Abstract
Background and Objectives: There is no biomarker to predict lithium response. This study used CellPrint™ enhanced flow cytometry to study 28 proteins representing a spectrum of cellular pathways in monocytes and CD4+ lymphocytes before and after lithium treatment in patients with bipolar disorder (BD). Materials and Methods: Symptomatic patients with BD type I or II received lithium (serum level ≥ 0.6 mEq/L) for 16 weeks. Patients were assessed with standard rating scales and divided into two groups, responders (≥50% improvement from baseline) and non-responders. Twenty-eight intracellular proteins in CD4+ lymphocytes and monocytes were analyzed with CellPrint™, an enhanced flow cytometry procedure. Data were analyzed for differences in protein expression levels. Results: The intent-to-treat sample included 13 lithium-responders (12 blood samples before treatment and 9 after treatment) and 11 lithium-non-responders (11 blood samples before treatment and 4 after treatment). No significant differences in expression between the groups was observed prior to lithium treatment. After treatment, the majority of analytes increased expression in responders and decreased expression in non-responders. Significant increases were seen for PDEB4 and NR3C1 in responders. A significant decrease was seen for NR3C1 in non-responders. Conclusions: Lithium induced divergent directionality of protein expression depending on the whether the patient was a responder or non-responder, elucidating molecular characteristics of lithium responsiveness. A subsequent study with a larger sample size is warranted.
1. Introduction
In acute and maintenance treatment of bipolar disorder (BD), lithium is still a first-line medication [1]. About 1/3 to 2/3 of patients may reach treatment response, defined as ≥50% symptom improvement from baseline, in the acute phase of treatment [2,3,4,5]. However, the mechanism of lithium treatment response remains unclear [6] and there is no reliable predictor for lithium treatment response. Demographic and clinical characteristics have been compared between lithium responders and non-responders [3,7], and some of them have been considered as guidance in clinical practice. Along with its requirement for repeated laboratory monitoring of renal and thyroid functions, the use of lithium in BD continues declining [8] although lithium has unique neuroprotective and anti-suicidal effects [9,10]. However, if there is a biomarker or a panel of biomarkers for predicting lithium response, the use of lithium can be maximized, and the inconvenience and potential side effects related to lithium use can be avoided.
The effort of searching for biomarkers to predict lithium treatment response has been ongoing for decades. Using blood samples, researchers have investigated potential predictors for lithium response at different levels including genomic [11,12,13,14], gene expression [15,16,17,18,19,20], protein levels [21,22,23,24,25], neurotransmitters, signal transduction and pathways, endocrine systems, cytokines and immune systems, circadian rhythm, and mitochondria [6]. Brain imaging and brain activity-related measures have also been used to study the predictors for lithium treatment response [6,26,27]. However, these efforts have yet to produce reliable predictors of lithium responsiveness [28,29] although a large study from the International Consortium on Lithium Genetics (ConLi*Gen) found that bipolar patients with low genetic loading for schizophrenia had better response to lithium than those with high genetic loading [14].
However, studies from human induced pluripotent stem cells (iPSC) and lymphoblastoid cell lines found that lithium responders and non-responders had different molecular biomarkers [30,31,32]. Lithium treatment was linked to differential gene expression and different electrophysiological activities [30,31,32]. More importantly, only neurons derived from lithium-responders responded to lithium [30,31], and neurons derived from lithium non-responders responded to lamotrigine, but not to lithium [30]. In addition, differential gene expression and protein phosphorylation in those two types of neurons were also observed [32]. Studies from the Pharmacogenomic Study of Bipolar Disorder (PGDB) found that lithium response was related to the architecture of circadian rhythms and lithium treatment stabilized circadian disruptions [33,34]. These data suggest that bipolar lithium responders and non-responders can be separated with identifiable biomarkers that may be used in routine clinical practice. However, a very small contribution of each gene SNP to a complex disease, a mismatch between mRNA and protein levels [35,36,37], and the inability to measure post translational protein modifications, such as phosphorylation or methylation events have challenged the usefulness of genomic and transcriptomic approaches in studying biomarkers for predicting treatment response of complex diseases like BD.
On the other hand, expressed protein levels and phosphorylation are highly correlated with cellular functions and phenotypes. Proteomic studies are not only likely to help us find different phenotypes based on lithium response and biomarkers, but also help us understand the pathology of BD. However, most plasma/serum-based technologies simultaneously interrogate the averaged productive capabilities of all cells in the body and thus are obligatorily low resolution and low sensitivity. Also, lithium can reach many organs, tissues, neurons, and non-neuronal cells, and act on different genes and pathways [6]. Therefore, it will be difficult to use low sensitive technologies to measure multiple proteins simultaneously in plasma/serum to find predictor(s) for lithium treatment response.
Flow cytometry can measure expression of multiple intracellular or surface-bound proteins including protein modifications in individual cells. This technology has been used in previous studies of BD [38,39,40,41,42]. However, flow cytometry has traditionally suffered poor signal to noise when measuring multiple markers at the same time or markers with low expression [43,44]. Enhanced flow cytometry developed by CellPrint Biotechnology, LLC (CellPrint™) is an innovative tyramide-based catalytic deposition labeling procedure, which improves dynamic range by 20 fold and signal to noise ratios by 10–100 fold compared to standard flow cytometric methods. The technology improves the capability of flow cytometers to report expression levels of low abundance analytes and intracellular molecules. As a result, this approach enables the detection and quantitative assessment of a wide variety of surface and intracellular proteins from numerous cell types [45,46,47,48,49,50,51,52]. In our previous analysis of 17 intracellular analytes, CellPrint™ was able to identify intracellular proteins in CD4+ lymphocytes and monocytes to differentiate bipolar lithium responders from non-responders [45]. The aim of this analysis was to assess differences in protein expression pre- and post- lithium for treatment responders and non-responders.
2. Methods
2.1. Study Design
The study design, study procedures, the inclusion and exclusion criteria, diagnosis, efficacy and safety assessments, and blood sample collection were detailed previously [45]. Briefly, this study was a 16-week open-label study of lithium monotherapy treatment of patients with BD type I or II who were at any phase of the illness and with at least mild symptoms (clinicaltrial.gov, NCT02909504). The diagnoses were ascertained with the MINI for DSM-5 and a structured research diagnostic interview. Standardized rating scales for depression, anxiety or mania were used for measuring symptom severity. Disability was measured with Sheehan Disability Rating scale and quality of life was measured with the Quality of Life, Enjoyment and Satisfaction Questionnaire. Rating scales were completed at baseline, week 1, 2, 4, 6, 8, 12, and 16. Eligible patients were treated with lithium for up to 16 weeks and ongoing unpermitted medications were tapered off by week 4. Blood samples of all patients were collected at the baseline and at the end of the study. Levels of intracellular proteins before and after lithium in monocytes and CD4+ lymphocytes of the lithium responders and lithium non-responders were measured.
2.2. Rationale of Using Monocytes and CD4+ Lymphocytes
The comparability of blood and brain have been investigated at different levels. At the DNA methylation level, the brain and blood are highly correlated. At the transcriptome level, whole blood [53,54] and peripheral blood mononuclear cells [55] had similar gene expression patterns as the brain tissues [56]. A systematic review has shown that neurotropic factors have similar changes in both central nervous system and peripheral blood system [25]. In addition, functional connections between brain and blood cells are through immune cells in the peripheral circulation with the brain [57,58].
Peripheral blood mononuclear cells (PBMC) have been used for genomic, genetic, gene expression, and protein expression studies of lithium treatment response in BD [16,59,60,61] as well as diagnostic and pathologic studies of BD [62,63,64,65,66,67]. Among the blood mononuclear cells, lymphocytes [38,68,69] and monocytes [69,70] have been extensively studied with flow cytometry for different purposes. In addition, collection of blood samples is relatively easy and cheap, and results from the PBMCs can be easily applied to routine clinical practice. Therefore, we chose monocytes and CD4+ lymphocytes as reporters of flow cytometry analysis in the current study.
2.3. Antibodies and Cytometric Analyses
CellPrint™ enhanced signal is generated by catalyzed reporter deposition and was used to measure the levels of intracellular protein in the study. The details of the procedure have been published elsewhere [45,46,47,48,49,50,51,52]. Briefly, antibodies targeting surface antigens to demarcate cell subtypes (CD4+ lymphocytes and monocytes) are employed using manufacturer standard protocols. Amplified signal for intracellular analytes is generated with commercially available antibodies to the target proteins or phospho-proteins. Once these primary antibodies are bound, peroxidase enzyme is bound via secondary antibodies. A tyramide-fluorophore conjugate substrate for peroxidase is then used to amplify the signal. Amplified signal is detected with a regular flow cytometer.
