Fast and Accurate Surrogate Virus Neutralization Test Based on Antibody-Mediated Blocking of the Interaction of ACE2 and SARS-CoV-2 Spike Protein RBD
Abstract
:1. Introduction
2. Materials and Methods
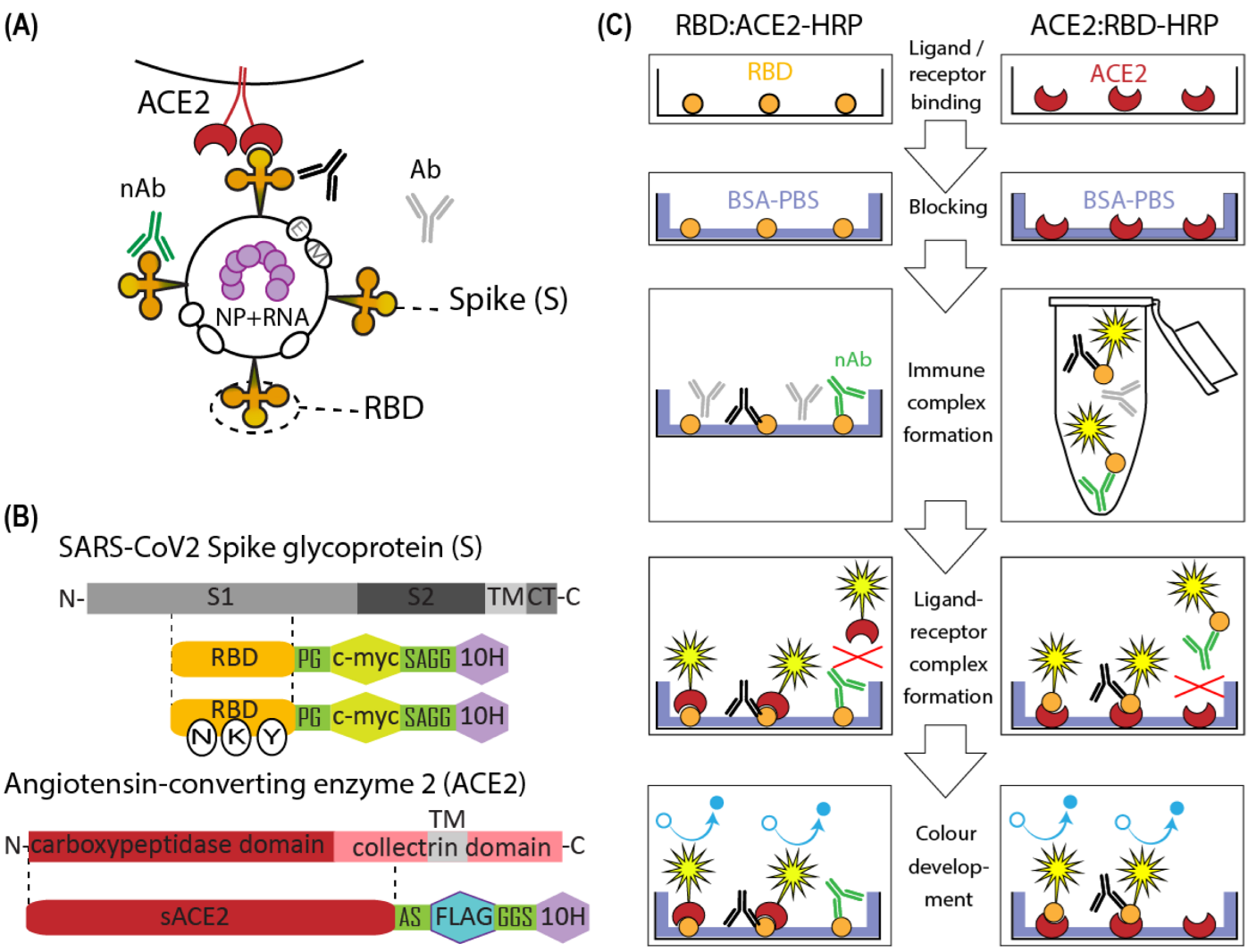
2.1. Molecular Cloning
2.2. Producer Cell Lines Generation and Cell Culture
2.3. Preparative Cell Cultures
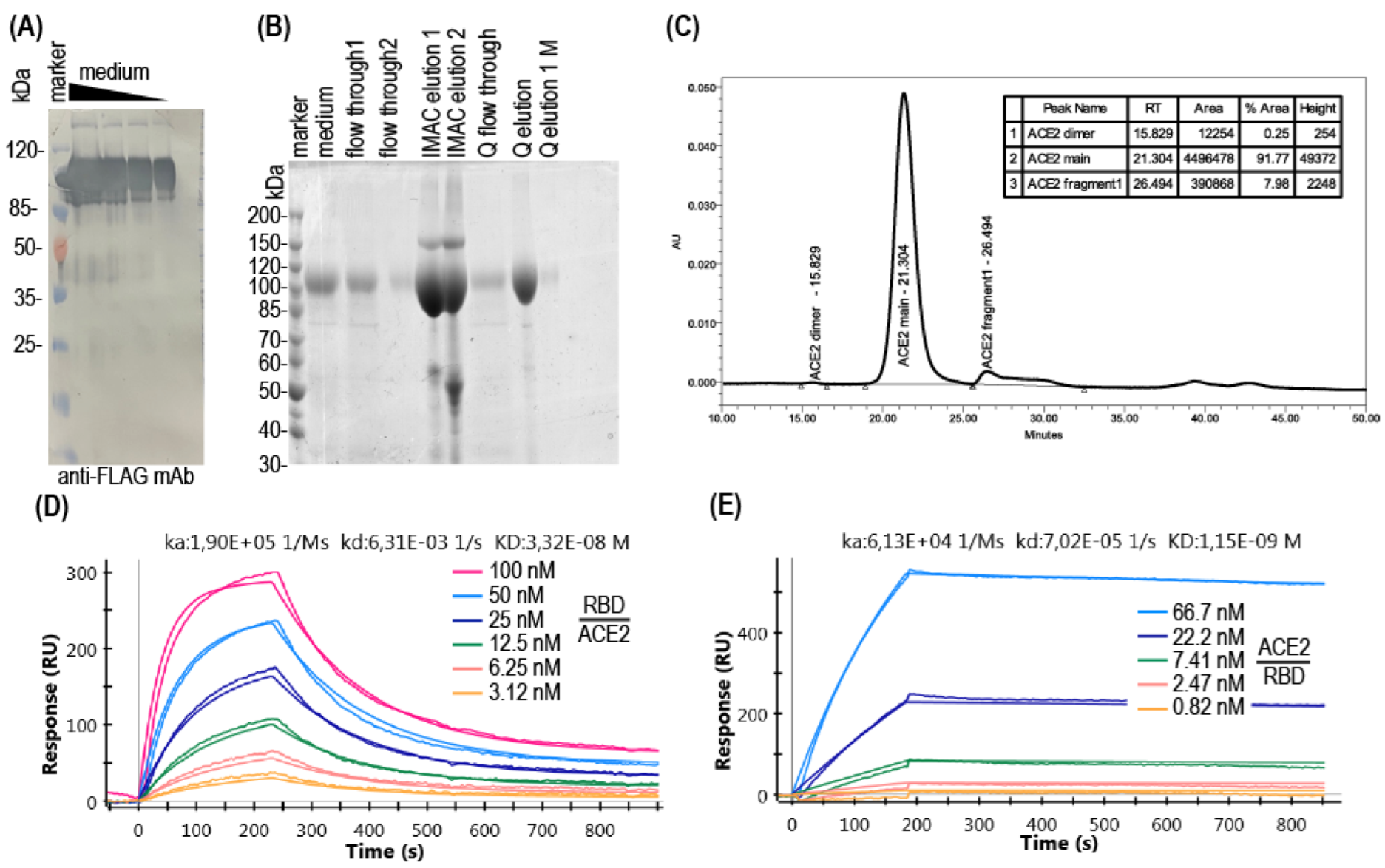
2.4. Western Blotting
