COVID-19 and Gastrointestinal Tract: From Pathophysiology to Clinical Manifestations
Abstract
:1. Introduction
2. Materials and Methods
3. Prevalence of Gastrointestinal Symptoms
4. Gastrointestinal Infection
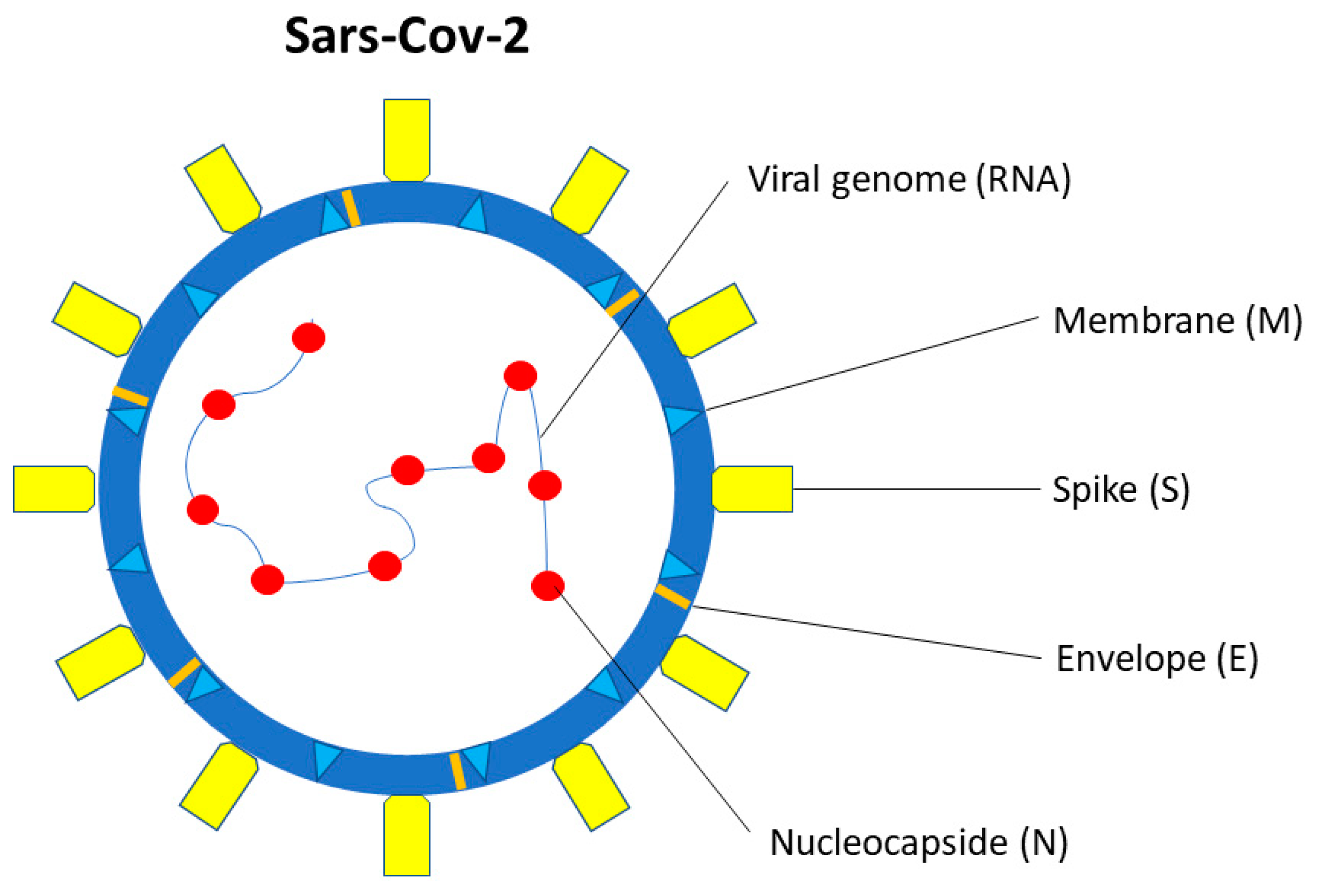
5. SARS-CoV-2 Structure and Interaction with the Host
6. Route of Gastrointestinal Infection
7. Gastrointestinal Tract–Virus Interaction
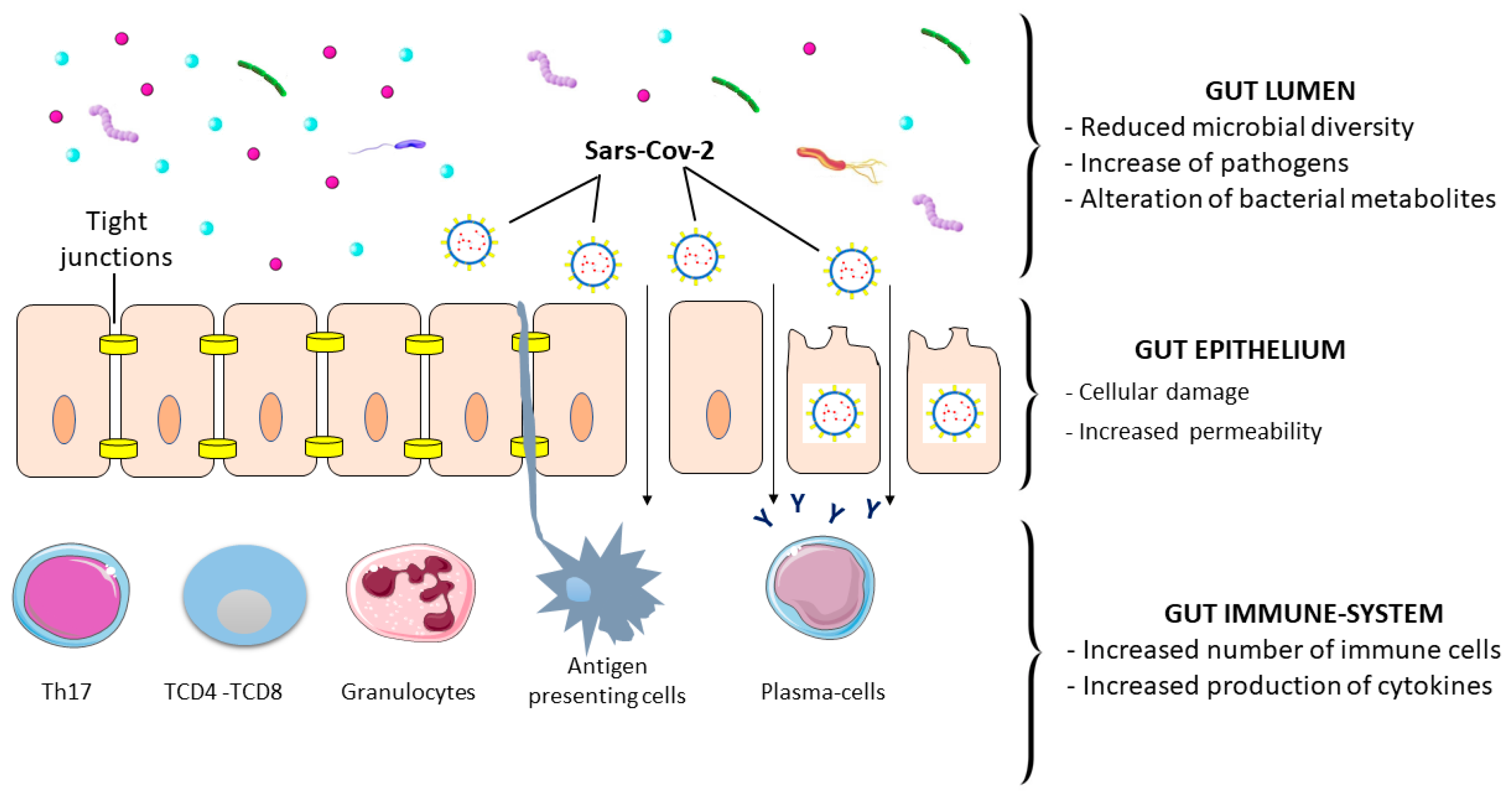
8. Gastrointestinal Damage, Inflammation, and Symptoms
9. SARS-CoV-2 and Gut Microbiota Alteration
10. Chronic GI Tract Diseases and SARS-CoV-2
11. Gastrointestinal Symptoms and Long COVID
12. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Coronavirus Disease (COVID-19). Available online: https://covid19.who.int/ (accessed on 4 March 2023).
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Ashktorab, H.; Russo, T.; Oskrochi, G.; Latella, G.; Massironi, S.; Luca, M.; Chirumamilla, L.G.; Laiyemo, A.O.; Brim, H. Clinical and Endoscopic Outcomes in Coronavirus Disease-2019 Patients with Gastrointestinal Bleeding. Gastro Hep. Adv. 2022, 1, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Pizuorno, A.; Brim, H.; Ashktorab, H. Gastrointestinal manifestations and SARS-CoV-2 infection. Curr. Opin. Pharmacol. 2021, 61, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Dyall, J.; Gross, R.; Kindrachuk, J.; Johnson, R.F.; Olinger, G.G., Jr.; Hensley, L.E.; Frieman, M.B.; Jahrling, P.B. Middle East Respiratory Syndrome and Severe Acute Respiratory Syndrome: Current Therapeutic Options and Potential Targets for Novel Therapies. Drugs 2017, 77, 1935–1966. [Google Scholar] [CrossRef]
- de Wit, E.; van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523–534. [Google Scholar] [CrossRef]
- Shehab, M.; Alrashed, F.; Shuaibi, S.; Alajmi, D.; Barkun, A. Gastroenterological and hepatic manifestations of patients with COVID-19, prevalence, mortality by country, and intensive care admission rate: Systematic review and meta-analysis. BMJ Open Gastroenterol. 2021, 8, e000571. [Google Scholar] [CrossRef]
- Bolia, R.; Dhanesh Goel, A.; Badkur, M.; Jain, V. Gastrointestinal Manifestations of Pediatric Coronavirus Disease and Their Relationship with a Severe Clinical Course: A Systematic Review and Meta-analysis. J. Trop. Pediatr. 2021, 67, fmab051. [Google Scholar] [CrossRef]
- Russo, T.; Pizuorno, A.; Oskrochi, G.; Latella, G.; Massironi, S.; Schettino, M.; Aghemo, A.; Pugliese, N.; Brim, H.; Ashktorab, H. Gastrointestinal Manifestations, Clinical Characteristics and Outcomes of COVID-19 in Adult and Pediatric Patients. SOJ Microbiol. Infect. Dis. 2021, 8, 109. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Zhang, Y.; Liu, Y.; Liu, Y. Are gastrointestinal symptoms associated with higher risk of Mortality in COVID-19 patients? A systematic review and meta-analysis. BMC Gastroenterol. 2022, 22, 106. [Google Scholar] [CrossRef]
- Hayashi, Y.; Wagatsuma, K.; Nojima, M.; Yamakawa, T.; Ichimiya, T.; Yokoyama, Y.; Kazama, T.; Hirayama, D.; Nakas, H. The characteristics of gastrointestinal symptoms in patients with severe COVID-19: A systematic review and meta-analysis. J. Gastroenterol. 2021, 56, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, F.; Fahriani, M.; Mamada, S.S.; Frediansyah, A.; Abubakar, A.; Maghfirah, D.; Fajar, J.K.; Maliga, H.A.; Ilmawan, M.; Emran, T.B.; et al. Global prevalence of prolonged gastrointestinal symptoms in COVID-19 survivors and potential pathogenesis: A systematic review and meta-analysis. F1000Research 2021, 10, 301. [Google Scholar] [CrossRef]
