A Phylogenetic Analysis Based on Whole Genome Re-Sequencing of 41 Dendrobium Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials, Sampling and Whole Genome Resequencing
2.2. Whole Genome Sequence Alignment, SNP Detection, and Annotation
2.3. Phylogenetic Analyses and Divergence Time Estimation of Dendrobium
3. Results
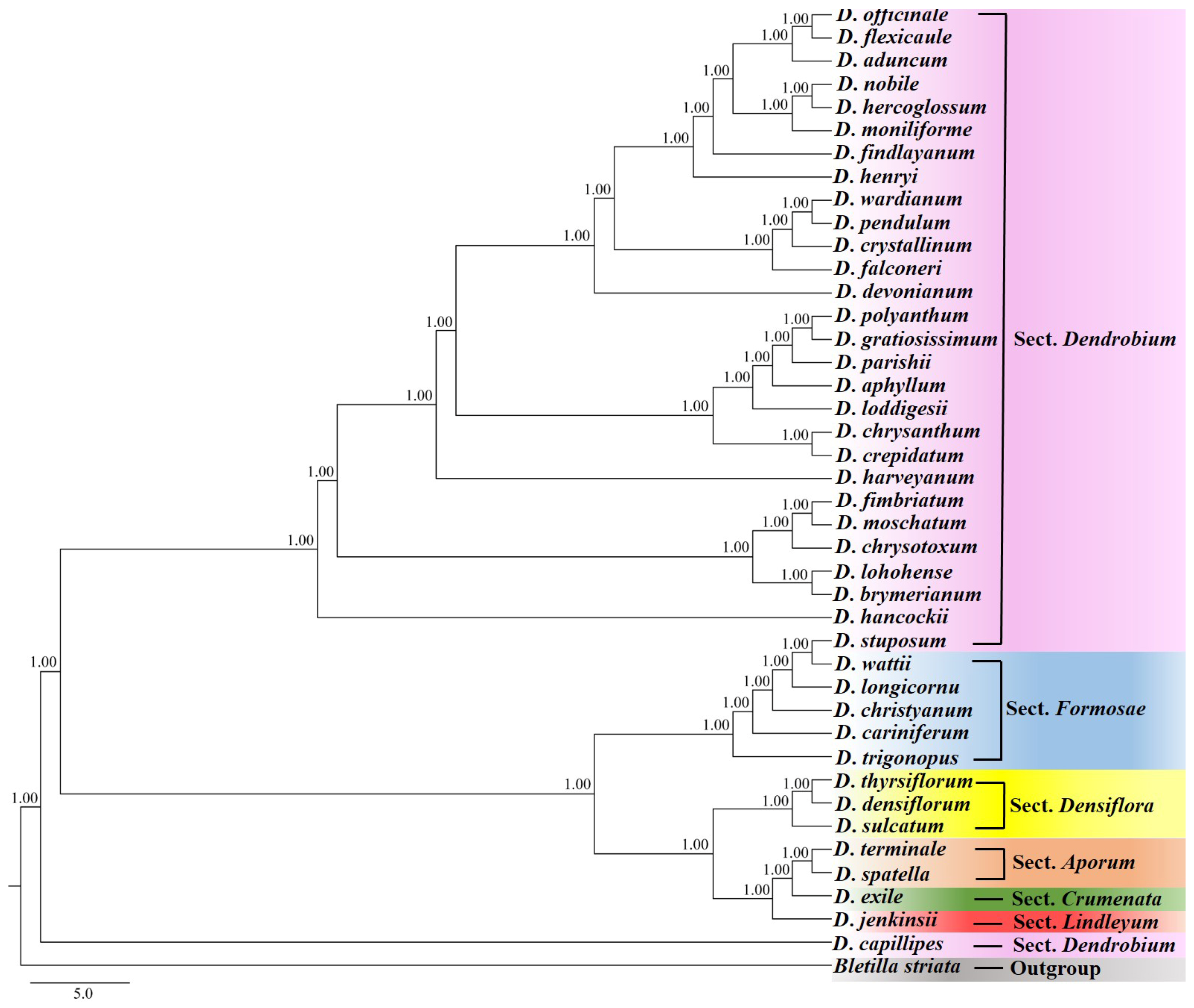
3.1. Species Discrimination of Dendrobium Based on Phylogenetic Tree
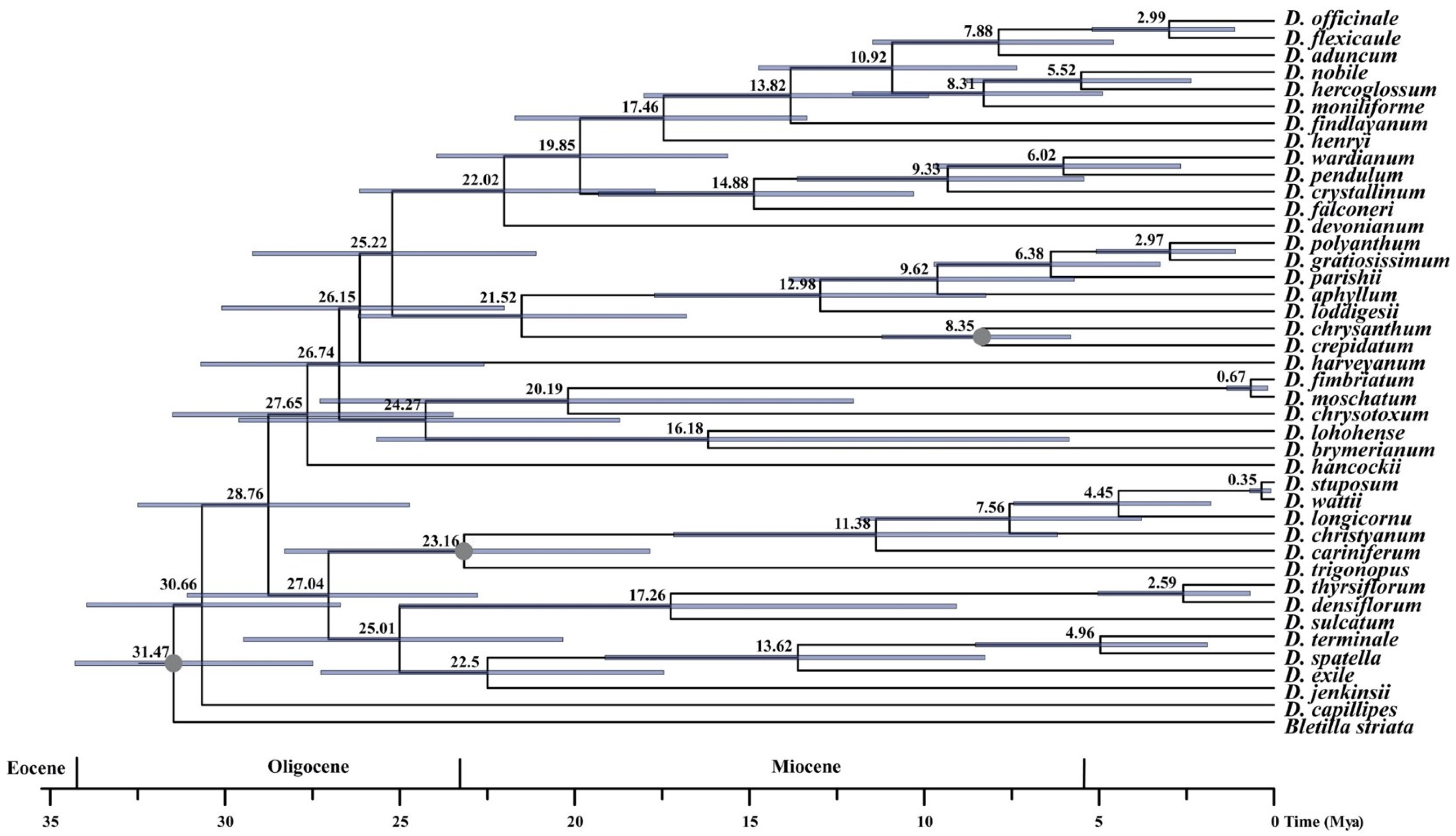
3.2. Divergence Time Estimation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mishra, P.; Kumar, A.; Nagireddy, A.; Mani, D.N.; Shukla, A.K.; Tiwari, R.; Sundaresan, V. DNA barcoding: An efficient tool to overcome authentication challenges in the herbal market. Plant Biotechnol. J. 2016, 14, 8–21. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.F.; Lu, T.Q.; Zhang, Z.R.; Cai, C.T.; Yang, J.B.; Tian, B. Authentication of traditional Chinese medicinal herb “Gusuibu” by DNA- based molecular methods. Ind. Crops Prod. 2019, 141, 111756. [Google Scholar] [CrossRef]
- Morris, M.W.; Stern, W.L.; Judd, W.S. Vegetative anatomy and systematics of subtribe Dendrobiinae (Orchidaceae). Bot. J. Linn. Soc. 1996, 120, 89–144. [Google Scholar] [CrossRef]
- Yukawa, T.; Uehara, K. Vegetative diversification and radiation in subtribe Dendrobiinae (Orchidaceae): Evidence from chloroplast DNA phylogeny and anatomical characters. Plant Syst. Evol. 1996, 201, 1–14. [Google Scholar] [CrossRef]
- Adams, P.B. Systematics of Dendrobiinae (Orchidaceae), with special reference to Australian taxa. Bot. J. Linn. Soc. 2011, 166, 105–126. [Google Scholar] [CrossRef]