2.5. Proteins Purification
2.6. Recombinant Proteins Analysis
2.7. HRP Conjugation
2.8. Sample Collection and VNT Assay
2.9. ELISA
2.10. Surrogate Virus Neutralization Test
2.11. Statistical Analysis
3. Results
3.1. Production of Soluble ACE2
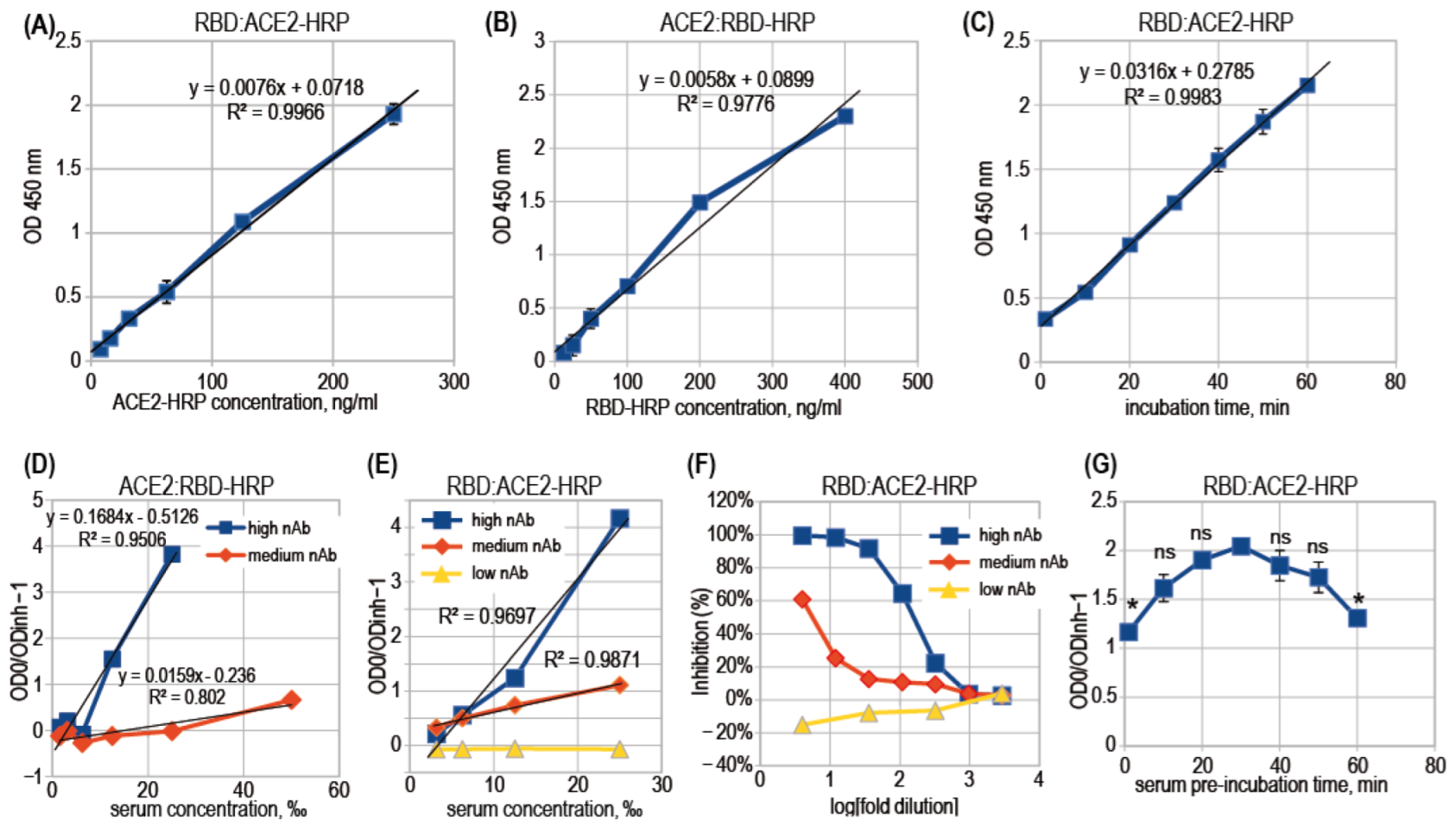
3.2. ACE2-RBD Interaction in Microplates
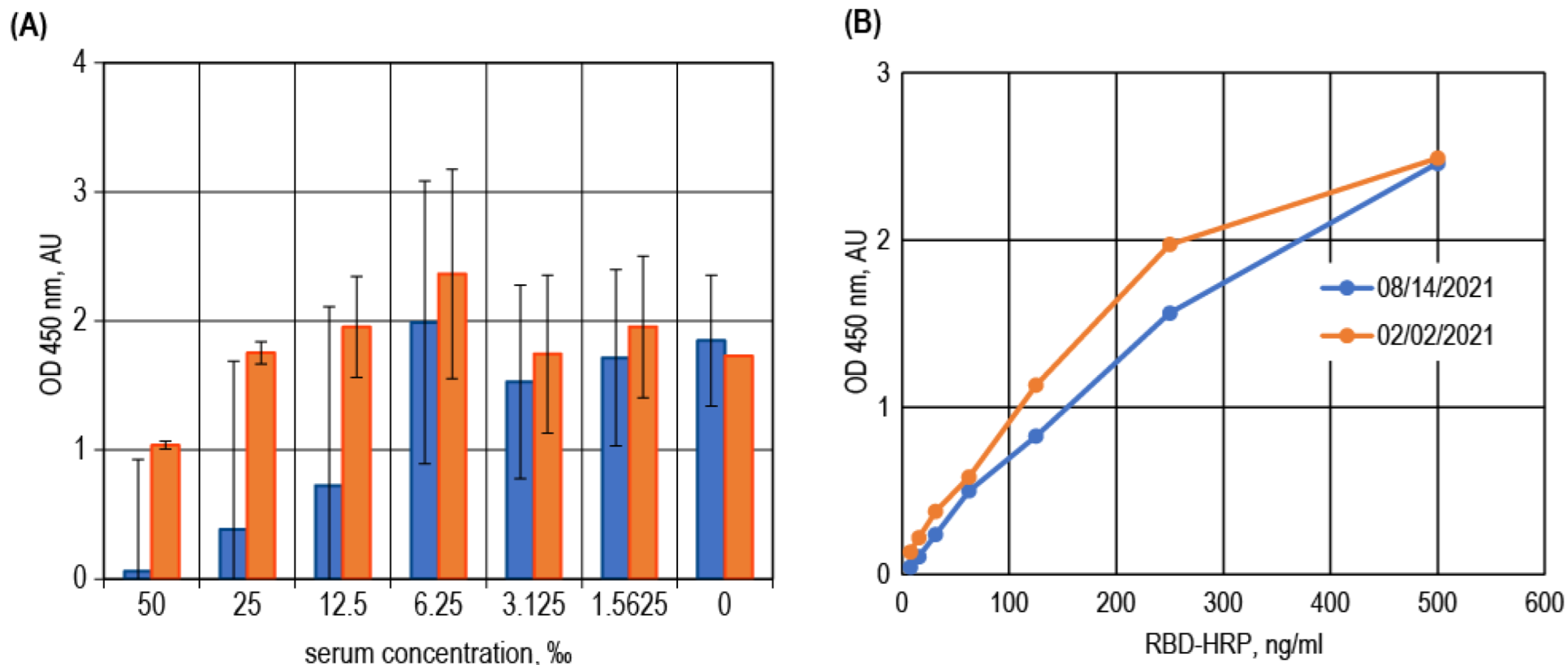
3.3. Surrogate VNT Assay Design Variants
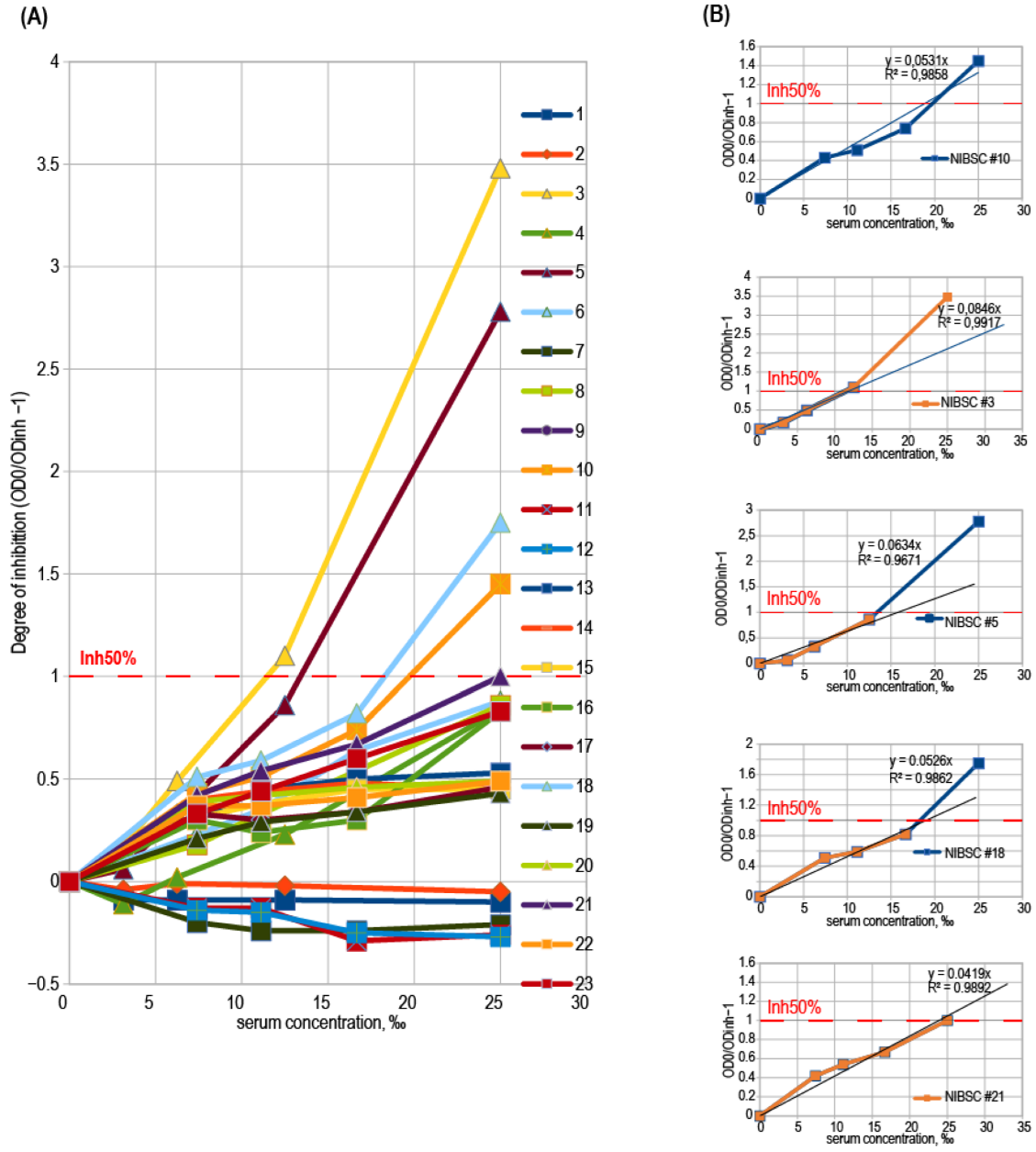
