Exome Sequencing Revealed a Novel Splice Site Variant in the CRB2 Gene Underlying Nephrotic Syndrome
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Genomic DNA Extraction and Biochemical Analysis
2.3. Whole Exome Sequencing and Variant Selection
2.4. Primer Designing and Sanger Sequencing
3. Results
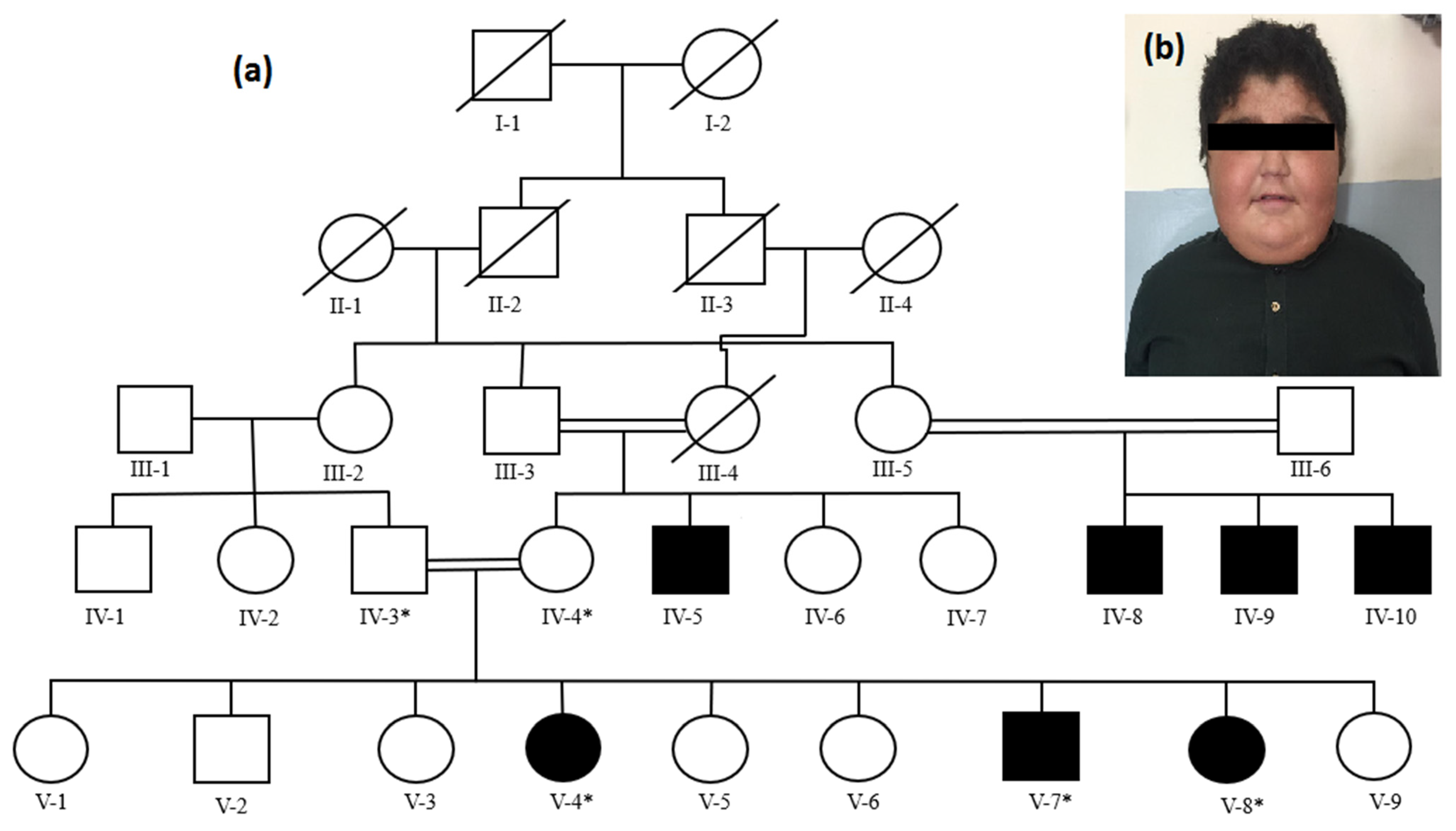
3.1. Clinical Features
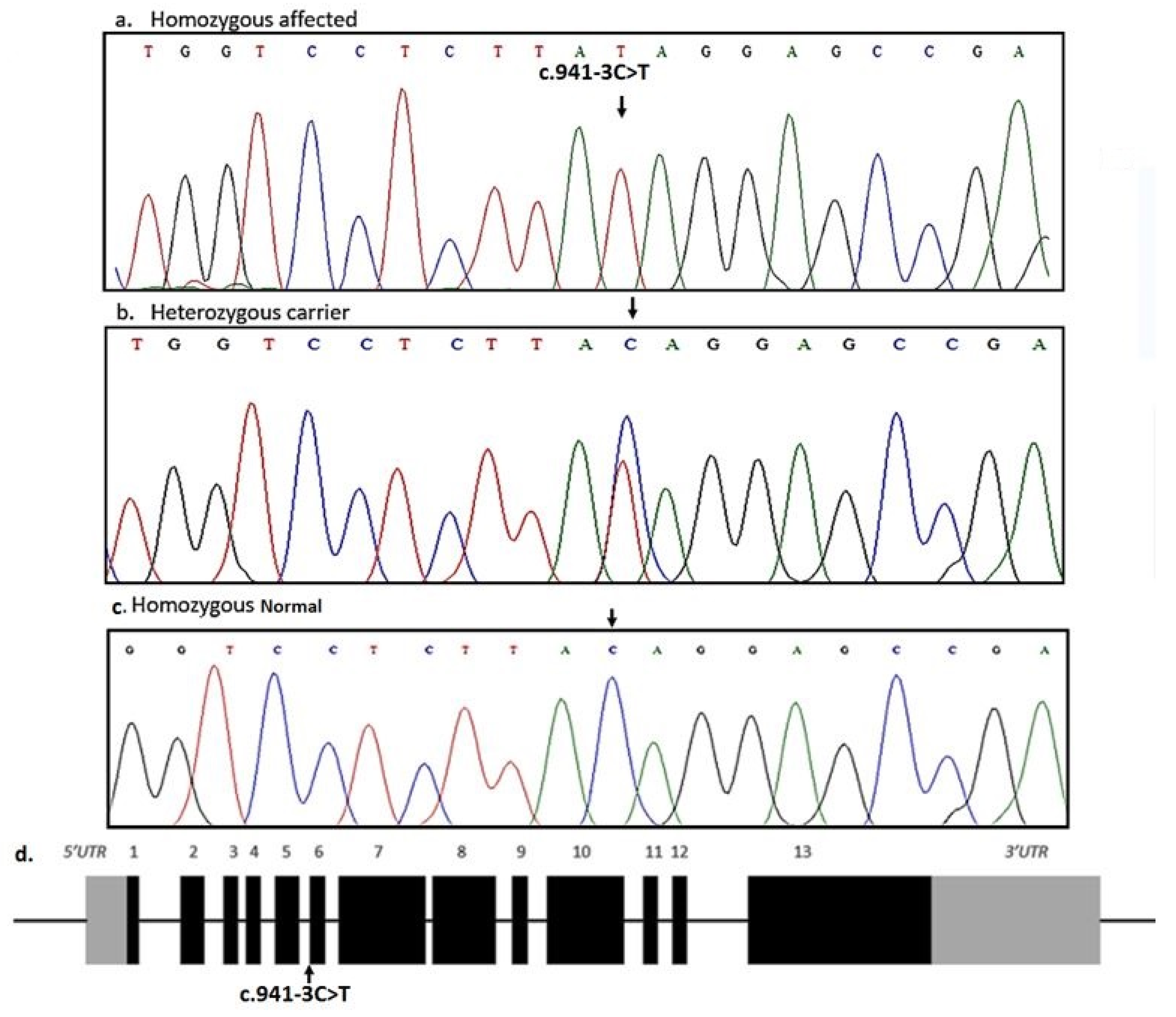
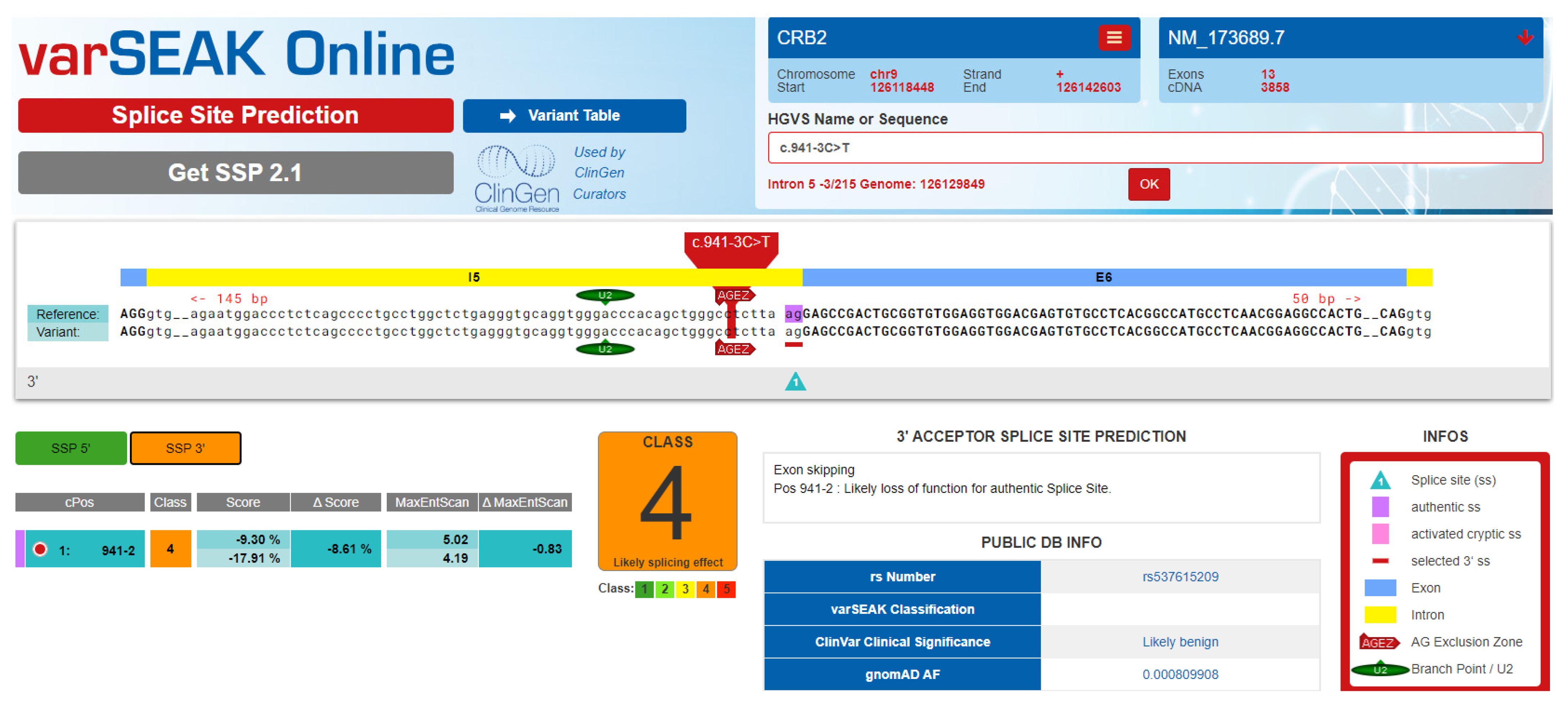
3.2. Genetic Description
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ebarasi, L.; Ashraf, S.; Bierzynska, A.; Gee, H.; McCarthy, H.; Lovric, S.; Sadowski, C.; Pabst, W.; Vega-Warner, V.; Fang, H.; et al. Defects of CRB2 cause steroid-resistant nephrotic syndrome. Am. J. Hum. Genet. 2015, 96, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Reiser, J.; Kriz, W.; Kretzler, M.; Mundel, P. The glomerular slit diaphragm is a modified adherens junction. J. Am. Soc. Nephrol. 2000, 11, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.I.; Kronbichler, A.; Oh, J.; Meijers, B. Nephrotic Syndrome: Genetics, Mechanism, and Therapies. Biomed. Res. Int. 2018, 2018, 6215946. [Google Scholar] [CrossRef] [PubMed]
- Wiggins, R.C. The spectrum of podocytopathies: A unifying view of glomerular diseases. Kidney Int. 2007, 71, 1205–1214. [Google Scholar] [CrossRef] [PubMed]
- Downie, M.L.; Gallibois, C.; Parekh, R.S.; Noone, D.G. Nephrotic syndrome in infants and children: Pathophysiology and management. Paediatr. Int. Child Health 2017, 37, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.Z.; Shah, S.J.; Anwar, N.; Hakeem, F. Clinical presentations of nephrotic syndrome in patients of a tertiary care hospital at Peshawar. J. Ayub. Med. Coll. Abbottabad 2013, 25, 31–34. [Google Scholar]