- Fuentes-Pardo, A.P.; Ruzzante, D.E. Whole-genome sequencing approaches for conservation biology: Advantages, limitations and practical recommendations. Mol. Ecol. 2017, 26, 5369–5406. [Google Scholar] [CrossRef]
- Jing, Z.; Cheng, K.; Shu, H.; Ma, Y.; Liu, P. Whole genome resequencing approach for conservation biology of endangered plants. Biodivers. Sci. 2023, 31, 22679. [Google Scholar] [CrossRef]
- Ding, G.; Zhang, D.Z.; Ding, X.Y.; Zhou, Q.; Zhang, W.C.; Li, X.X. Genetic variation and conservation of the endangered Chinese endemic herb Dendrobium officinale based on SRAP analysis. Plant Syst. Evol. 2008, 276, 149–156. [Google Scholar] [CrossRef]
- Asahina, H.; Shinozaki, J.; Masuda, K.; Morimitsu, Y.; Satake, M. Identification of medicinal Dendrobium species by phylogenetic analyses using matK and rbcL sequences. J. Nat. Med. 2010, 64, 133–138. [Google Scholar] [CrossRef]
- Xu, H.; Hou, B.W.; Zhang, J.Z.; Tian, M.; Yuan, Y.H.; Niu, Z.T.; Ding, X.Y. Detecting adulteration of Dendrobium officinale by real-time PCR coupled with ARMS. Int. J. Food Sci. Tech. 2012, 47, 1695–1700. [Google Scholar] [CrossRef]
- Ding, G.; Zhang, D.; Feng, Z.; Fan, W.; Ding, X.; Li, X. SNP, ARMS and SSH authentication of medicinal Dendrobium officinale KIMURA et MIGO and application for identification of Fengdou Drugs. Biol. Pharm. Bull. 2008, 31, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Clements, M.A. Molecular phylogenetic systematics in Dendrobieae (Orchidaceae). Aliso 2006, 22, 465–480. [Google Scholar] [CrossRef]
- Niu, Z.T.; Zhu, S.Y.; Pan, J.J.; Li, L.D.; Sun, J.; Ding, X.Y. Comparative analysis of Dendrobium plastomes and utility of plastomic mutational hotspots. Sci. Rep. 2017, 7, 2073. [Google Scholar]