After blood sample collection of the study was completed, the frozen samples were thawed for cell-specific molecular expression analysis by CellPrint Biotechnology, LLC (Cleveland, OH, USA). Acquisition of fluorescence levels was accomplished on a BD Accuri C6 flow cytometer and recorded as median fluorescence intensity (MFI). MFI for each analyte was normalized to the fluorescence minus one (FMO) control to generate a median fluorescence ratio (MFR), which is a quantitative measure of relative protein expression level. MFR = 1 means no detection of an analyte.
Antibodies to 28 proteins were used in the study. The commercially available antibodies were evaluated with proprietary quality controls established by the CellPrint Biotechnology team. The 28 intracellular analytes probed were involved in a spectrum of pathways/functions (Supplemental Table S1). The selection of these proteins was based on previous studies [24,25,38,39,40,41,42,69,71,72,73,74].
After the flow cytometric analyses were completed, the raw cytometric data were sent to the clinical investigation site. The clinical team provided the status of each patient as a responder or a non-responder to the statistics team. The statistics team conducted analyses to assess any differences between responders and non-responders, and between before and after lithium treatment.
2.4. Normalization of Raw Data
The MFR of each analyte for responders and non-responders, and before and after lithium was “normalized” with fold change/difference (FC). The FC of each analyte was calculated with a formula of for comparison between lithium responders and non-responders, or for before and after lithium comparison. Therefore, a positive value of the log2 (FC) is indicative of a higher level in non-responders or after lithium, and a negative value of log2 (FC) is indicative of a lower level in non-responders or after lithium.
2.5. Statistical Analysis
The statistical method is dependent on the variables being analyzed. Overall, the bivariate data were analyzed with Chi-square or Fisher Exact tests. Continuous variables were analyzed with T-test. Analyses of demographics and clinical characteristics between responders and non-responders were conducted as previously described [45]. Similarly, a ≥50% reduction in Montgomery Asberg Rating Scale (MADRS and/or Young Mania Rating Scale (YMRS) from baseline to the end of study were used to define a responder.
The flow cytometric data of each of the 28 analytes in monocytes and CD4+ lymphocytes between responders and non-responders at baseline were analyzed with unpaired t-test, and within group before and after lithium were analyzed with paired t-test. The log2 (FC) normalization data and the raw MFR for the analytes were used to study the changes in each analyte before and after lithium. Due to the explorative nature of the study, no adjustment was attempted for multiple comparisons. Data were analyzed using SAS software (SAS version 9.2, SAS Institute Inc., Cary, NC, USA).
3. Results
The demographics, historical correlates, and changes in depression and anxiety severities after lithium treatment were described previously [45]. Of the 25 patients who were treated with lithium, twenty-four had at least one post-baseline visit (intent to treat, ITT). Of the 24 patients, 13 were classified as treatment responders (R), and 11 were treatment non-responders (NR).
3.1. MFR of Analytes at Baseline: ITT-Responders vs. ITT-Non-Responders
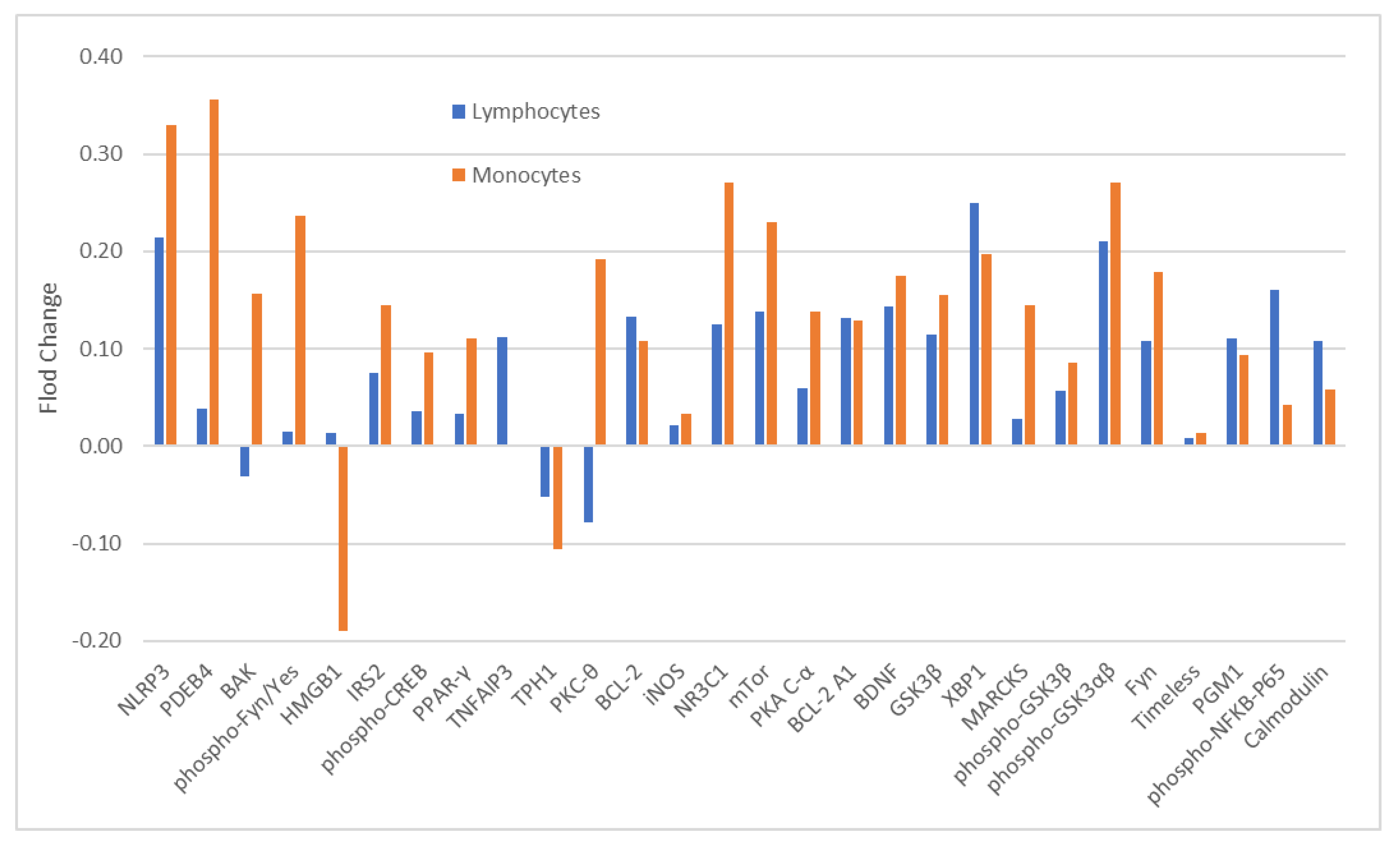
The blood samples of 12 of 13 ITT-responders (ITT-R-baseline) and all 11-ITT non-responders (ITT-NR-baseline) were available for baseline analysis of 28 analytes. The expression levels of these analytes between ITT-R-baseline and ITT-NR-baseline were not significantly different in both monocytes (Table 1) and CD4+ lymphocytes (Supplemental Table S2). With the exception of a few analytes, a majority of analytes had higher levels in ITT-NR-baseline than in ITT-R-baseline in both cell types although the magnitude of differences varied widely (Figure 1).

Table 1.
Comparison of protein levels in monocytes between lithium responders and non-responders before and lithium treatment.
Figure 1.