- Marasco, G.; Maida, M.; Morreale, G.C.; Licata, M.; Renzulli, M.; Cremon, C.; Stanghellini, V.; Barbara, G. Gastrointestinal Bleeding in COVID-19 Patients: A Systematic Review with Meta-Analysis. Can. J. Gastroenterol. Hepatol. 2021, 2021, 2534975. [Google Scholar] [PubMed]
- Vanella, G.; Capurso, G.; Burti, C.; Fanti, L.; Ricciardiello, L.; Souza Lino, A.; Boskoski, I.; Bronswijk, M.; Tyberg, A.; Krishna Kumar Nair, G.; et al. Gastrointestinal mucosal damage in patients with COVID-19 undergoing endoscopy: An international multicentre study. BMJ Open Gastroenterol. 2021, 8, e000578. [Google Scholar] [CrossRef]
- van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Tang, M.; Zheng, X.; Liu, Y.; Li, X.; Shan, H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology 2020, 158, 1831–1833. [Google Scholar] [CrossRef]
- Xing, Y.H.; Ni, W.; Wu, Q.; Li, W.J.; Li, G.J.; Wang, W.D.; Tong, J.N.; Song, X.F.; Wing-Kin Wong, G.; Xing, Q.S. Prolonged viral shedding in feces of pediatric patients with coronavirus disease 2019. J. Microbiol. Immunol. Infect. 2020, 53, 473–480. [Google Scholar] [CrossRef]
- Wu, Y.; Guo, C.; Tang, L.; Hong, Z.; Zhou, J.; Dong, X.; Yin, H.; Xiao, Q.; Tang, Y.; Qu, X.; et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020, 5, 434–435. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, L.; Deng, Q.; Wu, K.; Ni, L.; Yang, Y.; Liu, B.; Wang, W.; Wei, C.; Yang, J.; et al. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 2020, 92, 833–840. [Google Scholar] [CrossRef]
- Kang, M.; Wei, J.; Yuan, J.; Guo, J.; Zhang, Y.; Hang, J.; Qu, Y.; Qian, H.; Zhuang, Y.; Chen, X.; et al. Probable Evidence of Fecal Aerosol Transmission of SARS-CoV-2 in a High-Rise Building. Ann. Intern. Med. 2020, 173, 974–980. [Google Scholar] [CrossRef]
- Qian, Q.; Fan, L.; Liu, W.; Li, J.; Yue, J.; Wang, M.; Ke, X.; Yin, Y.; Chen, Q.; Jiang, C. Direct evidence of active SARS-CoV-2 replication in the intestine. Clin. Infect. Dis. 2021, 73, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C. Virological assessment of hospitalized patients with COVID-19. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef]
- Xu, Y.; Li, X.; Zhu, B.; Liang, H.; Fang, C.; Gong, Y.; Guo, Q.; Sun, X.; Zhao, D.; Shen, J.; et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020, 26, 502–505. [Google Scholar] [CrossRef]
- Zheng, S.; Fan, J.; Yu, F.; Feng, B.; Lou, B.; Zou, Q.; Xie, G.; Lin, S.; Wang, R.; Yang, X.; et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January–March 2020: Retrospective cohort study. BMJ 2020, 369, m1443. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.L.; Baluja, M.Q.; Graham, D.W.; Corbishley, A.; McDonald, J.E.; Malham, S.K.; Hillary, L.S.; Connor, T.R.; Gaze, W.H.; Moura, I.B.; et al. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020, 749, 141364. [Google Scholar] [CrossRef] [PubMed]
- Han, M.S.; Seong, M.W.; Heo, E.Y.; Park, J.H.; Kim, N.; Shin, S.; Cho, S.I.; Park, S.S.; Choi, E.H. Sequential analysis of viral load in a neonate and her mother infected with SARS-CoV-2. Clin. Infect. Dis. 2020, 71, 2236–2239. [Google Scholar] [CrossRef]