3.4. Binding Inhibition Curves Analysis
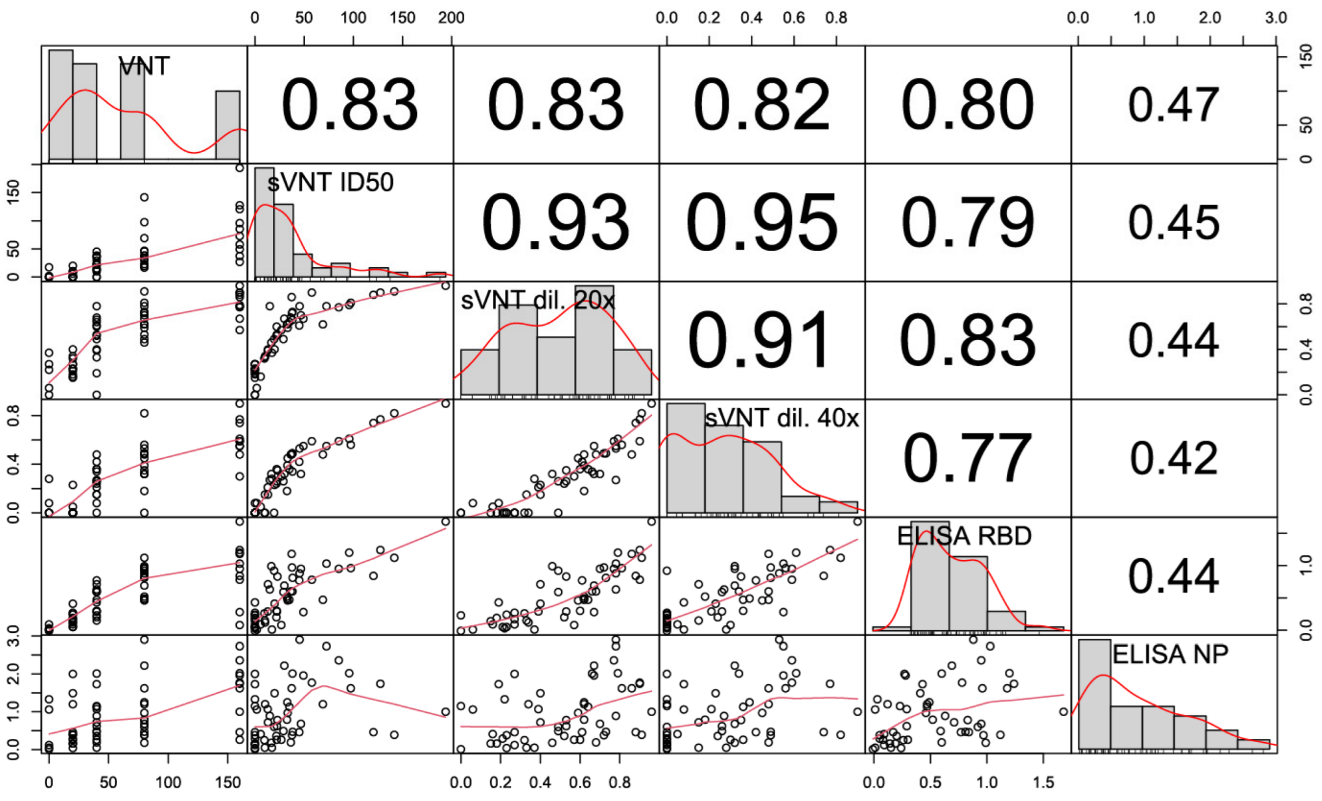
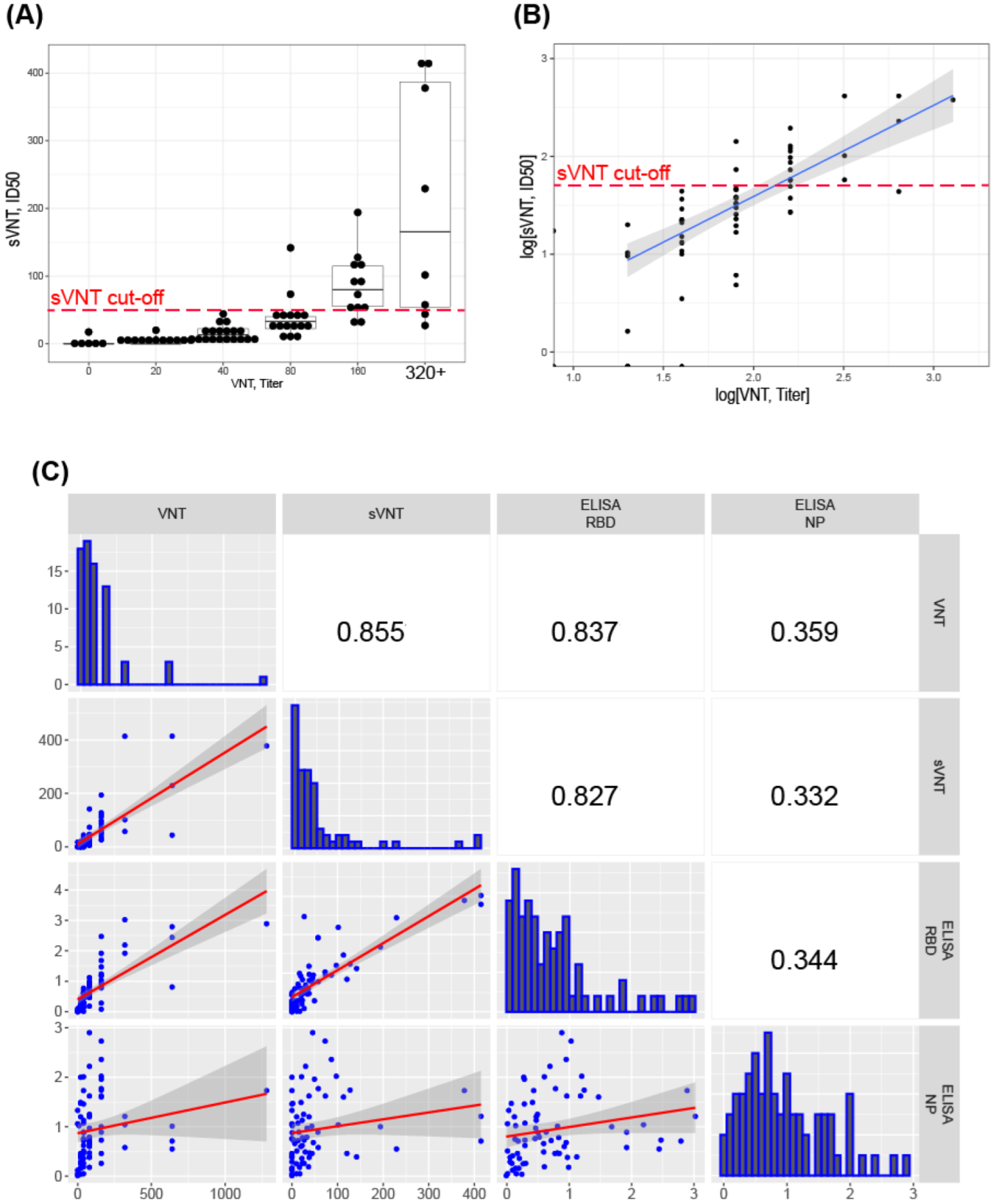
3.5. Correlation of sVNT and Live Virus VNT Values
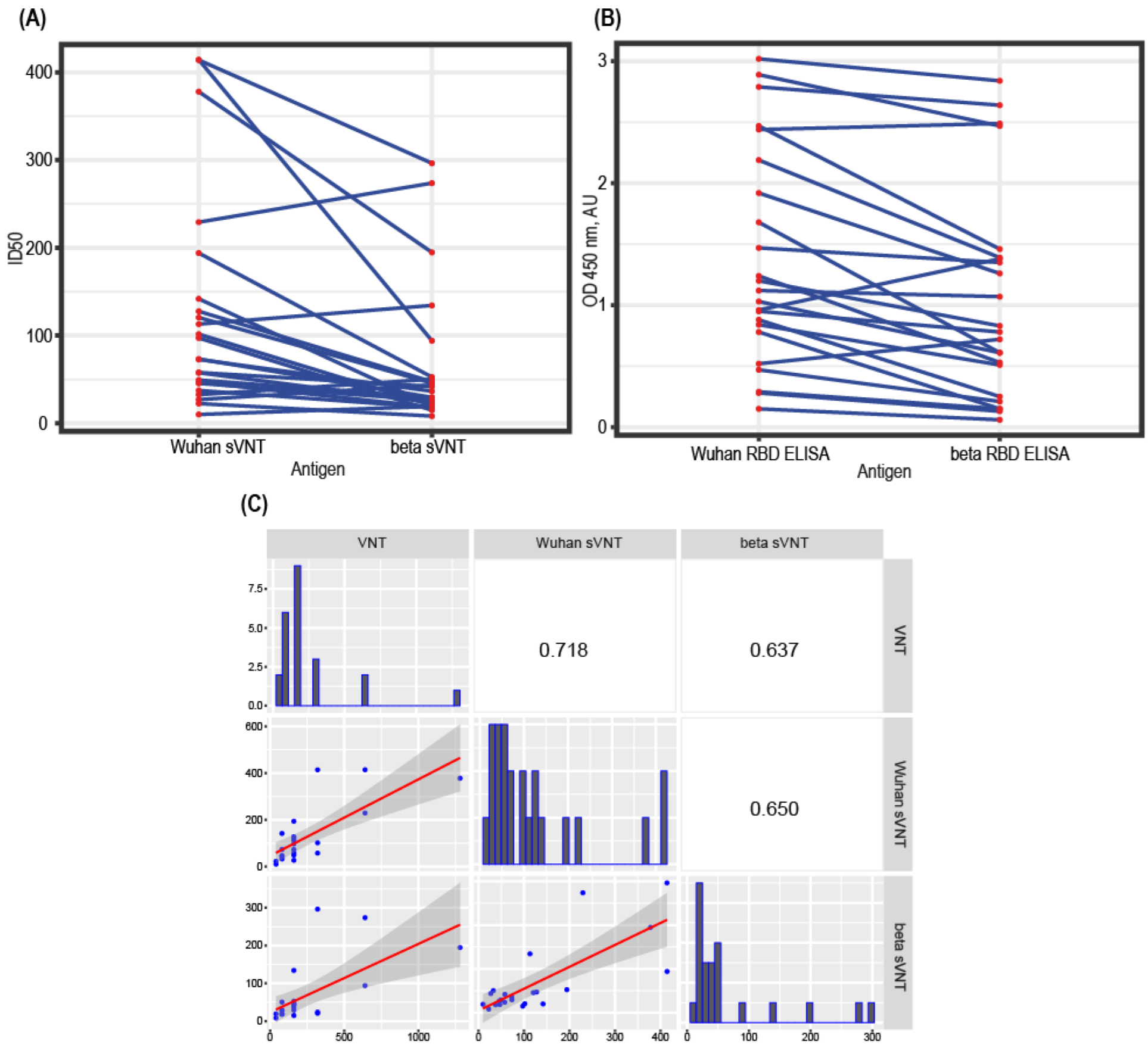
3.6. Test Design Adaptation for SARS-CoV-2 Variants
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Oligonucleotides Used for the pTF Vector Construction, 5′–3′ | |
|---|---|