- Zhu, S.Y.; Niu, Z.T.; Xue, Q.Y.; Wang, H.; Xie, X.Z.; Ding, X.Y. Accurate authentication of Dendrobium officinale and its closely related species by comparative analysis of complete plastomes. Acta Pharm. Sin. B 2018, 8, 969–980. [Google Scholar] [CrossRef]
- Schuiteman, A. Dendrobium (Orchidaceae): To split or not to split? Gard. Bull. Singap. 2011, 63, 245–257. [Google Scholar]
- Xiang, X.G.; Schuiteman, A.; Li, D.Z.; Huang, W.C.; Chung, S.W.; Li, J.W.; Zhou, H.L.; Jin, W.T.; Lai, Y.J.; Li, Z.Y.; et al. Molecular systematics of Dendrobium (Orchidaceae, Dendrobieae) from mainland Asia based on plastid and nuclear sequences. Mol. Phylogenet. Evol. 2013, 69, 950–960. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast allin-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.Y.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. Molecular Evolution: A Statistical Approach; Oxford University Press: Oxford, UK, 2014. [Google Scholar]
- Xiang, X.G.; Mi, X.C.; Zhou, H.L.; Li, J.W.; Chung, S.W.; Li, D.Z.; Huang, W.C.; Jin, W.T.; Li, Z.Y.; Huang, L.Q.; et al. Biogeographical diversification of mainland Asian Dendrobium (Orchidaceae) and its implications for the historical dynamics of evergreen broad-leaved forests. J. Biogeogr. 2016, 43, 1310–1323. [Google Scholar] [CrossRef]
- Gamisch, A.; Comes, H.P. Clade-age-dependent diversification under high species turnover shapes species richness disparities among tropical rainforest lineages of Bulbophyllum (Orchidaceae). BMC Evol. Biol. 2019, 19, 93. [Google Scholar] [CrossRef]
- Rambaut, A.; Suchard, M.; Xie, D.; Drummond, A. Tracer v1.6. 2014. Available online: http://beast.bio.ed.ac.uk/Tracer (accessed on 25 January 2017).