- Pan, Y.; Zhang, D.; Yang, P.; Poon, L.M.; Wang, Q. Viral load of SARSCoV-2 in clinical samples. Lancet Infect. Dis. 2020, 20, 411–412. [Google Scholar] [CrossRef]
- Walsh, K.A.; Jordan, K.; Clyne, B.; Rohde, D.; Drummond, L.; Byrne, P.; Ahern, S.; Carty, P.G.; O’Brien, K.K.; O’Murchu, E.; et al. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J. Infect. 2020, 81, 357–371. [Google Scholar] [CrossRef]
- Xiao, F.; Sun, J.; Xu, Y.; Li, F.; Huang, X.; Li, H.; Zhao, J.; Huang, J.; Zhao, J. Infectious SARS-CoV-2 in Feces of Patient with Severe COVID-19. Emerg. Infect. Dis. 2020, 26, 1920–1922. [Google Scholar] [CrossRef]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef]
- Natarajan, A.; Han, A.; Zlitni, S.; Brooks, E.F.; Vance, S.E.; Wolfe, M.; Singh, U.; Jagannathan, P.; Pinsky, B.A.; Boehm, A.; et al. Standardized preservation, extraction and quantification techniques for detection of fecal SARS-CoV-2 RNA. Nat. Commun. 2021, 12, 5753. [Google Scholar] [CrossRef] [PubMed]
- Miyakawa, K.; Machida, M.; Kawasaki, T.; Nishi, M.; Akutsu, H.; Ryo, A. Reduced replication efficacy of severe acute respiratory syndrome coronavirus 2 omicron variant in “mini-gut” organoids. Gastroenterology 2022, 163, 514–516. [Google Scholar] [CrossRef] [PubMed]
- Cheung, K.S.; Hung, I.F.N.; Chan, P.P.Y.; Lung, K.C.; Tso, E.; Liu, R.; Ng, Y.Y.; Chu, M.Y.; Chung, T.W.H.; Tam, A.R.; et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from the Hong Kong cohort and systematic review and meta-analysis. Gastroenterology 2020, 159, 81–95. [Google Scholar] [CrossRef] [PubMed]
- von Stillfried, S.; Villwock, S.; Bülow, R.D.; Djudjaj, S.; Buhl, E.M.; Maurer, A.; Ortiz-Brüchle, N.; Celec, P.; Klinkhammer, B.M.; Wong, D.W.L.; et al. SARS-CoV-2 RNA screening in routine pathology specimens. Microb. Biotechnol. 2021, 14, 1627–1641. [Google Scholar] [CrossRef] [PubMed]
- Livanos, A.E.; Jha, D.; Cossarini, F.; Gonzalez-Reiche, A.S.; Tokuyama, M.; Aydillo, T.; Parigi, T.L.; Ladinsky, M.S.; Ramos, I.; Dunleavy, K.; et al. Intestinal Host Response to SARS-CoV-2 Infection and COVID-19 Outcomes in Patients with Gastrointestinal Symptoms. Gastroenterology 2021, 160, 2435–2450. [Google Scholar] [CrossRef]
- Lamers, M.M.; Beumer, J.; van der Vaart, J.; Knoops, K.; Puschhof, J.; Breugem, T.I.; Ravelli, R.B.G.; van Schayck, J.P.; Mykytyn, A.Z.; Duimel, H.Q.; et al. SARS-CoV-2 productively infects human gut enterocytes. Science 2020, 369, 50–54. [Google Scholar] [CrossRef]
- Yang, D.; Perbtani, Y.B.; Loeb, J.; Liu, N.; Draganov, P.V.; Estores, D.E.; Lauzardo, M.; Maurelli, A.; Lednicky, J.A.; Morris, J.G. Detection of SARS-CoV-2 in the gastrointestinal tract among patients with negative nasopharyngeal COVID-19 testing prior to endoscopy. Endosc. Int. Open 2021, 9, E1276–E1282. [Google Scholar] [CrossRef]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2015; Volume 1282, pp. 1–23. [Google Scholar]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Guo, Y.R.; Cao, Q.D.; Hong, Z.S.; Tan, Y.Y.; Chen, S.D.; Jin, H.J.; Tan, K.S.; Wang, D.Y.; Yan, Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak. An update on the status. Mil. Med. Res. 2020, 7, 11. [Google Scholar] [CrossRef]
- Arya, R.; Kumari, S.; Pandey, B.; Mistry, H.; Bihani, S.C.; Das, A.; Prashar, V.; Gupta, G.D.; Panicker, L.; Kumar, M. Structural insights into SARS-CoV-2 proteins. J. Mol. Biol. 2021, 433, 166725. [Google Scholar] [CrossRef]