| AS-Flag-AbsF | TCGAGGCTGCAGCTAGCGATTACAAGGATGACGACGATAAGGGAGGATCCCATCACCATCACCATCACCATCATCACCACTAA |
| AS-Flag-AbsR | ACTAGTTAGTGGTGATGATGGTGATGGTGATGGTGATGGGATCCTCCCTTATCGTCGTCATCCTTGTAATCGCTAGCTGCAGCC |
| Oligonucleotides used for the pTF-ACE2s construction (restriction sites in bold, synthetic Kozak sequence in underlined) | |
| AD-ACE-AbsF | CCTCGAGGCCGCCACCATGTCAAGCTCTTCCTGGCTC |
| AD-ACE-NheR | GCTAGCGGAAACAGGGGGCTGGTT |
| Sequencing oligonucleotides | |
| SQ-ACE875-R | GCAGGGAGGCATCCAA |
| SQ-ACE592-F | TCTGAGGTCGGCAAGCA |
| SQ-ACE1170-F | AAAGGTGACAATGGACGACT |
| SQ-ACE1512-F | GTGGTGGGAGATGAAGCG |
| SQ-5CH6-F | GCCGCTGCTTCCTGTGAC |

References
- Foundation for Innovative New Diagnostics (FIND). COVID-19 Diagnostics Database. Available online: https://www.finddx.org/test-directory/?_type_of_technology=immunoassay (accessed on 26 November 2021).