- Chen, X.M.; Guo, S.X. Advances in the research of constituents and pharmacology of Dendrobium. Nat. Prod. Res. Dev. 2001, 3, 70–75. [Google Scholar]
- Zhang, T.; Wang, Z.T.; Xu, L.S.; Zhou, K.Y. Application of mitochondrial nad 1 intron 2 sequences to molecular identification of some species of Dendrobium Sw. Chin. Tradit. Herb. Drugs 2005, 36, 1059–1062. [Google Scholar]
- Jin, X.H.; Chen, S.H.; Luo, Y.B. Taxonomic revision of Dendrobium moniliforme complex (Orchidaceae). Sci. Hortic. 2009, 120, 143–145. [Google Scholar]
- Chen, S.L.; Yao, H.; Han, J.P.; Liu, C.; Song, J.Y.; Shi, L.C.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X.; et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef]
- Niu, Z.; Pan, J.; Xue, Q.; Zhu, S.; Liu, W.; Ding, X. Plastome-wide comparison reveals new SNV resources for the authentication of Dendrobium huoshanense and its corresponding medicinal slice (Huoshan Fengdou). Acta Pharmacol. Sin. 2018, 8, 466–477. [Google Scholar] [CrossRef]
- Zhang, Y.B.; Pui-Hay, B.P.; Wang, Z.T.; Shaw, P.C. Current approaches for the authentication of medicinal Dendrobium species and its products. Plant Genet. Resour. 2005, 3, 144–148. [Google Scholar] [CrossRef]
- Xu, S.Z.; Li, D.Z.; Li, J.W.; Xiang, X.G.; Jin, W.T.; Huang, W.C.; Jin, X.H.; Huang, L.Q. Evaluation of the DNA barcodes in Dendrobium (Orchidaceae) from mainland Asia. PLoS ONE 2015, 10, e0115168. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Chen, X.C.; Yang, P.; Wang, L.L.; Han, J.P. Barcoding the Dendrobium (Orchidaceae) species and analysis of the intragenomic variation based on the internal transcribed spacer 2. Biomed. Res. Int. 2017, 2017, 2734960. [Google Scholar] [CrossRef] [PubMed]
- Li, L.D.; Jiang, Y.; Liu, Y.Y.; Niu, Z.T.; Xue, Q.Y.; Liu, W.; Ding, X.Y. The large single copy (LSC) region functions as a highly effective and efficient molecular marker for accurate authentication of medicinal Dendrobium species. Acta Pharm. Sinica B 2020, 10, 1989–2001. [Google Scholar] [CrossRef] [PubMed]
- Campbelln, N.R.; Narum, S.R. Quantitative PCR assessment of microsatellite and SNP genotyping with variable quality DNA extracts. Conserv. Genet. 2009, 10, 779–784. [Google Scholar] [CrossRef]
- Cabezas, J.A.; Ibáñez, J.; Lijavetzky, D.; Dolores, V.; Bravo1, G.; Rodríguez, V.; Carreño, I.; Jermakow, A.M.; Carreño, J.; Ruiz-García, L.; et al. A 48 SNP set for grapevine cultivar identification. BMC Plant Boil. 2011, 11, 153. [Google Scholar] [CrossRef]
- Kapur, V.; Sischo, W.M.; Greer, R.S.; Whittam, T.S.; Musser, J.M. Molecular population genetic analysis of Staphylococcus aureus recovered from cows. J. Clin. Microbiol. 1995, 33, 376–380. [Google Scholar] [CrossRef]
- Shang, P.; Li, W.T.; Tan, Z.K.; Zhang, J.; Dong, S.X.; Wang, K.J.; Chambam, Y. Population genetic analysis of ten geographically isolated Tibetan pig populations. Animals 2020, 10, 1297. [Google Scholar] [CrossRef]
- Shi, P.; Zhou, J.; Song, H.L.; Wu, Y.J.; Lan, L.; Tang, X.Y.; Ma, Z.G.; Vossbrinck, C.R.; Vossbrinck, B.; Zhou, Z.Y.; et al. Genomic analysis of Asian honeybee populations in China reveals evolutionary relationships and adaptation to abiotic stress. Ecol. Evol. 2020, 10, 13427–13438. [Google Scholar] [CrossRef]