- Schoeman, D.; Fielding, B.C. Coronavirus envelope protein: Current knowledge. Virol. J. 2019, 16, e69. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef] [PubMed]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef]
- Zang, R.; Gomez Castro, M.F.; McCune, B.T.; Zeng, Q.; Rothlauf, P.W.; Sonnek, N.M.; Liu, Z.; Brulois, K.F.; Wang, X.; Greenberg, H.B.; et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020, 5, eabc3582. [Google Scholar] [CrossRef]
- Hirose, R.; Nakaya, T.; Naito, Y.; Daidoji, T.; Watanabe, Y.; Yasuda, H.; Konishi, H.; Itoh, Y. Mechanism of human influenza virus RNA persistence and virion survival in feces: Mucus protects virions from acid and digestive juices. J. Infect. Dis. 2017, 216, 105–109. [Google Scholar] [CrossRef]
- Holmes, K.V. Enteric infections with coronaviruses and toroviruses. In Novartis Foundation Symposium; John Wiley: Chichester, UK, 2001. [Google Scholar]
- Zhou, J.; Li, C.; Zhao, G.; Chu, H.; Wang, D.; Yan, H.H.; Poon, V.K.; Wen, L.; Wong, B.H.; Zhao, X.; et al. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci. Adv. 2017, 3, eaao4966. [Google Scholar] [CrossRef]
- Simsek, C.; Erul, E.; Balaban, H.Y. Role of gastrointestinal system on transmission and pathogenesis of SARS-CoV-2. World J. Clin. Cases 2021, 9, 5427–5434. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Feng, C.; Zhang, X.; Hu, S.; Zhang, Y.; Min, M.; Liu, B.; Ying, X.; Liu, Y. Susceptibility Factors of Stomach for SARS-CoV-2 and Treatment Implication of Mucosal Protective Agent in COVID-19. Front. Med. 2021, 7, 597967. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Yamaoka, Y.; Shirai, N.; Furuta, T. Role of renin-angiotensin system in gastric oncogenesis. J. Gastroenterol. Hepatol. 2012, 27, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Balamtekin, N.; Artuk, C.; Arslan, M.; Gülşen, M. The Effect of Helicobacter pylori on the Presentation and Clinical Course of Coronavirus Disease 2019 Infection. J. Pediatr. Gastroenterol. Nutr. 2021, 72, 511–513. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; Ha, E.K.; Yeniova, A.Ö.; Moon, S.Y.; Kim, S.Y.; Koh, H.Y.; Yang, J.M.; Jeong, S.J.; Moon, S.J.; Cho, J.Y.; et al. Severe clinical outcomes of COVID-19 associated with proton pump inhibitors: A nationwide cohort study with propensity score matching. Gut 2021, 70, 76–84. [Google Scholar] [CrossRef]
- Zippi, M.; Fiorino, S.; Budriesi, R.; Micucci, M.; Corazza, I.; Pica, R.; de Biase, D.; Gallo, C.G.; Hong, W. Paradoxical relationship between proton pump inhibitors and COVID-19: A systematic review and meta-analysis. World J. Clin. Cases 2021, 9, 2763–2777. [Google Scholar] [CrossRef]
- Israelsen, S.B.; Ernst, M.T.; Lundh, A.; Lundbo, L.F.; Sandholdt, H.; Hallas, J.; Benfield, T. Proton Pump Inhibitor Use Is Not Strongly Associated With SARS-CoV-2 Related Outcomes: A Nationwide Study and Meta-analysis. Clin. Gastroenterol. Hepatol. 2021, 19, 1845–1854. [Google Scholar] [CrossRef]
- Hamming, I.; Timens, W.; Bulthuis, M.; Lely, A.T.; Navis, G.J.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS Coronavirus. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef]
- Qi, F.; Qian, S.; Zhang, S.; Zhang, Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020, 526, 135–140. [Google Scholar] [CrossRef]
- Zou, X.; Chen, K.; Zou, J.; Han, P.; Hao, J.; Han, Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020, 14, 185–192. [Google Scholar] [CrossRef]
