72 | 5 min | 1 | 72 | 300 | 1 |
| Product size (bp) | 375 | 309 | ||||
| Mutation Number | Exon | Position | Mutation Type | Amino Acid Change | Codon Change | Recorded Genotype (N) |
|---|---|---|---|---|---|---|
| rs1180873760 | 3 | c.36G > A | Synonymous Variant | Leu12 | CTG > CTA | G/A |
| rs371534192 | 3 | c.6C > T | Synonymous Variant | Ser2 | AGC > AGT | C/T |
| rs2140948571 | 4 | c.168T > A | Synonymous Variant | Ser56 | TCT > TCA | T/A |
| rs370287382 | 4 | c.120G > A | Synonymous Variant | Glu40 | GAG > GAA | G/A |
| rs2140949629 | 4 | c.80T > A | Missense Variant | Phe27Tyr | TTC > TAC | T/A * |
| rs749781543 | 4 | c.81C > A | Missense Variant | Phe27Leu | TTC > TTA | C/A * |
| rs1892948718 | 4 | c.172G > A | Missense Variant | Ala58Thr | GCG > ACG | C/A |
| rs2140948579 | 4 | c.167C > T | Missense Variant | Ser56Phe | TCT > TTT | C/T |
| rs753765116 | 4 | c.121C > T | Missense Variant | Arg41Trp | CGG > TGG | T/T |
| rs2140949369 | 4 | c.103T > A | Missense Variant | Phe35Leu | TTC > ATC | T/A |
| rs201636005 | 4 | c.96T > A | Missense Variant | Asp32Glu | GAT > GAA | T/A * |
| rs2140949360 | 4 | c.104T > C | Missense Variant | Phe35Tyr | TTC > TAC | T/C * |
| COSV62573749 COSM6196752 | 4 | c.127C > A | Missense Variant | Gln43Lys | CAG > AAG | C/A |
| Exon | Position | Amino Acid Change | Codon Change | Recorded Genotype | Poly-Phen2 Prediction * |
|---|---|---|---|---|---|
| 4 | c.157A > T | Asn53Tyr | TCA > TCT | A/T | Probably damaging |
| 4 | c.175C > A | Gln59Lys | CAG > AAG | C/A | Benign |
| Variables | |||||||
|---|---|---|---|---|---|---|---|
| Prostate-Specific Antigen (PSA) ng/mL | p Value | ||||||
| <4 | 4–10 | 10–20 | 20–100 | >100 | No Data | ||
| Age group | 0.15 | ||||||
| <70 years | 4 | 5 | 2 | 6 | 3 | 11 | |
| 70–80 years | 4 | 5 | 2 | 9 | 2 | 16 | |
| >80 years | 0 | 2 | 3 | 2 | 2 | 6 | |
| Gleason score | 0.24 | ||||||
| <7 | 1 | 4 | 1 | 2 | 0 | 7 | |
| =7 | 3 | 4 | 2 | 3 | 1 | 12 | |
| >7 | 4 | 2 | 4 | 11 | 6 | 14 | |
| No data | 1 | 1 | 0 | 0 | 0 | 1 | |
| Parameter | Category | AKT1 Exon 4 Mutation Status | p Value | ||
|---|---|---|---|---|---|
| N | M | W | |||
| Age years | <70 | 30 | 7 | 23 | 0.9645 |
| 70–80 | 39 | 9 | 30 | ||
| >80 | 15 | 3 | 12 | ||
| PSA level ng/mL | <4 | 8 | 1 | 7 | 0.7306 |
| 4–20 | 19 | 5 | 14 | ||
| >20 | 23 | 5 | 18 | ||
| Gleason score | <7 | 15 | 2 | 13 | 0.1313 |
| =7 | 25 | 9 | 16 | ||
| >7 | 41 | 7 | 34 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alasmar, A.; Al-Alami, Z.; Zein, S.; Al-Smadi, A.; Al Bashir, S.; Alorjani, M.S.; Al-Zoubi, R.M.; Al Zoubi, M. Novel Mutations in AKT1 Gene in Prostate Cancer Patients in Jordan. Curr. Issues Mol. Biol. 2024, 46, 9856-9866. https://doi.org/10.3390/cimb46090586
Alasmar A, Al-Alami Z, Zein S, Al-Smadi A, Al Bashir S, Alorjani MS, Al-Zoubi RM, Al Zoubi M. Novel Mutations in AKT1 Gene in Prostate Cancer Patients in Jordan. Current Issues in Molecular Biology. 2024; 46(9):9856-9866. https://doi.org/10.3390/cimb46090586
Chicago/Turabian StyleAlasmar, Ala’a, Zina Al-Alami, Sima Zein, Asmaa Al-Smadi, Samir Al Bashir, Mohammed S. Alorjani, Raed M. Al-Zoubi, and Mazhar Al Zoubi. 2024. "Novel Mutations in AKT1 Gene in Prostate Cancer Patients in Jordan" Current Issues in Molecular Biology 46, no. 9: 9856-9866. https://doi.org/10.3390/cimb46090586