Fold change (FC) of 28 analytes in monocytes and CD4+ lymphocytes at baseline between lithium responders and non-responders. Note: FC = log2 (median fluorescent ratio of non-responders/median fluorescent ratio of responders). Positive value is indicative of a higher level of protein expression in lithium non-responders than in lithium responders. Negative value is indicative of a lower level of protein expression in lithium non-responders than in lithium responders. Abbreviations: BAK: BAX, BCL2-Associated × Protein; BCL-2: B-cell lymphoma 2; BCL-2 A1: Bcl-2-related protein A1; BDNF: brain-derived neurotrophic factor; Calmodulin: calciummodulated protein; Fyn: a tyrosine kinase belongs to the Src family of tyrosine kinases including src, fyn, and yes; GSK3β: glycogen synthase kinase 3 beta; HMGB1: High mobility group box 1 protein; iNOS: inducible isoform nitric oxide synthases; IRS2: Insulin receptor substrate 2; MARCKS: myristoylated alaninerich C-kinase substrate; mTor: mammalian target of rapamycin; NLRP3: NACHT, LRR and PYD domains-containing protein 3; NR3C1: nuclear receptor subfamily 3, group C, member 1; phospho-CREB: phosphorylated cAMP response element-binding protein (Ser133); phospho-Fyn/Yes: phosphorylated Fyn(Y530)/Yes(Y537); phospho-GSK3αβ: phosphorylated glycogen synthase kinase 3 alpha(Tyr279) beta(Tyr216); phospho-GSK 3β: phospho-glycogen synthase kinase 3 beta(Tyr216); phospho-NFkB-P65: phosphorylated nuclear factor NF-kappa-B p65(Ser536) subunit; PDEB4: cAMP-specific 3′,5′-cyclic phosphodiesterase 4B; PGM1: phosphoglucomutase 1; PKA C-α: protein kinase A catalytic subunit; PKC-θ: protein kinase C theta; PPAR-γ: peroxisome proliferator-activated receptor gamma; Timeless: a protein is necessary of proper functioning of circadian rhythm; TNFAIP3: tumor necrosis factor, alpha-induced protein 3; TPH1: tryptophan hydroxylase 1; XBP1: X-box binding protein 1.
3.2. MFR of Analytes at Baseline and at the End of Study: Completed Responders vs. Completed Non-Responders
Nine of 12 ITT-R completed the study (C-R) and 4 of 11 ITT-NR completed the study (C-NR). All these completers had blood samples available at both baseline and the end of study (EOS). As with the analysis in the ITT sample, the protein levels between C-R-baseline and C-NR-baseline were not significantly different in both monocytes and CD4+ lymphocytes (Supplemental Table S3). However, after lithium treatment, there were significant differences in a number of protein levels between C-R-EOS and C-NR-EOS in monocytes (Table 1) and CD4+ lymphocytes (Supplemental Table S2). The levels of iNOS, NLRP3, phospho-GSK3β, and PGM1were significantly lower in C-NR-EOS than in C-R-EOS in both cell types (Table 1 and Supplemental Table S2).
3.3. MFR of Analytes before and after Lithium in Completed Responders and Completed Non-Responders
Of the 9 C-R, the expression levels of 28 analytes in monocytes (Table 2) and CD4+ lymphocytes (Supplemental Table S4) before and after lithium treatment were compared. In the monocytes, PDEB4 (p = 0.03) and NRC31 (p = 0.03) were significantly increased (Table 2). In the CD4+ lymphocytes, PDEB4 was significantly increased (p = 0.05) (Supplemental Table S4).

Table 2.
Comparison of protein levels in monocytes of lithium completed responders and non-responders between before and after lithium treatment.
Of the 4 C-NR, all 28 analytes before and after lithium treatment were also compared. In the monocytes, NRC31 levels were significantly decreased with lithium (p = 0.04) (Table 2). However, in the CD4+ lymphocytes, none of the changes in analytes after lithium treatment was significantly different from the levels before lithium (Supplemental Table S4).
3.4. Fold Change in MFR of Analytes before and after Lithium in Responders and Non-Responders
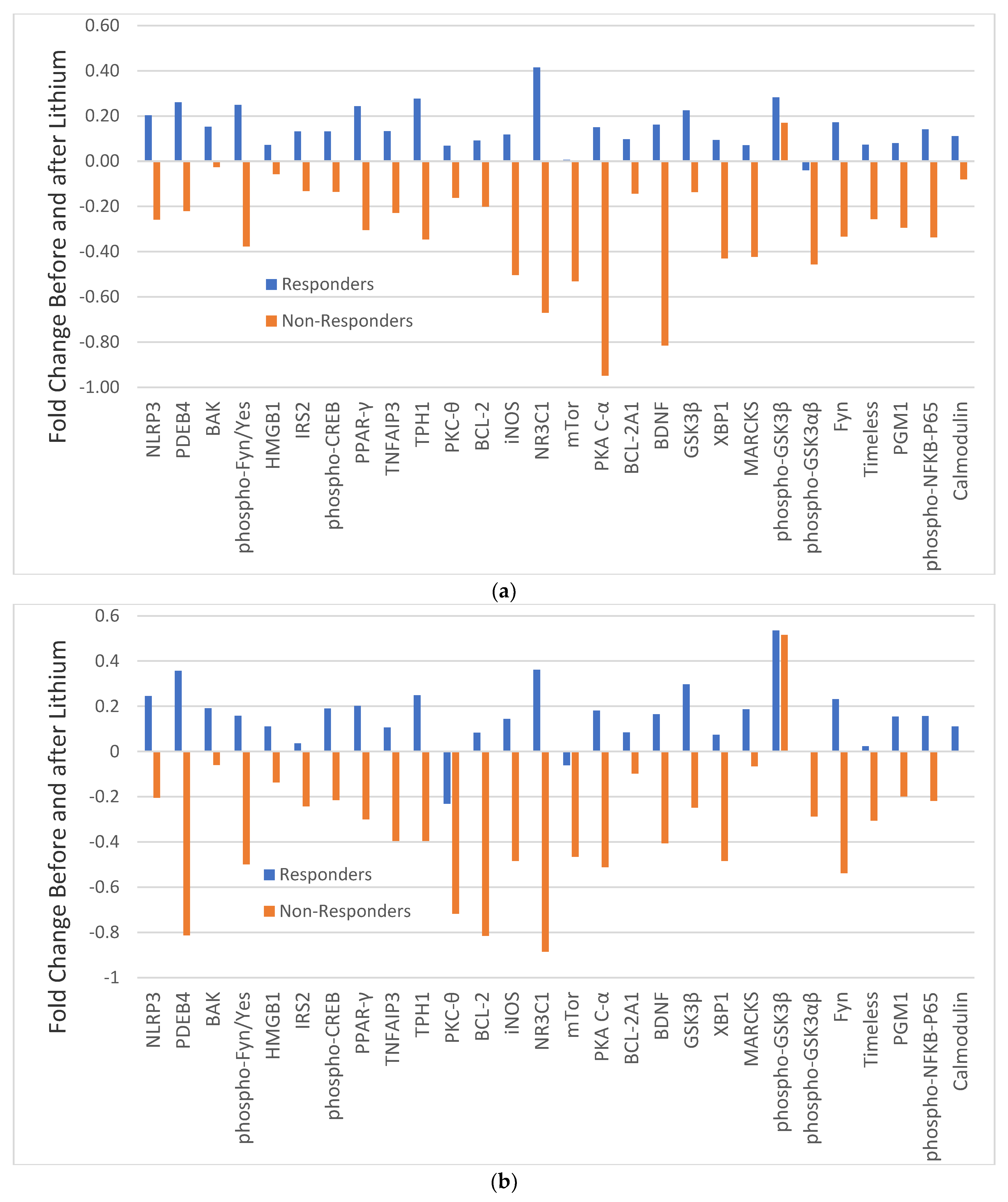
In C-R and C-NR, the changes in 28 analytes before and after lithium treatment varied widely as manifested with different magnitudes of FC (Figure 2). In monocytes, with the exception of PKC-θ and possibly mTor, all other analytes in C-R were increased with different magnitudes (blue bars in Figure 2a), but only PDEB4 and NRC31 were significantly increased (Table 2). For C-NR, with the exception of phospho-GSK3β, all other analytes were decreased with different magnitudes (brown bars in Figure 2a), but only the change in NRC31 was significantly different (Table 2). The changes in most analytes before and after lithium were in opposite directions between C-R and C-NR.
Figure 2.