- Logunov, D.Y.; Dolzhikova, I.V.; Shcheblyakov, D.V.; Tukhvatulin, A.I.; Zubkova, O.V.; Dzharullaeva, A.S.; Kovyrshina, A.V.; Lubenets, N.L.; Grousova, D.M.; Erokhova, A.S.; et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: An interim analysis of a randomised controlled phase 3 trial in Russia. Lancet 2021, 397, 671–681. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Bao, L.; Mao, H.; Wang, L.; Xu, K.; Yang, M.; Li, Y.; Zhu, L.; Wang, N.; Lv, Z.; et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 2020, 369, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Tostanoski, L.H.; Peter, L.; Mercado, N.B.; Mcmahan, K.; Mahrokhian, S.H.; Nkolola, J.P.; Liu, J.; Li, Z.; Chandrashekar, A.; et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 2020, 369, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Widge, A.T.; Rouphael, N.G.; Jackson, L.A.; Anderson, E.J.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J.; et al. Durability of Responses after SARS-CoV-2 mRNA-1273 Vaccination. N. Engl. J. Med. 2021, 384, 80–82. [Google Scholar] [CrossRef] [PubMed]
- Seow, J.; Graham, C.; Merrick, B.; Acors, S.; Pickering, S.; Steel, K.J.A.; Hemmings, O.; O’Byrne, A.; Kouphou, N.; Galao, R.P.; et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol. 2020, 5, 1598–1607. [Google Scholar] [CrossRef]
- Bohn, M.K.; Lippi, G.; Horvath, A.; Sethi, S.; Koch, D.; Ferrari, M.; Wang, C.-B.; Mancini, N.; Steele, S.; Adeli, K. Molecular, serological, and biochemical diagnosis and monitoring of COVID-19: IFCC taskforce evaluation of the latest evidence. Clin. Chem. Lab. Med. 2020, 58, 1037–1052. [Google Scholar] [CrossRef]
- Robbiani, D.F.; Gaebler, C.; Muecksch, F.; Lorenzi, J.C.C.; Wang, Z.; Cho, A.; Agudelo, M.; Barnes, C.O.; Gazumyan, A.; Finkin, S.; et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 2020, 584, 437–442. [Google Scholar] [CrossRef]
- Billon-Denis, E.; Ferrier-Rembert, A.; Garnier, A.; Cheutin, L.; Vigne, C.; Tessier, E.; Denis, J.; Badaut, C.; Rougeaux, C.; Wuille, A.D.; et al. Differential serological and neutralizing antibody dynamics after an infection by a single SARS-CoV-2 strain. Infection 2021, 49, 781–783. [Google Scholar] [CrossRef]
- Ju, B.; Zhang, Q.; Ge, J.; Wang, R.; Sun, J.; Ge, X.; Yu, J.; Shan, S.; Zhou, B.; Song, S.; et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020, 584, 115–119. [Google Scholar] [CrossRef]
- Bewley, K.R.; Coombes, N.S.; Gagnon, L.; McInroy, L.; Baker, N.; Shaik, I.; St-Jean, J.R.; St-Amant, N.; Buttigieg, K.R.; Humphries, H.E.; et al. Quantification of SARS-CoV-2 neutralizing antibody by wild-type plaque reduction neutralization, microneutralization and pseudotyped virus neutralization assays. Nat. Protoc. 2021, 16, 3114–3140. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Vandergaast, R.; Carey, T.; Reiter, S.; Lathrum, C.; Lech, P.; Gnanadurai, C.; Haselton, M.; Buehler, J.; Narjari, R.; Schnebeck, L.; et al. IMMUNO-COV v2.0: Development and Validation of a High-Throughput Clinical Assay for Measuring SARS-CoV-2-Neutralizing Antibody Titers. mSphere 2021, 6, e0017021. [Google Scholar] [CrossRef]
- Tan, C.W.; Chia, W.N.; Qin, X.; Liu, P.; Chen, M.I.-C.; Tiu, C.; Hu, Z.; Chen, V.C.-W.; Young, B.E.; Sia, W.R.; et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2–spike protein–protein interaction. Nat. Biotechnol. 2020, 38, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Perera, R.A.P.M.; Ko, R.; Tsang, O.T.Y.; Hui, D.S.C.; Kwan, M.Y.M.; Brackman, C.J.; To, E.M.W.; Yen, H.-L.; Leung, K.; Cheng, S.M.S.; et al. Evaluation of a SARS-CoV-2 Surrogate Virus Neutralization Test for Detection of Antibody in Human, Canine, Cat, and Hamster Sera. J. Clin. Microbiol. 2021, 59, e02504-20. [Google Scholar] [CrossRef] [PubMed]
- Meyer, B.; Reimerink, J.; Torriani, G.; Brouwer, F.; Godeke, G.-J.; Yerly, S.; Hoogerwerf, M.; Vuilleumier, N.; Kaiser, L.; Eckerle, I.; et al. Validation and clinical evaluation of a SARS-CoV-2 surrogate virus neutralisation test (sVNT). Emerg. Microbes Infect. 2020, 9, 2394–2403. [Google Scholar] [CrossRef]