- Vuille-dit-Bille, R.N.; Camargo, S.M.; Emmenegger, L.; Sasse, T.; Kummer, E.; Jando, J.; Hamie, Q.M.; Meier, C.F.; Hunziker, S.; Forras-Kaufmann, Z.; et al. Human intestine luminal ACE2 and amino acid transporter expression increased by ACE-inhibitors. Amino Acids 2015, 47, 693–705. [Google Scholar] [CrossRef]
- Sungnak, W.; Huang, N.; Bécavin, C.; Berg, M. HCA Lung Biological Network. SARS-CoV-2 Entry Genes Are Most Highly Expressed in Nasal Goblet and Ciliated Cells within Human Airways. arXiv 2020, arXiv:2003.06122v1. [Google Scholar]
- Zhang, H.; Li, H.B.; Lyu, J.R.; Lei, X.M.; Li, W.; Wu, G.; Lyu, J.; Dai, Z.M. Specific ACE2 expression in small intestinal enterocytes may cause gastrointestinal symptoms and injury after 2019-nCoV infection. Int. J. Infect. Dis. 2020, 96, 19–24. [Google Scholar] [CrossRef]
- Camargo, S.M.; Singer, D.; Makrides, V.; Huggel, K.; Pos, K.M.; Wagner, C.A.; Kuba, K.; Danilczyk, U.; Skovby, F.; Kleta, R.; et al. Tissue-specific amino acid transporter partners ACE2 and collectrin differentially interact with hartnup mutations. Gastroenterology 2009, 136, 872–882. [Google Scholar] [CrossRef]
- Gao, N.; Dou, X.; Yin, T.; Yang, Y.; Yan, D.; Ma, Z.; Bi, C.; Shan, A. Tryptophan Promotes Intestinal Immune Defense through Calcium-Sensing Receptor (CaSR)-Dependent Metabolic Pathways. J. Agric. Food Chem. 2021, 69, 13460–13473. [Google Scholar] [CrossRef]
- Qi, J.; Zhou, Y.; Hua, J.; Zhang, L.; Bian, J.; Liu, B.; Zhao, Z.; Jin, S. The scRNA-seq Expression Profiling of the Receptor ACE2 and the Cellular Protease TMPRSS2 Reveals Human Organs Susceptible to SARS-CoV-2 Infection. Int. J. Environ. Res. Public Health 2021, 18, 284. [Google Scholar] [CrossRef]
- Pearce, S.C.; Suntornsaratoon, P.; Kishida, K.; Al-Jawadi, A.; Guardia, J.; Nadler, I.; Flores, J.; Shiarella, R.; Auvinen, M.; Yu, S.; et al. Expression of SARS-CoV-2 entry factors, electrolyte, and mineral transporters in different mouse intestinal epithelial cell types. Physiol. Rep. 2021, 9, e15061. [Google Scholar] [CrossRef]
- Singh, M.; Bansal, V.; Feschotte, C. A Single-Cell RNA Expression Map of Human Coronavirus Entry Factors. Cell Rep. 2020, 32, 108175. [Google Scholar] [CrossRef]
- Zhou, L.; Niu, Z.; Jiang, X.; Zhang, Z.; Zheng, Y.; Wang, Z.; Zhu, Y.; Gao, L.; Huang, H.; Wang, X.; et al. SARS-CoV-2 Targets by the pscRNA Profiling of ACE2, TMPRSS2 and Furin Proteases. iScience 2020, 23, 101744. [Google Scholar] [CrossRef]
- Johnson, B.A.; Xie, X.; Bailey, A.L.; Kalveram, B.; Lokugamage, K.G.; Muruato, A.; Zou, J.; Zhang, X.; Juelich, T.; Smith, J.K.; et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 2021, 591, 293–299. [Google Scholar] [CrossRef]
- Papa, G.; Mallery, D.L.; Albecka, A.; Welch, L.G.; Cattin-Ortolá, J.; Luptak, J.; Paul, D.; McMahon, H.T.; Goodfellow, I.G.; Carteret, A.; et al. Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion. PLoS Pathog. 2021, 17, e1009246. [Google Scholar] [CrossRef] [PubMed]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.E.; Williamson, M.K.; Antón-Plágaro, C.; Shoemark, D.K.; Simón-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Berni Canani, R.; Comegna, M.; Paparo, L.; Cernera, G.; Bruno, C.; Strisciuglio, C.; Zollo, I.; Gravina, A.G.; Miele, E.; Cantone, E.; et al. Age-Related Differences in the Expression of Most Relevant Mediators of SARS-CoV-2 Infection in Human Respiratory and Gastrointestinal Tract. Front. Pediatr. 