Fold change (FC) of 28 analytes in monocytes (a) and CD4+ lymphocytes (b) in responders (blue bars) and non-responders (brown bars) before and after lithium. Note: FC = log2 (median fluorescent ratio of after lithium/median fluorescent ratio of before lithium Positive value is indicative of a higher level of protein expression after lithium than before lithium. Negative value is indicative of a lower level of protein expression after lithium than before lithium. Abbreviations: BAK: BAX, BCL2-Associated × Protein; BCL-2: B-cell lymphoma 2; BCL-2 A1: Bcl-2-related protein A1; BDNF: brain-derived neurotrophic factor; Calmodulin: calcium-modulated protein; Fyn: a tyrosine kinase belongs to the Src family of tyrosine kinases including src, fyn, and yes; GSK3β: glycogen synthase kinase 3 beta; HMGB1: High mobility group box 1 protein; iNOS: inducible isoform nitric oxide synthases; IRS2: Insulin receptor substrate 2; MARCKS: myristoylated alanine-rich C-kinase substrate; mTor: mammalian target of rapamycin; NLRP3: NACHT, LRR and PYD domains-containing protein 3; NR3C1: nuclear receptor subfamily 3, group C, member 1; phospho-CREB: phosphorylated cAMP response element-binding protein (Ser133); phospho-Fyn/Yes: phosphorylated Fyn(Y530)/Yes(Y537); phospho-GSK3αβ: phosphorylated glycogen synthase kinase 3 alpha(Tyr279) beta(Tyr216); phospho-GSK 3β: phospho-glycogen synthase kinase 3 beta(Tyr216); phospho-NFkB-P65: phosphorylated nuclear factor NF-kappa-B p65(Ser536) subunit; PDEB4: cAMP-specific 3′,5′-cyclic phosphodiesterase 4B; PGM1: phosphoglucomutase 1; PKA C-α: protein kinase A catalytic subunit; PKC-θ: protein kinase C theta; PPAR-γ: peroxisome proliferator-activated receptor gamma; Timeless: a protein is necessary of proper functioning of circadian rhythm;TNFAIP3: tumor necrosis factor, alpha-induced protein 3; TPH1: tryptophan hydroxylase 1; XBP1: X-box binding protein 1.
Similarly, in CD4+ lymphocytes, all analytes in C-R were also increased with different magnitude, but only the increase in PDEB4 (p = 0.051) was significantly different (blue bars in Figure 2b, Supplemental Table S4). In contrast, almost all analytes in C-NR were decreased with different magnitude (brown bars in Figure 2b), with PKA C-α having the largest difference. However, individually, none was significantly different (Supplemental Table S4).
4. Discussion
In this pilot study, we employed a sensitive enhanced flow cytometric analysis to quantify protein expression changes in specific circulating mononuclear cells from BP I or II disorder lithium responders and non-responders. We found that lithium induced protein expression level changes in CD4+ lymphocytes and monocytes of bipolar patients. The direction of changes, upregulation or downregulation, moved in opposite directions depending on whether the patient was responsive or nonresponsive to lithium. Patients responsive to lithium showed a general increase in expression of the 28 analytes tested, whereas nonresponsive patients showed a general decrease in expression.
Although none of the specific analytes showed statistically significant differences between the two groups at baseline, lithium non-responders generally had higher levels of protein expression levels than lithium responders, which is consistent with our previous analysis of 17 intracellular analytes [45]. The upregulation of a majority of proteins in lithium responders and downregulation in non-responders further support that baseline levels of intracellular proteins may determine the treatment responsiveness to lithium. The change in expression levels after lithium in responders suggests that an upregulation may be a component of the lithium response. However, the meaning of the downregulation after lithium treatment in non-responders remains unclear.
Among the 28 analytes in the current study, only GSK3β and phospho-GSK3β were the subject of previous prospective studies [71,73,75], and only one study used lithium monotherapy [71]. These studies indicate that lithium monotherapy or combination therapy with other psychotropics was able to increase phospho-GSK3β levels, but not the total GSK3β levels. However, the relationship between the increase in phospho-GSK3β levels and improvement in the symptom severity of depression or mania was inconsistent. Since we found that lithium responders and non-responders had different changes after lithium treatment (Figure 2), the previous inconsistencies may have arisen because patients in prior studies were not first stratified by lithium responsiveness.
In our current analysis, phospho-GSK3β levels in monocytes were increased after lithium treatment in both responders and non-responders (Figure 2) with a trended significance in responders (Table 2). This result is consistent with previous studies where patients were analyzed without stratification by lithium responsiveness [71,73,75]. Therefore, the outcome of increased phospho-GSK3β with lithium for improving depression, mania, or both remains unclear. Our protein-protein network analysis of 17 analytes [45] indicated that most analytes in the current analysis are in the same protein network as GSK3β. Therefore, the inhibitory effect of phospho-GSK3β could trigger a cascade effect on downstream targets and pathways that are involved in inflammation, energy metabolism, and immune dysfunction [76,77].
One of the downstream targets of GSK3β is NFkB [45,78]. Previous studies have shown that lithium can decrease NFkB expression through inhibition of GSK3β and lithium has an anti-inflammatory effect [79,80,81,82,83]. However, in our current study, phospho-NFkB caused opposite changes in responders and non-responders after lithium in both cell types (Figure 2). Since the NFkB system is involved in immune development, immune response and inflammation [84,85], and cytokines may play a role in the pathophysiology in BD and lithium treatment response [25], it may not be surprising that factors related to inflammation such as NLRP3 and NR3C1 showed larger increases in lithium responders (Table 2). However, the meaning of the increase in NLRP3 and NR3C1 in lithium responders after lithium remains unclear.
The decreased expression levels induced by lithium of multiple analytes in non-responders is consistent with prior reports of the downregulation of genes in non-responders given lithium [15,30]. Since we only measured the change in symptom severity as a “benefit” of lithium treatment, other potential benefits or harm could have been neglected. Antisuicidal effects of lithium without mood stabilization has been reported [86]. Neurons derived from lithium non-responders did respond to lamotrigine [30], but lithium plus lamotrigine in bipolar depression was more effective than lithium alone [4]. The downregulation of some genes and/or proteins may be necessary for lithium non-responders to respond to lamotrigine adjunctive therapy to lithium. The “benefit” from lithium in lithium non-responders is worthy of further exploration.
In our previous study [45], baseline levels of 17 proteins including BCL2, BDNF, calmodulin, Fyn, phospho-Fyn/phospho-Yes, GSK3β, phospho-GSK3αβ, HMGB1, iNOS, IRS2, mTor, NLPR3, PGM1, PKA C-α, PPARγ, phosphorylated nuclear factor NF-kappa-B p65(Ser536) subunit (phospho-RelA), and TPH1 in monocytes and CD4+ lymphocytes were measured with Cellprint™. The levels of the majority of analytes in lithium responders were lower than in non-responders in both cell types as in the current study, but the level of GSK3β in monocytes was significantly different (p = 0.034). Among the 17 analytes assessed in both studies, most FCs of analytes between responders and non-responders in both cell types were larger in the previous study than in the current one. The FCs of GSK3β, phospho-GSK3αβ, and phospho-RelA in monocytes between non-responders and responders were 0.72, 0.47, and 0.73, respectively. In CD4+ lymphocytes, the FCs of phospho-GSK3αβ and GSK3β between two groups were 0.57 and 0.53, respectively. However, in the current study, none of the FCs was over 0.4 (Figure 1). The main differences between these two analyses were: (1) the number of the analytes in the current study was 28 versus 17 in the previous one; (2) the analysis of the previous study was performed after completing the analysis of the current study and proprietary improvement of the CellPrint™.
We also found that the combination of GSK3β and phospho-GSK3αβ levels in monocytes was able to correctly classified 11/11 responders and 5/8 non-responders. The combination of GSK3β, phospho-RelA, TPH1 and PGM1 correctly predicted 10/11 responders and 6/7 non-responders, both with a likelihood of ≥85%. In addition, signaling pathways of BDNF, neurotrophin, prolactin, leptin, and epidermal growth factor/epidermal growth factor receptors were found to be involved in the lithium treatment response. Similarly, in the current study, BDNF, neurotrophin, prolactin, and leptin pathways were involved in lithium response (data not shown).
Taken together, measuring multiple intracellular proteins with high sensitive flow cytometry such as the CellPrint™ may help us find biomarkers for predicting lithium treatment response in BD. A multiple approach model including clinical phenotypes, omics, neuroimaging, neuropsychological profiles, and neurophysiological characteristics may be necessary to achieve this goal [87].
Limitations
Although our results are promising, we should be cautious when interpreting the results. The sample size was small, and only four patients in the non-response group had pre- and post—lithium blood collection. Some markers might have significant differences between lithium responders and non-responders, and/or between before and after lithium with a larger sample size. This is the first study using the significantly more sensitive technique of the CellPrint™ to measure intracellular proteins in CD4+ lymphocytes and monocytes in patients with BD. Although more studies are needed to assess the utility of CellPrint™ in bipolar research, our current and previous analyses [45] along with the use of the CellPrint™ platform for different diseases [46,47,48,49,50,51,52] suggest that this technology may help the field to elucidate protein biomarkers in blood cells for predicting lithium response and its mechanism.