- Abe, K.T.; Li, Z.; Samson, R.; Samavarchi-Tehrani, P.; Valcourt, E.J.; Wood, H.; Budylowski, P.; Dupuis, A.P., 2nd; Girardin, R.C.; Rathod, B.; et al. A simple protein-based surrogate neutralization assay for SARS-CoV-2. JCI Insight 2020, 5, e142362. [Google Scholar] [CrossRef]
- Sinegubova, M.V.; Orlova, N.A.; Kovnir, S.V.; Dayanova, L.K.; Vorobiev, I.I. High-level expression of the monomeric SARS-CoV-2 S protein RBD 320-537 in stably transfected CHO cells by the EEF1A1-based plasmid vector. PLoS ONE 2021, 16, e0242890. [Google Scholar] [CrossRef]
- Urlaub, G.; Chasin, L.A. Isolation of Chinese hamster cell mutants deficient in dihydrofolate reductase activity. Proc. Natl. Acad. Sci. USA 1980, 77, 4216–4220. [Google Scholar] [CrossRef] [Green Version]
- Kolesov, D.E.; Sinegubova, M.V.; Safenkova, I.V.; Vorobiev, I.I.; Orlova, N.A. Antigenic properties of the SARS-CoV-2 nucleoprotein are altered by the RNA admixture. PeerJ 2022, 10, e12751. [Google Scholar] [CrossRef]
- Kostin, A.I.; Lundgren, M.N.; Bulanov, A.Y.; Ladygina, E.A.; Chirkova, K.S.; Gintsburg, A.L.; Logunov, D.Y.; Dolzhikova, I.V.; Shcheblyakov, D.V.; Borovkova, N.V.; et al. Impact of pathogen reduction methods on immunological properties of the COVID-19 convalescent plasma. Vox Sang. 2021, 116, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Lardeux, F.; Torrico, G.; Aliaga, C. Calculation of the ELISA’s cut-off based on the change-point analysis method for detection of Trypanosoma cruzi infection in Bolivian dogs in the absence of controls. Mem. Inst. Oswaldo Cruz 2016, 111, 501–504. [Google Scholar] [CrossRef] [Green Version]
- Surrogate Virus Neutralization Test data analysis and visualization. Available online: https://github.com/d-kolesiko/sVNT (accessed on 2 February 2022).
- Orlova, A.N.; Kovnir, S.V.; Hodak, J.A.; Vorobiev, I.I.; Gabibov, A.G.; Skryabin, K.G. Improved elongation factor-1 alpha-based vectors for stable high-level expression of heterologous proteins in Chinese hamster ovary cells. BMC Biotechnol. 2014, 14, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guy, J.L.; Jackson, R.M.; Acharya, K.R.; Sturrock, E.D.; Hooper, A.N.M.; Turner, A.J. Angiotensin-Converting Enzyme-2 (ACE2): Comparative Modeling of the Active Site, Specificity Requirements, and Chloride Dependence. Biochemistry 2003, 42, 13185–13192. [Google Scholar] [CrossRef] [PubMed]
- McMillan, P.; Dexhiemer, T.; Neubig, R.R.; Uhal, B.D. COVID-19—A Theory of Autoimmunity Against ACE-2 Explained. Front. Immunol. 2021, 12, 582166. [Google Scholar] [CrossRef]
- Casciola-Rosen, L.; Thiemann, D.R.; Andrade, F.; Trejo Zambrano, M.I.; Hooper, J.E.; Leonard, E.; Spangler, J.; Cox, A.L.; Machamer, C.; Sauer, L.; et al. IgM autoantibodies recognizing ACE2 are associated with severe COVID-19. medRxiv 2020. [Google Scholar] [CrossRef]
- Arthur, J.M.; Forrest, J.C.; Boehme, K.W.; Kennedy, J.L.; Owens, S.; Herzog, C.; Liu, J.; Harville, T.O. Development of ACE2 autoantibodies after SARS-CoV-2 infection. PLoS ONE 2021, 16, e0257016. [Google Scholar] [CrossRef]
- Levin, E.G.; Lustig, Y.; Cohen, C.; Fluss, R.; Indenbaum, V.; Amit, S.; Doolman, R.; Asraf, K.; Mendelson, E.; Ziv, A.; et al. Waning Immune Humoral Response to BNT162b2 COVID-19 Vaccine over 6 Months. N. Engl. J. Med. 2021, 385, e84. [Google Scholar] [CrossRef]
- Zhang, H.; Jia, Y.; Ji, Y.; Cong, X.; Liu, Y.; Yang, R.; Kong, X.; Shi, Y.; Zhu, L.; Wang, Z.; et al. Studies on the level of neutralizing antibodies produced by inactivated COVID-19 vaccines in the real world. medRxiv 2021, 2214. [Google Scholar] [CrossRef]
- Cele, S.; Jackson, L.; Khan, K.; Khoury, D.S.; Moyo-Gwete, T.; Tegally, H.; Scheepers, C.; Amoako, D.; Karim, F.; Bernstein, M.; et al. SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. medRxiv 2021. [Google Scholar] [CrossRef]
- The US Food and Drug Administration. Convalescent Plasma EUA Letter of Authorization 06032021. 2021. Available online: https://img-cdn.tinkoffjournal.ru/-/ccp-eua-loatableamendment_6221.pdf (accessed on 2 February 2022).