- Rheault, M.N.; Zhang, L.; Selewski, D.T.; Kallash, M.; Tran, C.L.; Seamon, M.; Katsoufis, C.; Ashoor, I.; Hernandez, J.; Supe-Markovina, K.; et al. AKI in Children Hospitalized with Nephrotic Syndrome. Clin. J. Am. Soc. Nephrol. 2015, 10, 2110–2118. [Google Scholar] [CrossRef] [PubMed]
- Rheault, M.N.; Gbadegesin, R.A. The Genetics of Nephrotic Syndrome. J. Pediatr. Genet. 2015, 5, 15–24. [Google Scholar] [PubMed]
- Eddy, A.A.; Symons, J.M. Nephrotic Syndrome in Childhood. Lancet 2003, 362, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Basit, S.; Al-Edressi, H.M.; Sairafi, M.H.; Hashmi, J.A.; Alharby, E.; Safar, R.; Ramzan, K. Centromere protein I (CENPI) is a candidate gene for X-linked steroid sensitive nephrotic syndrome. J. Nephrol. 2020, 33, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Hashmi, J.A.; Safar, R.A.; Afzal, S.; Albalawi, A.M.; Abdu-Samad, F.; Iqbal, Z.; Basit, S. Whole exome sequencing identification of a novel insertion mutation in the phospholipase C ε-1 gene in a family with steroid resistant inherited nephrotic syndrome. Mol. Med. Rep. 2018, 18, 5095–5100. [Google Scholar] [CrossRef] [PubMed]
- Lipska-Ziętkiewicz, B.S.; Ozaltin, F.; Hölttä, T.; Bockenhauer, D.; Bérody, S.; Levtchenko, E.; Vivarelli, M.; Webb, H.; Haffner, D.; Schaefer, F.; et al. Genetic aspects of congenital nephrotic syndrome: A consensus statement from the ERKNet–ESPN inherited glomerulopathy working group. Eur. J. Hum. Genet. 2020, 28, 1368–1378. [Google Scholar] [CrossRef] [PubMed]