5. Conclusions
The preliminary results of the current study suggest that enhanced flow cytometry can be used to measure multiple intracellular proteins in the peripheral blood cells of patients with BD. Differences between lithium responders and non-responders at baseline may help us find biomarkers for predicting lithium treatment response. Differences between before and after lithium may shed light on the mechanism of lithium response. Large studies are needed to explore the utility of enhanced flow cytometry such as the CellPrint™ in searching for predictors of lithium treatment response.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/medicina59010120/s1. References are cited in [88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103]. Table S1. Abbreviation of 28 proteins in the discovery analysis in the pilot study; Table S2. Protein levels between lithium responders and non-responders in lymphocytes before and after lithium treatment; Table S3. Comparison of protein levels in lymphocytes and monocytes at baseline be-tween lithium completed responders and completed non-responders; Table S4. Comparison of protein levels in CD4+ lymphocytes between completed responder and non-responders before and after lithium.
Author Contributions
K.G., N.M.K., J.R.C. and D.K. wrote, revised, and finalized the study protocol; K.G. and J.R.C. collected the clinical data and blood samples. N.M.K., E.C. and D.K. performed cytometric analyses. K.G., M.A., N.M.K. and M.K. conducted statistical analysis. K.G. drafted the manuscript. The N.M.K., M.A., M.K., J.R.C., E.C., H.M.L., D.K. revised and finalized the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Brain and Behavior Research Foundation with an independent investigator grant to the first author.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of the University Hospitals Cleveland Medical Centger (UHCMC IRB number: 07-16-05 and date of approval was 26 September 2016).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study before any research procedure was performed.
Data Availability Statement
Data are available upon request.
Conflicts of Interest
Drs. Kaye, Christian, Kaplan, and Lazarus are the employees or the owners of the CellPrint Biotechnology, LLC, Cleveland, OH, USA. Other authors do not have conflict of interest to disclose.
References
- Yatham, L.N.; Kennedy, S.H.; Parikh, S.V.; Schaffer, A.; Bond, D.J.; Frey, B.N.; Sharma, V.; Goldstein, B.I.; Rej, S.; Beaulieu, S.; et al. Canadian Network for Mood and Anxiety Treatments (CANMAT) and International Society for Bipolar Disorders (ISBD) 2018 guidelines for the management of patients with bipolar disorder. Bipolar Disord. 2018, 20, 97–170. [Google Scholar] [CrossRef]
- Gao, K.; Goto, T.; Yuan, C.; Brownrigg, B.; Conroy, C.; Chan, P.K.; Serrano, M.B.; Ganocy, S.J.; Fang, F.; Calabrese, J.R. A pilot study of the effectiveness of lithium versus quetiapine immediate release monotherapy in patients with bipolar spectrum disorders. J. Clin. Psychopharmacol. 2018, 38, 422–434. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Maihofer, A.X.; Stapp, E.; Ritchey, M.; Alliey-Rodriguez, N.; Anand, A.; Balaraman, Y.; Berrettini, W.H.; Bertram, H.; Bhattacharjee, A.; et al. Clinical predictors of non-response to lithium treatment in the Pharmacogenomics of Bipolar Disorder (PGBD) study. Bipolar Disord. 2021, 23, 821–831. [Google Scholar] [CrossRef] [PubMed]
- van der Loos, M.L.; Mulder, P.G.; Hartong, E.G.; Blom, M.B.; Vergouwen, A.C.; de Keyzer, H.J.; Notten, P.J.; Luteijn, M.L.; Timmermans, M.A.; Vieta, E.; et al. Efficacy and safety of lamotrigine as add-on treatment to lithium in bipolar depression: A multicenter, double-blind, placebo-controlled trial. J. Clin. Psychiatry 2009, 70, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Young, A.H.; McElroy, S.L.; Bauer, M.; Philips, N.; Chang, W.; Olausson, B.; Paulsson, B.; Brecher, M.; EMBOLDEN I (Trial 001) Investigators. A double-blind, placebo-controlled study of quetiapine and lithium monotherapy in adults in the acute phase of bipolar depression (EMBOLDEN I). J. Clin. Psychiatry 2010, 71, 150–162. [Google Scholar] [CrossRef]
- Gao, K.; Calabrese, J.R. The mechanisms of action of lithium in bipolar disorder. In Neurobiology of Bipolar Disorder Road to Novel Therapeutics; Quevedo, J., Carvalho, A.F., Vieta, E., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 357–364. [Google Scholar]
- Hui, T.P.; Kandola, A.; Shen, L.; Lewis, G.; Osborn, D.P.J.; Geddes, J.R.; Hayes, J.F. A systematic review and meta-analysis of clinical predictors of lithium response in bipolar disorder. Acta Psychiatr. Scand. 2019, 140, 94–115. [Google Scholar] [CrossRef]
- Rybakowski, J.K. Challenging the negative perception of lithium and optimizing its long-term administration. Front. Mol. Neurosci. 2018, 11, 349. [Google Scholar] [CrossRef]
- Can, A.; Schulze, T.G.; Gould, T.D. Molecular actions and clinical pharmacogenetics of lithium therapy. Pharmacol. Biochem. Behav. 2014, 123, 3–16. [Google Scholar] [CrossRef]
- Tondo, L.; Baldessarini, R.J. Antisuicidal effects in mood disorders: Are they unique to lithium? Pharmacopsychiatry 2018, 51, 177–188. [Google Scholar] [CrossRef]
- Rybakowski, J.K. Response to lithium in bipolar disorder: Clinical and genetic findings. ACS Chem. Neurosci. 2014, 5, 413–421. [Google Scholar] [CrossRef]
- Budde, M.; Degner, D.; Brockmöller, J.; Schulze, T. Pharmacogenomic aspects of bipolar disorder: An update. Eur. Neuropsychopharmacol. 2017, 27, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Amare, A.T.; Schubert, K.O.; Baune, B.T. Pharmacogenomics in the treatment of mood disorders: Strategies and opportunities for personalized psychiatry. EPMA J. 2017, 8, 211–227. [Google Scholar] [CrossRef]
- International Consortium on Lithium Genetics (ConLi+Gen); Amare, A.T.; Schubert, K.O.; Hou, L.; Clark, S.R.; Papiol, S.; Heilbronner, U.; Degenhardt, F.; Tekola-Ayele, F.; Hsu, Y.H.; et al. Association of polygenic score for schizophrenia and HLA antigen and inflammation genes with response to lithium in bipolar affective disorder: A genome-wide association study. JAMA Psychiatry 2018, 75, 65–74. [Google Scholar]
- Beech, R.D.; Leffert, J.J.; Lin, A.; Sylvia, L.G.; Umlauf, S.; Mane, S.; Zhao, H.; Bowden, C.; Calabrese, J.R.; Friedman, E.S.; et al. Gene-expression differences in peripheral blood between lithium responders and non-responders in the Lithium Treatment-Moderate dose Use Study (LiTMUS). Pharm. J. 2013, 14, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Anand, A.; McClintick, J.N.; Murrell, J.; Karne, H.; Nurnberger, J.I.; Edenberg, H.J. Effects of lithium monotherapy for bipolar disorder on gene expression in peripheral lymphocytes. Complex Psychiatry 2016, 2, 115–123. [Google Scholar] [CrossRef]
- Kittel-Schneider, S.; Hilscher, M.; Scholz, C.-J.; Weber, H.; Grünewald, L.; Schwarz, R.; Chiocchetti, A.G.; Reif, A. Lithium-induced gene expression alterations in two peripheral cell models of bipolar disorder. World J. Biol. Psychiatry 2019, 20, 462–475. [Google Scholar] [CrossRef]