- Yuan, M.; Liu, H.; Wu, N.C.; Wilson, I.A. Recognition of the SARS-CoV-2 receptor binding domain by neutralizing antibodies. Biochem. Biophys. Res. Commun. 2020, 538, 192–203. [Google Scholar] [CrossRef]
- Nayak, K.; Gottimukkala, K.; Kumar, S.; Reddy, E.S.; Edara, V.V.; Kauffman, R.; Floyd, K.; Mantus, G.; Savargaonkar, D.; Goel, P.K.; et al. Characterization of neutralizing versus binding antibodies and memory B cells in COVID-19 recovered individuals from India. Virology 2021, 558, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Putcharoen, O.; Wacharapluesadee, S.; Ni Chia, W.; Paitoonpong, L.; Tan, C.W.; Suwanpimolkul, G.; Jantarabenjakul, W.; Ruchisrisarod, C.; Wanthong, P.; Sophonphan, J.; et al. Early detection of neutralizing antibodies against SARS-CoV-2 in COVID-19 patients in Thailand. PLoS ONE 2021, 16, e0246864. [Google Scholar] [CrossRef] [PubMed]
- Valcourt, E.J.; Manguiat, K.; Robinson, A.; Chen, J.C.-Y.; Dimitrova, K.; Philipson, C.; Lamoureux, L.; McLachlan, E.; Schiffman, Z.; Drebot, M.A.; et al. Evaluation of a commercially-available surrogate virus neutralization test for severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). Diagn. Microbiol. Infect. Dis. 2021, 99, 115294. [Google Scholar] [CrossRef]
- Jiang, S.; Hillyer, C.; Du, L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol. 2020, 41, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, G.; Li, J.; Nie, Y.; Shi, X.; Lian, G.; Wang, W.; Yin, X.; Zhao, Y.; Qu, X.; et al. Identification of an Antigenic Determinant on the S2 Domain of the Severe Acute Respiratory Syndrome Coronavirus Spike Glycoprotein Capable of Inducing Neutralizing Antibodies. J. Virol. 2004, 78, 6938–6945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Li, W.; Drabek, D.; Okba, N.M.A.; van Haperen, R.; Osterhaus, A.D.M.E.; van Kuppeveld, F.J.M.; Haagmans, B.L.; Grosveld, F.; Bosch, B.-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020, 11, 2251. [Google Scholar] [CrossRef]
- Premkumar, L.; Segovia-Chumbez, B.; Jadi, R.; Martinez, D.R.; Raut, R.; Markmann, A.; Cornaby, C.; Bartelt, L.; Weiss, S.; Park, Y.; et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020, 5, eabc8413. [Google Scholar] [CrossRef]
- Embregts, C.W.; Verstrepen, B.; Langermans, J.A.; Böszörményi, K.P.; Sikkema, R.S.; de Vries, R.D.; Hoffmann, D.; Wernike, K.; Smit, L.A.; Zhao, S.; et al. Evaluation of a multi-species SARS-CoV-2 surrogate virus neutralization test. One Health 2021, 13, 100313. [Google Scholar] [CrossRef]
- Danh, K.; Karp, D.G.; Robinson, P.V.; Seftel, D.; Stone, M.; Simmons, G.; Bagri, A.; Schreibman, M.; Buser, A.; Holbro, A.; et al. Detection of SARS-CoV-2 neutralizing antibodies with a cell-free PCR assay. medRxiv 2020. [Google Scholar] [CrossRef]
- Luo, Y.R.; Yun, C.; Chakraborty, I.; Wu, A.H.B.; Lynch, K.L. A SARS-CoV-2 Label-Free Surrogate Virus Neutralization Test and a Longitudinal Study of Antibody Characteristics in COVID-19 Patients. J. Clin. Microbiol. 2021, 59, e0019321. [Google Scholar] [CrossRef] [PubMed]
- Rockstroh, A.; Wolf, J.; Fertey, J.; Kalbitz, S.; Schroth, S.; Lübbert, C.; Ulbert, S.; Borte, S. Correlation of humoral immune responses to different SARS-CoV-2 antigens with virus neutralizing antibodies and symptomatic severity in a German COVID-19 cohort. Emerg. Microbes Infect. 2021, 10, 774–781. [Google Scholar] [CrossRef] [PubMed]
- Papenburg, J.; Cheng, M.P.; Corsini, R.; Caya, C.; Mendoza, E.; Manguiat, K.; Lindsay, L.R.; Wood, H.; Drebot, M.A.; Dibernardo, A.; et al. Evaluation of a Commercial Culture-Free Neutralization Antibody Detection Kit for Severe Acute Respiratory Syndrome-Related Coronavirus-2 and Comparison with an Antireceptor-Binding Domain Enzyme-Linked Immunosorbent Assay. Open Forum Infect. Dis. 2021, 8, ofab220. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kolesov, D.E.; Sinegubova, M.V.; Dayanova, L.K.; Dolzhikova, I.V.; Vorobiev, I.I.; Orlova, N.A. Fast and Accurate Surrogate Virus Neutralization Test Based on Antibody-Mediated Blocking of the Interaction of ACE2 and SARS-CoV-2 Spike Protein RBD. Diagnostics 2022, 12, 393. https://doi.org/10.3390/diagnostics12020393
Kolesov DE, Sinegubova MV, Dayanova LK, Dolzhikova IV, Vorobiev II, Orlova NA. Fast and Accurate Surrogate Virus Neutralization Test Based on Antibody-Mediated Blocking of the Interaction of ACE2 and SARS-CoV-2 Spike Protein RBD. Diagnostics. 2022; 12(2):393. https://doi.org/10.3390/diagnostics12020393
Chicago/Turabian StyleKolesov, Denis E., Maria V. Sinegubova, Lutsia K. Dayanova, Inna V. Dolzhikova, Ivan I. Vorobiev, and Nadezhda A. Orlova. 2022. "Fast and Accurate Surrogate Virus Neutralization Test Based on Antibody-Mediated Blocking of the Interaction of ACE2 and SARS-CoV-2 Spike Protein RBD" Diagnostics 12, no. 2: 393. https://doi.org/10.3390/diagnostics12020393
APA StyleKolesov, D. E., Sinegubova, M. V., Dayanova, L. K., Dolzhikova, I. V., Vorobiev, I. I., & Orlova, N. A. (2022). Fast and Accurate Surrogate Virus Neutralization Test Based on Antibody-Mediated Blocking of the Interaction of ACE2 and SARS-CoV-2 Spike Protein RBD. Diagnostics, 12(2), 393. https://doi.org/10.3390/diagnostics12020393