- Tepass, U.; Theres, C.; Knust, E. Crumb encodes an EGF-like protein expressed on apical membranes of Drosophila epithelial cells and required for organization of epithelia. Cell 1990, 61, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Van den Hurk, J.A.J.M.; Rashbass, P.; Roepman, R.; Davis, J.; Voesenek, K.E.J.; Arends, M.L.; Zonneveld, M.N.; van Roekel, M.H.G.; Cameron, K.; Rohrschneider, K.; et al. Characterization of the Crumbs homolog 2 (CRB2) gene and analysis of its role in retinitis pigmentosa and Leber congenital amaurosis. Mol. Vis. 2005, 11, 263–273. [Google Scholar] [PubMed]
- Jaron, R.; Rosenfeld, N.; Zahdeh, F.; Carmi, S.; Beni-Adani, L.; Doviner, V.; Picard, E.; Segel, R.; Zeligson, S.; Carmel, L.; et al. Expanding the phenotype of CRB2 mutations—A new ciliopathy syndrome? Clin. Genet. 2016, 90, 540–544. [Google Scholar] [CrossRef] [PubMed]
- Slavotinek, A.M. The family of crumbs genes and human disease. Mol. Syndromol. 2016, 7, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Fu, R.; Ren, F.; He, J.; Wang, S.; Gou, M. A case report of CRB2 mutation identified in a Chinese boy with focal segmental glomerulosclerosis. Medicine 2018, 97, e12362. [Google Scholar] [CrossRef] [PubMed]
- Lamont, R.E.; Tan, W.; Innes, A.M.; Parboosingh, J.S.; Schneidman-Duhovny, D.; Rajkovic, A.; Pappas, J.; Altschwager, P.; De Ward, S.; Fulton, A.; et al. Expansion of phenotype and genotypic data in CRB2-related syndrome. Eur. J. Hum. Genet. 2016, 24, 1436–1444. [Google Scholar] [CrossRef] [PubMed]