- Papadima, E.M.; Niola, P.; Melis, C.; Pisanu, C.; Congiu, D.; Cruceanu, C.; Lopez, J.P.; Turecki, G.; Ardau, R.; Severino, G.; et al. Evidence towards RNA Binding Motif (RNP1, RRM) Protein 3 (RBM3) as a potential biomarker of lithium response in bipolar disorder patients. J. Mol. Neurosci. 2017, 62, 304–308. [Google Scholar] [CrossRef]
- Costa, M.; Squassina, A.; Piras, I.S.; Pisanu, C.; Congiu, D.; Niola, P.; Angius, A.; Chillotti, C.; Ardau, R.; Severino, G.; et al. Preliminary transcriptome analysis in lymphoblasts from cluster headache and bipolar disorder patients implicates dysregulation of circadian and serotonergic genes. J. Mol. Neurosci. 2015, 56, 688–695. [Google Scholar] [CrossRef]
- Pisanu, C.; Papadima, E.M.; Melis, C.; Congiu, D.; Loizedda, A.; Orrù, N.; Calza, S.; Carcassi, C.; Severino, G.; Ardau, R.; et al. Whole genome expression analyses of miRNAs and mRNAs suggest the involvement of miR-320a and miR-155-3p and their targeted genes in lithium response in bipolar disorder. Int. J. Mol. Sci. 2019, 20, 6040. [Google Scholar] [CrossRef] [PubMed]
- Soeiro-De-Souza, M.G.; Gold, P.W.; Brunoni, A.R.; de Sousa, R.T.; Zanetti, M.V.; Carvalho, A.F.; Gattaz, W.F.; Machado-Vieira, R.; Teixeira, A.L. Lithium decreases plasma adiponectin levels in bipolar depression. Neurosci. Lett. 2014, 564, 111–114. [Google Scholar] [CrossRef]
- Teixeira, A.L.; de Sousa, R.T.; Zanetti, M.V.; Brunoni, A.R.; Busatto, G.F.; Zarate, C.A., Jr.; Gattaz, W.F.; Machado-Vieira, R. Increased plasma levels of soluble TNF receptors 1 and 2 in bipolar depression and impact of lithium treatment. Hum. Psychopharmacol. 2015, 30, 52–56. [Google Scholar] [CrossRef]
- Loch, A.A.; Zanetti, M.V.; de Sousa, R.T.; Chaim, T.M.; Serpa, M.H.; Gattaz, W.F.; Teixeira, A.L.; Machado-Vieira, R. Elevated neurotrophin-3 and neurotrophin 4/5 levels in unmedicated bipolar depression and the effects of lithium. Prog. Neuropsychopharmacol. Biol. Psychiatry 2015, 56, 243–246. [Google Scholar] [CrossRef]
- van den Ameele, S.; van Diermen, L.; Staels, W.; Coppens, V.; Dumont, G.; Sabbe, B.; Morrens, M. The effect of mood-stabilizing drugs on cytokine levels in bipolar disorder: A systematic review. J. Affect. Disord. 2016, 203, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, S.D.; Williams, A.J.; Blacker, C.J.; Voort, J.L.V.; Schak, K.M.; Nemeroff, C.B.; Widge, A.S.; Tohen, M. Putative biological predictors of treatment response in bipolar disorders. Pers. Med. Psychiatry 2017, 1, 39–58. [Google Scholar] [CrossRef]
- Hibar, D.P.; The ENIGMA Bipolar Disorder Working Group; Westlye, L.T.; Doan, N.T.; Jahanshad, N.; Cheung, J.W.; Ching, C.R.K.; Versace, A.; Bilderbeck, A.C.; Uhlmann, A.; et al. Cortical abnormalities in bipolar disorder: An MRI analysis of 6503 individuals from the ENIGMA Bipolar Disorder Working Group. Mol. Psychiatry 2018, 23, 932–942. [Google Scholar] [CrossRef] [PubMed]
- Atagün, M.I. Brain oscillations in bipolar disorder and lithium-induced changes. Neuropsychiatr. Dis. Treat. 2016, 12, 589–601. [Google Scholar] [CrossRef]
- Gao, K.; Calabrese, J.R. Pharmacogenetics of lithium response: Close to clinical practice? Lancet 2016, 387, 1034–1036. [Google Scholar] [CrossRef] [PubMed]
- Vieta, E.; Berk, M.; Schulze, T.G.; Carvalho, A.F.; Suppes, T.; Calabrese, J.R.; Gao, K.; Miskowiak, K.W.; Grande, I. Bipolar disorders. Nat. Rev. Dis. Prim. 2018, 4, 18008. [Google Scholar] [CrossRef]
- Mertens, J.; Wang, Q.W.; Kim, Y.; Yu, D.X.; Pham, S.; Yang, B.; Zheng, Y.; Diffenderfer, K.E.; Zhang, J.; Soltani, S.; et al. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature 2015, 527, 95–99. [Google Scholar] [CrossRef]
- Stern, S.; Santos, R.; Marchetto, M.C.; Mendes, A.P.D.; Rouleau, G.A.; Biesmans, S.; Wang, Q.-W.; Yao, J.; Charnay, P.; Bang, A.G.; et al. Neurons derived from patients with bipolar disorder divide into intrinsically different sub-populations of neurons, predicting the patients’ responsiveness to lithium. Mol. Psychiatry 2017, 23, 1453–1465. [Google Scholar] [CrossRef] [PubMed]
- Tobe, B.T.D.; Crain, A.M.; Winquist, A.M.; Calabrese, B.; Makihara, H.; Zhao, W.N.; Lalonde, J.; Nakamura, H.; Konopaske, G.; Sidor, M.; et al. Proc Probing the lithium-response pathway in hiPSCs implicates the phosphoregulatory set-point for a cytoskeletal modulator in bipolar pathogenesis. Proc. Natl. Acad. Sci. USA 2017, 114, E4462–E4471. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, M.J.; Wei, H.; Nievergelt, C.M.; Stautland, A.; Maihofer, A.X.; Welsh, D.K.; Shilling, P.; Alda, M.; Alliey-Rodriguez, N.; Anand, A.; et al. Chronotype and cellular circadian rhythms predict the clinical response to lithium maintenance treatment in patients with bipolar disorder. Neuropsychopharmacology 2019, 44, 620–628. [Google Scholar] [CrossRef] [PubMed]
- Federoff, M.; McCarthy, M.J.; Anand, A.; Berrettini, W.H.; Bertram, H.; Bhattacharjee, A.; Calkin, C.V.; Conroy, C.; Coryell, W.H.; D’Arcangelo, N.; et al. Correction of depression-associated circadian rhythm abnormalities is associated with lithium response in bipolar disorder. Bipolar Disord. 2022, 24, 521–529. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, J.; Wang, X.; Zhu, J.; Liu, Q.; Shi, Z.; Chambers, M.C.; Zimmerman, L.J.; Shaddox, K.F.; Kim, S.; et al. Proteogenomic characterization of human colon and rectal cancer. Nature 2014, 513, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Hukelmann, J.L.; Anderson, K.E.; Sinclair, L.V.; Grzes, K.M.; Murillo, A.B.; Hawkins, P.T.; Stephens, L.R.; Lamond, A.I.; Cantrell, D.A. The cytotoxic T cell proteome and its shaping by the kinase mTOR. Nat. Immunol. 2016, 17, 104–112. [Google Scholar] [CrossRef]
- Darmanis, S.; Gallant, C.J.; Marinescu, V.D.; Niklasson, M.; Segerman, A.; Flamourakis, G.; Fredriksson, S.; Assarsson, E.; Lundberg, M.; Nelander, S.; et al. Simultaneous multiplexed measurement of RNA and proteins in single cells. Cell. Rep. 2016, 14, 380–389. [Google Scholar] [CrossRef]
- Barbosa, I.G.; Nogueira, C.R.; Rocha, N.P.; Queiroz, A.L.; Vago, J.P.; Tavares, L.P.; Assis, F.; Fagundes, C.T.; Huguet, R.B.; Bauer, M.E.; et al. Altered intracellular signaling cascades in peripheral blood mononuclear cells from BD patients. J. Psychiatr. Res. 2013, 47, 1949–1954. [Google Scholar] [CrossRef] [PubMed]
- Brietzke, E.; Stertz, L.; Fernandes, B.S.; Kauer-Sant’Anna, M.; Mascarenhas, M.; Vargas, A.E.; Chies, J.A.; Kapczinski, F. Comparison of cytokine levels in depressed, manic and euthymic patients with bipolar disorder. J. Affect. Disord. 2009, 116, 214–217. [Google Scholar] [CrossRef] [PubMed]
- do Prado, C.H.; Rizzo, L.B.; Wieck, A.; Lopes, R.P.; Teixeira, A.L.; Grassi-Oliveira, R.; Bauer, M.E. Reduced regulatory T cells are associated with higher levels of Th1/TH17 cytokines and activated MAPK in type 1 bipolar disorder. Psychoneuroendocrinology 2013, 38, 667–676. [Google Scholar] [CrossRef]
- Guloksuz, S.; Cetin, E.A.; Cetin, T.; Deniz, G.; Oral, E.T.; Nutt, D.J. Cytokine levels in euthymic bipolar patients. J. Affect. Disord. 2010, 126, 458–462. [Google Scholar] [CrossRef]
- Wieck, A.; Grassi-Oliveira, R.; Prado, C.H.D.; Rizzo, L.B.; de Oliveira, A.S.; Kommers-Molina, J.; Viola, T.W.; Teixeira, A.L.; Bauer, M.E. Differential neuroendocrine and immune responses to acute psychosocial stress in women with type 1 bipolar disorder. Brain Behav. Immun. 2013, 34, 47–55. [Google Scholar] [CrossRef]