- Lin, Y.; Xiao, Q.; Lin, Y.F.; Li, X.X.; Yin, W.L.; Li, H.; Chen, L. Development of SNP Markers for Pomacea canaliculata and Application of Species Identification in P. maculate. J. Nanjing Norm. Univ. 2021, 44, 96–102. [Google Scholar]
- Luo, R.; Chen, Y.; Zhang, H.M. Research progress on whole-genome resequencing in Brassica. Biodivers. Sci. 2023, 31, 23237. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, Z.T.; Hu, Y.H.; Zhang, H.J.; Zhang, M.; Fu, T.; Liu, X.; Li, Z.G.; Qi, X.L.; Wang, E.Y.; et al. Genetic diversity and population structure analysis of jiaxian red cattle based on whole genome sequencing. China Anim. Husb. Vet. Med. 2024, 51, 2933–2942. [Google Scholar]
- Pearce, N.R.; Cribb, P.J. The Orchids of Bhutan; Charlesworth Group: Huddersfield, UK, 2002. [Google Scholar]
- Zhu, G.H.; Ji, Z.H.; Wood, J.J.; Wood, H.P. Dendrobium. In Flora of China; Wu, C.Y., Raven, P.H., Hong, D.Y., Eds.; Scientific Press: Beijing, China, 2009; pp. 367–397. [Google Scholar]
- Conran, J.G.M.; Bannister, J.M.; Lee, D.E. Earliest orchid macrofossils: Early Miocene Dendrobium and Earina (Orchidaceae: Epidendroideae) from New Zealand. Am. J. Bot. 2009, 96, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, J.A. A Paleobotanical interpretation of tertiary climates in the Northern Hemisphere: Data from fossil plants make it possible to reconstruct Tertiary climatic changes, which may be correlated with changes in the inclination of the earth’s rotational axis. Am. Sci. 1978, 66, 694–703. [Google Scholar]
- Keller, G.; Barron, J.A. Paleoceanographic implications of Miocene deep-seahiatuses. Geol. Soc. Am. Bull. 1983, 94, 590–613. [Google Scholar] [CrossRef]
- Miller, K.G.; Fairbanks, R.G. Evidence for Oligocene-middle Miocene abyssal circulation changes in the western North Atlantic. Nature 1983, 306, 250–253. [Google Scholar] [CrossRef]
- Zachos, J. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 2001, 292, 686–693. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, F.-P.; Fu, X.-W.; Li, H.-R.; Zhang, S.-B. A Phylogenetic Analysis Based on Whole Genome Re-Sequencing of 41 Dendrobium Species. Curr. Issues Mol. Biol. 2025, 47, 276. https://doi.org/10.3390/cimb47040276
Zhang F-P, Fu X-W, Li H-R, Zhang S-B. A Phylogenetic Analysis Based on Whole Genome Re-Sequencing of 41 Dendrobium Species. Current Issues in Molecular Biology. 2025; 47(4):276. https://doi.org/10.3390/cimb47040276
Chicago/Turabian StyleZhang, Feng-Ping, Xue-Wei Fu, Han-Run Li, and Shi-Bao Zhang. 2025. "A Phylogenetic Analysis Based on Whole Genome Re-Sequencing of 41 Dendrobium Species" Current Issues in Molecular Biology 47, no. 4: 276. https://doi.org/10.3390/cimb47040276
APA StyleZhang, F.-P., Fu, X.-W., Li, H.-R., & Zhang, S.-B. (2025). A Phylogenetic Analysis Based on Whole Genome Re-Sequencing of 41 Dendrobium Species. Current Issues in Molecular Biology, 47(4), 276. https://doi.org/10.3390/cimb47040276