- Adutwum, M.; Hurst, A.; Mirzaa, G.; Kushner, J.D.; Rogers, C.; Khalek, N.; Cristancho, A.G.; Burrill, N.; Seifert, M.E.; Scarano, M.I.; et al. Six new cases of CRB2-related syndrome and a review of clinical findings in 28 reported patients. Clin. Genet. 2022. [Google Scholar] [CrossRef] [PubMed]
| Parameters | Patient V-4 | Patient V-7 | Patient V-8 | Reference Range |
|---|---|---|---|---|
| Age | 13 | 11 | 8 | |
| Weight | 63 kg | 36 kg | 29 kg | |
| Height | 154 cm | 139 cm | 124 cm | |
| Blood routine tests | ||||
| Erythrocytes, m/mm3 | 5.3 | 4.9 | 5.7 | 4.7–6.1 |
| Leucocyte, 109/L | 6.73 | 7.32 | 5.92 | 4.0–10.5 |
| Platelet count, 109/L | 345 | 298 | 166 | 150–400 |
| Hemoglobin, g/L | 13.2 | 11 | 10.4 | 12.5–16.1 |
| Neutrophil, (%) | 63 | 73 | 59 | 54–62 |
| Lymphocyte (%) | 33 | 27 | 30 | 25–33 |
| Urine routine tests | ||||
| Specific gravity | 1.010 | 1.006 | 1.009 | 1.005–1.025 |
| Urine protein | 300 mg/dl | 700 mg/dl | 650 mg/dl | <150 mg/dl |
| Urinary occult blood | 3 | 0 | 2 | <4 RBC/HPF |
| Urine glucose | <130 | <130 | <130 | <130 mg/dl |
| Urine ketone bodies | Nil | nil | Nil | nil |
| Urine creatinine, mmol/L | 7 | 8.2 | 6.9 | 6–13 |
| 24-h UPE, mg/d | <100 | <100 | <100 | <100 mg/m2/day |
| Urinary β2- microglobulin, mg/L | 0.1 | 0.2 | 0.13 | <0.3 |
| Immunology | ||||
| Immunoglobulin A, g/L | 104 | 188 | 204 | 33–236 |
| Immunoglobulin G, g/L | 788 | 1023 | 987 | 608–1572 |
| Immunoglobulin M, g/L | 69 | 123 | 215 | 52–242 |
| Complement C3, g/L | 98 | 102 | 173 | 88–201 |
| Complement C4, g/L | 32 | 39 | 22 | 15–45 |
| Serum chemistry | ||||
| Total protein, g/L | 6.8 | 7.2 | 6.3 | 6.0–8.3 |
| Albumin, g/L | 2.5 | 2.3 | 2.4 | 3.5–5.6 |
| Globulin, g/L | 2.5 | 2.9 | 3.3 | 2.0–3.5 |
| Serum creatinine, mmol/L | 0.36 | 0.64 | 0.73 | 0.31–0.88 |