- Han, Y.; Gu, Y.; Zhang, A.C.; Lo, Y.-H. Review: Imaging technologies for flow cytometry. Lab Chip 2016, 16, 4639–4647. [Google Scholar] [CrossRef]
- McKinnon, K.M. Flow Cytometry: An Overview. Curr. Protoc. Immunol. 2018, 120, 5.1.1–5.1.11. [Google Scholar] [CrossRef]
- Gao, K.; Ayati, M.; Koyuturk, M.; Calabrese, J.R.; Ganocy, S.J.; Kaye, N.M.; Lazarus, H.M.; Christian, E.; Kaplan, D. Protein biomarkers in monocytes and CD4 + Lymphocytes for predicting lithium treatment response of bipolar disorder: A feasibility study with tyramine-based signal-amplified flow cytometry. Psychopharmacol. Bull. 2022, 52, 8–35. [Google Scholar]
- Kaplan, D.; Smith, D. Enzymatic amplification staining for flow cytometric analysis of cell surface molecules. Cytometry 2000, 40, 81–85. [Google Scholar] [CrossRef]
- Kaplan, D.; Husel, W.; Meyerson, H. Immunophenotypic analysis with enhanced sensitivity of detection by enzymatic amplification staining. Clin. Lab. Med. 2001, 21, 763–778. [Google Scholar]
- Kaplan, D.; Meyerson, H.; Lewandowska, K. High resolution immunophenotypic analysis of chronic lymphocytic leukemic cells by enzymatic amplification staining. Am. J. Clin. Pathol. 2001, 116, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, D. Enzymatic amplification staining for single cell analysis: Applied to in situ hybridization. J. Immunol. Methods 2003, 283, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, D. Enzymatic Amplification Staining for Cell Surface Antigens. Curr. Protoc. Cytom. 2003, 23, 6.14.1–6.14.10. [Google Scholar] [CrossRef]
- Kaplan, D.; Meyerson, H.; Husel, W.; Lewandowska, K.; MacLennan, G. D cyclins in lymphocytes. Cytom. Part A 2004, 63A, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Meyerson, H.J.; MacLennan, G.; Husel, W.; Tse, W.; Lazarus, H.M.; Kaplan, D. D Cyclins in CD5+ B-Cell lymphoproliferative disorders: Cyclin D1 and Cyclin D2 identify diagnostic groups and Cyclin D1 correlates with ZAP-70 expression in chronic lymphocytic leukemia. Am. J. Clin. Pathol. 2006, 125, 241–250. [Google Scholar] [CrossRef]
- Liew, C.-C.; Ma, J.; Tang, H.-C.; Zheng, R.; Dempsey, A.A. The peripheral blood transcriptome dynamically reflects system wide biology: A potential diagnostic tool. J. Lab. Clin. Med. 2006, 147, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, P.F.; Fan, C.; Perou, C. Evaluating the comparability of gene expression in blood and brain. Am. J. Med Genet. Part B Neuropsychiatr. Genet. 2006, 141B, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Rollins, B.; Martin, M.V.; Morgan, L.; Vawter, M.P. Analysis of whole genome biomarker expression in blood and brain. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2010, 153B, 919–936. [Google Scholar] [CrossRef] [PubMed]
- Tylee, D.S.; Kawaguchi, D.M.; Glatt, S.J. On the outside, looking in: A review and evaluation of the comparability of blood and brain “-omes”. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2013, 162B, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Spiliotaki, M.; Salpeas, V.; Malitas, P.; Alevizos, V.; Moutsatsou, P. Altered glucocorticoid receptor signaling cascade in lymphocytes of bipolar disorder patients. Psychoneuroendocrinology 2006, 31, 748–760. [Google Scholar] [CrossRef]
- Bei, E.; Salpeas, V.; Pappa, D.; Anagnostara, C.; Alevizos, V.; Moutsatsou, P. Phosphorylation status of glucocorticoid receptor, heat shock protein 70, cytochrome c and Bax in lymphocytes of euthymic, depressed and manic bipolar patients. Psychoneuroendocrinology 2009, 34, 1162–1175. [Google Scholar] [CrossRef]
- Hunsberger, J.G.; Chibane, F.L.; Elkahloun, A.G.; Henderson, R.; Singh, R.; Lawson, J.; Cruceanu, C.; Nagarajan, V.; Turecki, G.; Squassina, A.; et al. Novel integrative genomic tool for interrogating lithium response in bipolar disorder. Transl. Psychiatry 2015, 5, e504. [Google Scholar] [CrossRef] [PubMed]
- Marie-Claire, C.; Lejeune, F.X.; Mundwiller, E.; Ulveling, D.; Moszer, I.; Bellivier, F.; Etain, B. A DNA methylation signature discriminates between excellent and non-response to lithium in patients with bipolar disorder type 1. Sci. Rep. 2020, 10, 12239. [Google Scholar] [CrossRef]
- Eugene, A.R.; Masiak, J.; Eugene, B. Predicting lithium treatment response in bipolar patients using gender-specific gene expression biomarkers and machine learning. F1000Research 2018, 7, 474. [Google Scholar] [CrossRef]
- Munkholm, K.; Peijs, L.; Vinberg, M.; Kessing, L.V. A composite peripheral blood gene expression measure as a potential diagnostic biomarker in bipolar disorder. Transl. Psychiatry 2015, 5, e614. [Google Scholar] [CrossRef] [PubMed]
- So, J.; Warsh, J.J.; Li, P.P. Impaired endoplasmic reticulum stress response in B-Lymphoblasts from patients with Bipolar-I disorder. Biol. Psychiatry 2007, 62, 141–147. [Google Scholar] [CrossRef]
- Wieck, A.; Grassi-Oliveira, R.; Prado, C.H.D.; Viola, T.W.; Petersen, L.E.; Porto, B.; Teixeira, A.L.; Bauer, M.E. Toll-like receptor expression and function in type I bipolar disorder. Brain Behav. Immun. 2016, 54, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Amoruso, A.; Bardelli, C.; Cattaneo, C.I.; Fresu, L.G.; Manzetti, E.; Brunelleschi, S. Neurokinin (NK)-1 receptor expression in monocytes from bipolar disorder patients: A pilot study. J. Affect. Disord. 2015, 178, 188–192. [Google Scholar] [CrossRef]
- Pfaffenseller, B.; Wollenhaupt-Aguiar, B.; Fries, G.R.; Colpo, G.D.; Burque, R.K.; Bristot, G.; Ferrari, P.; Ceresér, K.M.M.; Rosa, A.R.; Klamt, F.; et al. Impaired endoplasmic reticulum stress response in bipolar disorder: Cellular evidence of illness progression. Int. J. Neuropsychopharmacol. 2014, 17, 1453–1463. [Google Scholar] [CrossRef]
- Barbosa, I.G.; Rocha, N.P.; Assis, F.; Vieira, E.L.M.; Soares, J.C.; Bauer, M.E.; Teixeira, A.L. Monocyte and lymphocyte activation in bipolar disorder: A new piece in the puzzle of immune dysfunction in mood disorders. Int. J. Neuropsychopharmacol. 2014, 18, pyu021. [Google Scholar] [CrossRef]
- Maecker, H.T.; McCoy, J.P.; Nussenblatt, R. Standardizing immunophenotyping for the Human Immunology Project. Nat. Rev. Immunol. 2012, 12, 191–200. [Google Scholar] [CrossRef]
- Miklowitz, D.J.; Portnoff, L.C.; Armstrong, C.C.; Keenan-Miller, D.; Breen, E.C.; Muscatell, K.A.; Eisenberger, N.I.; Irwin, M.R. Inflammatory cytokines and nuclear factor-kappa B activation in adolescents with bipolar and major depressive disorders. Psychiatry Res. 2016, 241, 315–322. [Google Scholar] [CrossRef]