2021, 9, 697390. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Li, W.; Feng, J.; Ramos da Silva, S.; Ju, E.; Zhang, H.; Chang, Y.; Moore, P.S.; Guo, H.; Gao, S.J. SARS-CoV-2 pseudovirus infectivity and expression of viral entry-related factors ACE2, TMPRSS2, Kim-1, and NRP-1 in human cells from the respiratory, urinary, digestive, reproductive, and immune systems. J. Med. Virol. 2021, 93, 6671–6685. [Google Scholar] [CrossRef] [PubMed]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef]
- Helal, M.A.; Shouman, S.; Abdelwaly, A.; Elmehrath, A.O.; Essawy, M.; Sayed, S.M.; Saleh, A.H.; El-Badri, N. Molecular basis of the potential interaction of SARS-CoV-2 spike protein to CD147 in COVID-19 associated-lymphopenia. J. Biomol. Struct. Dyn. 2022, 40, 1109–1119. [Google Scholar] [CrossRef]
- Fenizia, C.; Galbiati, S.; Vanetti, C.; Vago, R.; Clerici, M.; Tacchetti, C.; Daniele, T. SARS-CoV-2 entry: At the crossroads of CD147 and ACE2. Cells 2021, 10, 1434. [Google Scholar] [CrossRef]
- Shilts, J.; Crozier, T.W.M.; Greenwood, E.J.D.; Lehner, P.J.; Wright, G.J. No evidence for basigin/CD147 as a direct SARS-CoV-2 spike binding receptor. Sci. Rep. 2021, 11, 413. [Google Scholar] [CrossRef]
- Bortolotti, D.; Simioni, C.; Neri, L.M.; Rizzo, R.; Semprini, C.M.; Occhionorelli, S.; Laface, I.; Sanz, J.M.; Schiuma, G.; Rizzo, S.; et al. Relevance of VEGF and CD147 in different SARS-CoV-2 positive digestive tracts characterized by thrombotic damage. FASEB J. 2021, 35, e21969. [Google Scholar] [CrossRef]
- Wang, H.; Ye, J.; Liu, R.; Chen, G.; Zhao, J.; Huang, L.; Yang, F.; Li, M.; Zhang, S.; Xie, J.; et al. Clinical significance of CD147 in children with inflammatory bowel disease. Biomed. Res. Int. 2020, 2020, 7647181. [Google Scholar] [CrossRef]
- Tamhane, T.; Arampatzidou, M.; Gerganova, V.; Tacke, M.; Illukkumbura, R.; Dauth, S.; Schaschke, N.; Peters, C.; Reinheckel, T.; Brix, K. The activity and localization patterns of cathepsins B and X in cells of the mouse gastrointestinal tract differ along its length. Biol. Chem. 2014, 395, 1201–1219. [Google Scholar] [CrossRef]
- Tamhane, T.; Lllukkumbura, R.; Lu, S.; Maelandsmo, G.M.; Haugen, M.H.; Brix, K. Nuclear cathepsin L activity is required for cell cycle progression of colorectal carcinoma cells. Biochimie 2016, 122, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Noda, T.; Okabe, K.; Yanagida, S.; Nishida, M.; Kanda, Y. SARS-CoV-2 induces barrier damage and inflammatory responses in the human iPSC-derived intestinal epithelium. J. Pharmacol. Sci. 2022, 149, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Li, H.; Xu, J.; Yang, M.; Ma, C.; Li, J.; Zhao, S.; Wang, H.; Yang, Y.; Yu, W.; et al. The Gastrointestinal Tract Is an Alternative Route for SARS-CoV-2 Infection in a Nonhuman Primate Model. Gastroenterology 2021, 160, 1647–1661. [Google Scholar] [CrossRef] [PubMed]
- Zupin, L.; Fontana, F.; Clemente, L.; Ruscio, M.; Ricci, G.; Crovella, S. Effect of Short Time of SARS-CoV-2 Infection in Caco-2 Cells. Viruses 2022, 14, 704. [Google Scholar] [CrossRef]
- Lehmann, M.; Allers, K.; Heldt, C.; Meinhardt, J.; Schmidt, F.; Rodriguez-Sillke, Y.; Kunkel, D.; Schumann, M.; Böttcher, C.; Stahl-Hennig, C.; et al. Human small intestinal infection by SARS-CoV-2 is characterized by a mucosal infiltration with activated CD8 + T cells. Mucosal Immunol. 2021, 14, 1381–1392. [Google Scholar] [CrossRef]