| Blood coagulation tests | ||||
| Prothrombin time | 11.3 | 10.7 | 11.9 | 10.1–12 |
| Activated partial thromboplastin time, seconds | 33 | 35 | 29 | 26–36 |
| Prothrombin time-international normalized ratio | 0.81 | 0.91 | 0.89 | 0.8–1.1 |
| Fibrinogenic, g/L | 159 | 236 | 342 | 156–400 |
| Renal biopsy findings | focal segmental glomerulosclerosis | - | - | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simaab, A.; Krishin, J.; Alaradi, S.R.; Haider, N.; Shah, M.; Ullah, A.; Abdullah, A.; Ahmad, W.; Hansen, T.; Basit, S. Exome Sequencing Revealed a Novel Splice Site Variant in the CRB2 Gene Underlying Nephrotic Syndrome. Medicina 2022, 58, 1784. https://doi.org/10.3390/medicina58121784
Simaab A, Krishin J, Alaradi SR, Haider N, Shah M, Ullah A, Abdullah A, Ahmad W, Hansen T, Basit S. Exome Sequencing Revealed a Novel Splice Site Variant in the CRB2 Gene Underlying Nephrotic Syndrome. Medicina. 2022; 58(12):1784. https://doi.org/10.3390/medicina58121784
Chicago/Turabian StyleSimaab, Anam, Jai Krishin, Sultan Rashid Alaradi, Nighat Haider, Muqadar Shah, Asmat Ullah, Abdullah Abdullah, Wasim Ahmad, Torben Hansen, and Sulman Basit. 2022. "Exome Sequencing Revealed a Novel Splice Site Variant in the CRB2 Gene Underlying Nephrotic Syndrome" Medicina 58, no. 12: 1784. https://doi.org/10.3390/medicina58121784
APA StyleSimaab, A., Krishin, J., Alaradi, S. R., Haider, N., Shah, M., Ullah, A., Abdullah, A., Ahmad, W., Hansen, T., & Basit, S. (2022). Exome Sequencing Revealed a Novel Splice Site Variant in the CRB2 Gene Underlying Nephrotic Syndrome. Medicina, 58(12), 1784. https://doi.org/10.3390/medicina58121784