- Lambert, C.; Preijers, F.W.M.B.; Yanikkaya Demirel, G.; Sack, U. Monocytes and macrophages in flow: An ESCCA initiative on advanced analyses of monocyte lineage using flow cytometry. Cytom. B Clin. Cytom. 2017, 92, 180–188. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, R.T.; Zanetti, M.V.; Talib, L.L.; Serpa, M.H.; Chaim, T.M.; Carvalho, A.F.; Brunoni, A.R.; Busatto, G.F.; Gattaz, W.F.; Machado-Vieira, R. Lithium increases platelet serine-9 phosphorylated GSK-3β levels in drug-free bipolar disorder during depressive episodes. J. Psychiatr. Res. 2015, 62, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Ladeira, R.B.; Joaquim, H.P.; Talib, L.L.; Nunes, P.V.; Forlenza, O.V. Higher proportion of inactive Gsk3β in platelets of elderly patients with bipolar disorder: An effect of treatment? Braz. J. Psychiatry 2013, 35, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, M.; Cai, Z.; Wang, G.; Li, X. Regulation of glycogen synthase kinas 3 during bipolar mania treatment. Bipolar Disord. 2010, 12, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Friedman, A.B.; Zhu, W.; Wang, L.; Boswell, S.; May, R.S.; Davis, L.L.; Jope, R.S. Lithium regulates glycogen synthase kinase-3beta in human peripheral blood mononuclear cells: Implication in the treatment of bipolar disorder. Biol. Psychiatry 2007, 61, 216–222. [Google Scholar] [CrossRef]
- Pandey, G.N.; Ren, X.; Rizavi, H.S.; Dwivedi, Y.J. Glycogen synthase kinase-3beta in the platelets of patients with mood disorders: Effect of treatment. J. Psychiatr. Res. 2010, 44, 143–148. [Google Scholar] [CrossRef]
- Hoeflich, K.P.; Luo, J.; Rubie, E.A.; Tsao, M.S.; Jin, O.; Woodgett, J.R. Requirement for glycogen synthase kinase-3beta in cell survival and NF-kappaB activation. Nature 2000, 406, 86–90. [Google Scholar] [CrossRef]
- Hofmann, C.; Dunger, N.; Schölmerich, J.; Falk, W.; Obermeier, F. Glycogen synthase kinase 3-β: A master regulator of toll-like receptor-mediated chronic intestinal inflammation. Inflamm. Bowel. Dis. 2010, 16, 1850–1858. [Google Scholar] [CrossRef] [PubMed]
- Sakrajda, K.; Szczepankiewicz, A. Inflammation-related changes in mood disorders and the immunomodulatory role of lithium. Int. J. Mol. Sci. 2021, 22, 1532. [Google Scholar] [CrossRef]
- Wu, T.-N.; Lee, C.-S.; Wu, B.-J.; Sun, H.-J.; Chang, C.-H.; Chen, C.-Y.; Chen, C.-K.; Wu, L.S.-H.; Cheng, A.T.-A. Immunophenotypes associated with bipolar disorder and lithium treatment. Sci. Rep. 2019, 9, 17453. [Google Scholar] [CrossRef]
- Li, H.; Huang, K.; Liu, X.; Liu, J.; Lu, X.; Tao, K.; Wang, G.; Wang, J. Lithium chloride suppresses colorectal cancer cell survival and proliferation through ROS/GSK-3beta/NF-kappaB signaling pathway. Oxidative Med. Cell. Longev. 2014, 2014, 241864. [Google Scholar] [CrossRef]
- Nassar, A.; Azab, A.N. Effects of Lithium on Inflammation. ACS Chem. Neurosci. 2014, 5, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Troib, A.; Azab, A.N. Effects of psychotropic drugs on Nuclear Factor kappa B. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 1198–1208. [Google Scholar] [PubMed]
- Xia, Y.; Rao, J.; Yao, A.; Zhang, F.; Li, G.; Wang, X.; Lu, L. Lithium exacerbates hepatic ischemia/reperfusion injury by inhibiting GSK-3β/NF-κB-mediated protective signaling in mice. Eur. J. Pharmacol. 2012, 697, 117–125. [Google Scholar] [CrossRef]
- Christian, F.; Smith, E.L.; Carmody, R.J. The regulation of NF-κB subunits by phosphorylation. Cells 2016, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, S.; Vargas, J.; Hoffmann, A. Signaling via the NFκB system. Wiley Interdiscip. Rev. Syst. Biol. Med. 2016, 8, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Sarai, S.K.; Mekala, H.M.; Lippmann, S. Lithium suicide prevention: A brief review and reminder. Innov. Clin. Neurosci. 2018, 15, 30–32. [Google Scholar]
- Scott, J.; Hidalgo-Mazzei, D.; Strawbridge, R.; Young, A.; Resche-Rigon, M.; Etain, B.; Andreassen, O.A.; Bauer, M.; Bennabi, D.; Blamire, A.M.; et al. Prospective cohort study of early biosignatures of response to lithium in bipolar-I-disorders: Overview of the H2020-funded R-LiNK initiative. Int. J. Bipolar Disord. 2019, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Tolliver, B.K.; Kemp, D.E.; Verduin, M.L.; Ganocy, S.J.; Bilali, S.; Brady, K.T.; Shim, S.S.; Findling, R.L.; Calabrese, J.R. Differential interactions between comorbid anxiety disorders and substance use disorder in rapid cycling bipolar I or II disorder. J. Affect. Disord. 2008, 110, 167–173. [Google Scholar]
- Sheehan, D.V. M.I.N.I. Mini International Neuropsychiatric Interview, Version 7.02, for DSM-5. 2016. Available online: https://harmresearch.org/index.php/mini-international-neuropsychiatric-interview-mini/#How%20to%20Cite%20the%20MINI (accessed on 1 December 2022).
- Spearing, M.K.; Post, R.M.; Leverich, G.S.; Brandt, D.; Nolen, W. Modification of the Clinical Global Impressions (CGI) scale for use in bipolar illness (BP): The CGI-BP. Psychiatry Res. 1997, 73, 159–171. [Google Scholar]
- Rush, A.; Trivedi, M.H.; Ibrahim, H.M.; Carmody, T.J.; Arnow, B.; Klein, D.N.; Markowitz, J.C.; Ninan, P.T.; Kornstein, S.; Manber, R.; et al. The 16-Item quick inventory of depressive symptomatology (QIDS), clinician rating (QIDS-C), and self-report (QIDS-SR): A psychometric evaluation in patients with chronic major depression. Biol. Psychiatry 2003, 54, 573–583. [Google Scholar]
- Linn, B.S.; Linn, M.W.; Gurel, L. Cumulative Illness Rating Scale. J. Am. Geriatr. Soc. 1968, 16, 622–626. [Google Scholar]
- Montgomery, S.A.; Åsberg, M. A New Depression Scale Designed to be Sensitive to Change. Br. J. Psychiatry 1979, 134, 382–389. [Google Scholar]
- Young, R.C.; Biggs, J.T.; Ziegler, V.E.; Meyer, D.A. A Rating Scale for Mania: Reliability, Validity and Sensitivity. Br. J. Psychiatry 1978, 133, 429–435. [Google Scholar]
- Guy, W. ECDEU Assessment Manuel for Psychopharmacology, Revised; U.S. Government Printing Office: Washington, DC, USA, 1976.
- Hamilton, M. The Assessment of Anxiety States by Rating. Psychol. Psychother. Theory Res. Pr. 1959, 32, 50–55. [Google Scholar]
- Snaith, R.P.; Hamilton, M.; Morley, S.; Humayan, A.; Hargreaves, D.; Trigwell, P. A Scale for the Assessment of Hedonic Tone the Snaith–Hamilton Pleasure Scale. Br. J. Psychiatry 1995, 167, 99–103. [Google Scholar]
- Leon, A.C.; Olfson, M.; Portera, L.; Farber, L.; Sheehan, D.V. Assessing Psychiatric Impairment in Primary Care with the Sheehan Disability Scale. Int. J. Psychiatry Med. 1997, 27, 93–105. [Google Scholar]
- Endicott, J.; Nee, J.; Harrison, W.; Blumenthal, R. Quality of Life Enjoyment and Satisfaction Questionnaire: A new measure. Psychopharmacol. Bull. 1993, 29, 321–326. [Google Scholar]
- Hartz, A.; Bentler, S.; Watson, D. Measuring fatigue severity in primary care patients. J. Psychosom. Res. 2003, 54, 515–521. [Google Scholar]
- Sobell, L.C.; Sobell, M.B. Timeline Followback: A technique for assessing self-reported alcohol consumption. In Measuring Alcohol Consumption: Psychosocial and Biological Methods; Litten, R.Z., Allen, J.P., Eds.; Humana Press: Totowa, NJ, USA, 1996; pp. 41–72. [Google Scholar]
- Wisniewski, S.R.; Rush, A.J.; Balasubramani, G.K.; Trivedi, M.H.; Nierenberg, A.A. STARD Investigators. Self-rated global measure of the frequency, intensity, and burden of side effects. J. Psychiatr. Pract. 2006, 12, 71–79. [Google Scholar]
- Posner, K.; Oquendo, M.A.; Gould, M.; Stanley, B.; Davies, M. Columbia Classification Algorithm of Suicide Assessment (C-CASA): Classification of Suicidal Events in the FDA’s Pediatric Suicidal Risk Analysis of Antidepressants. Am. J. Psychiatry 2007, 164, 1035–1043. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).