- Wang, C.; Xu, J.; Wang, S.; Pan, S.; Zhang, J.; Han, Y.; Huang, M.; Wu, D.; Yang, Q.; Yang, X.; et al. Imaging Mass Cytometric Analysis of Postmortem Tissues Reveals Dysregulated Immune Cell and Cytokine Responses in Multiple Organs of COVID-19 Patients. Front. Microbiol. 2020, 11, 600989. [Google Scholar] [CrossRef]
- Lee, S.; Channappanavar, R.; Kanneganti, T.D. Coronaviruses: Innate Immunity, Inflammasome Activation, Inflammatory Cell Death, and Cytokines. Trends Immunol. 2020, 41, 1083–1099. [Google Scholar] [CrossRef]
- Lowery, S.A.; Sariol, A.; Perlman, S. Innate immune and inflammatory responses to SARS-CoV-2: Implications for COVID-19. Cell Host Microbe 2021, 29, 1052–1062. [Google Scholar] [CrossRef]
- Forsyth, C.B.; Zhang, L.; Bhushan, A.; Swanson, B.; Zhang, L.; Mamede, J.I.; Voigt, R.M.; Shaikh, M.; Engen, P.A.; Keshavarzian, A. The SARS-CoV-2 S1 Spike Protein Promotes MAPK and NF-kB Activation in Human Lung Cells and Inflammatory Cytokine Production in Human Lung and Intestinal Epithelial Cells. Microorganisms 2022, 10, 1996. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Y.; Wang, R.; Liu, X.; Zhao, J.; Tsuda, M.; Li, Y. SARSCoV-2 infection of intestinal epithelia cells sensed by RIG-I and DHX-15 evokes innate immune response and immune cross-talk. Front. Cell Infect. Microbiol. 2023, 12, 1035711. [Google Scholar] [CrossRef] [PubMed]
- Tao, W.; Zhang, G.; Wang, X.; Guo, M.; Zeng, W.; Xu, Z.; Cao, D.; Pan, A.; Wang, Y.; Zhang, K.; et al. Analysis of the intestinal microbiota in COVID-19 patients and its correlation with the inflammatory factor IL-18. Med. Microecol. 2020, 5, 100023. [Google Scholar] [CrossRef] [PubMed]
- Nasser, S.M.T.; Rana, A.A.; Doffinger, R.; Kafizas, A.; Khan, T.A.; Nasser, S. Elevated free interleukin-18 associated with severity and mortality in prospective cohort study of 206 hospitalised COVID-19 patients. Intensive Care Med. Exp. 2023, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Stanifer, M.L.; Kee, C.; Cortese, M.; Zumaran, C.M.; Triana, S.; Mukenhirn, M.; Kraeusslich, H.G.; Alexandrov, T.; Bartenschlager, R.; Boulant, S. Critical Role of Type III Interferon in Controlling SARS-CoV-2 Infection in Human Intestinal Epithelial Cells. Cell Rep. 2020, 32, 107863. [Google Scholar] [CrossRef]
- Effenberger, M.; Grabherr, F.; Mayr, L.; Schwaerzler, J.; Nairz, M.; Seifert, M.; Hilbe, R.; Seiwald, S.; Scholl-Buergi, S.; Fritsche, G.; et al. Faecal calprotectin indicates intestinal inflammation in COVID-19. Gut 2020, 69, 1543–1544. [Google Scholar] [CrossRef]
- Uzzan, M.; Soudan, D.; Peoc’h, K.; Weiss, E.; Corcos, O.; Treton, X. Patients with COVID-19 present with low plasma citrulline concentrations that associate with systemic inflammation and gastrointestinal symptoms. Dig. Liver Dis. 2020, 52, 1104–1105. [Google Scholar] [CrossRef]
- Crenn, P.; Messing, B.; Cynober, L. Citrulline as a biomarker of intestinal failure due to enterocyte mass reduction. Clin. Nutr. 2008, 27, 328–339. [Google Scholar] [CrossRef]
- Ha, S.; Jin, B.; Clemmensen, B.; Park, P.; Mahboob, S.; Gladwill, V.; Lovely, F.M.; Gottfried-Blackmore, A.; Habtezion, A.; Verma, S.; et al. Serotonin is elevated in COVID-19-associated diarrhoea. Gut 2021, 70, 2015–2017. [Google Scholar] [CrossRef]
- Gershon, M.D.; Tack, J. The serotonin signaling system: From basic understanding to drug development for functional Gi disorders. Gastroenterology 2007, 132, 397–414. [Google Scholar] [CrossRef]
- Ojetti, V.; Saviano, A.; Covino, M.; Acampora, N.; Troiani, E.; Franceschi, F.; Gemelli against COVID-19 Group. COVID-19 and intestinal inflammation: Role of fecal calprotectin. Dig. Liver Dis. 2020, 52, 1231–1233. [Google Scholar] [CrossRef]
- Britton, G.J.; Chen-Liaw, A.; Cossarini, F.; Livanos, A.E.; Spindler, M.P.; Plitt, T.; Eggers, J.; Mogno, I.; Gonzalez-Reiche, A.S.; Siu, S.; et al. Limited intestinal inflammation despite diarrhea, fecal viral RNA and SARS-CoV-2-specific IgA in patients with acute COVID-19. Sci. Rep. 2021, 11, 13308. [Google Scholar] [CrossRef]
- Isho, B.; Abe, K.T.; Zuo, M.; Jamal, A.J.; Rathod, B.; Wang, J.H.; Li, Z.; Chao, G.; Rojas, O.L.; Bang, Y.M.; et al. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci. Immunol. 2020, 5, eabe5511. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wei, J.; Zhu, R.; Chen, L.; Ding, F.; Zhou, R.; Ge, L.; Xiao, J.; Zhao, Q. Contribution of CD4+ T cell-mediated inflammation to diarrhea in patients with COVID-19. Int. J. Infect. Dis. 2022, 120, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Zhang, Y.; Li, Y.; Wu, Q.; Wu, J.; Park, S.K.; Guo, C.; Lu, J. Meta-analysis of 16S rRNA microbial data identified alterations of the gut microbiota in COVID-19 patients during the acute and recovery phases. BMC Microbiol. 2022, 22, 274. [Google Scholar] [CrossRef]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.C.K.; Cheung, C.P.; Chen, N.; et al. Alterations in Gut Microbiota of Patients with COVID-19 During Time of Hospitalization. Gastroenterology 2020, 159, 944–955.e8. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Chen, Y.; Wu, Z.; Chen, Y.; Gao, H.; Lv, L.; Guo, F.; Zhang, X.; Luo, R.; Huang, C.; et al. Alterations of the Gut Microbiota in Patients With Coronavirus Disease 2019 or H1N1 Influenza. Clin. Infect. Dis. 2020, 71, 2669–2678. [Google Scholar] [CrossRef]
- Blackett, J.W.; Sun, Y.; Purpura, L.; Margolis, K.G.; Elkind, M.S.V.; O’Byrne, S.; Wainberg, M.; Abrams, J.A.; Wang, H.H.; Chang, L.; et al. Decreased Gut Microbiome Tryptophan Metabolism and Serotonergic Signaling in Patients with Persistent Mental Health and Gastrointestinal Symptoms After COVID-19. Clin. Transl. Gastroenterol. 2022, 13, e00524. [Google Scholar] [CrossRef]
- Hashimoto, T.; Perlot, T.; Rehman, A.; Trichereau, J.; Ishiguro, H.; Paolino, M.; Sigl, V.; Hanada, T.; Hanada, R.; Lipinski, S.; et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 2012, 487, 477–481. [Google Scholar] [CrossRef]
- Ye, Q.; Wang, B.; Zhang, T.; Xu, J.; Shang, S. The mechanism and treatment of gastrointestinal symptoms in patients with COVID-19. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 319, G245–G252. [Google Scholar] [CrossRef]
- Zhang, F.; Wan, Y.; Zuo, T.; Yeoh, Y.K.; Liu, Q.; Zhang, L.; Zhan, H.; Lu, W.; Xu, W.; Lui, G.C.Y.; et al. Prolonged Impairment of Short-Chain Fatty Acid and L-Isoleucine Biosynthesis in Gut Microbiome in Patients with COVID-19. Gastroenterology 2022, 162, 548–561.e4. [Google Scholar] [CrossRef]
- Li, J.; Richards, E.M.; Handberg, E.M.; Pepine, C.J.; Raizada, M.K. Butyrate regulates COVID-19-relevant genes in gut epithelial organoids from normotensive rats. Hypertension 2021, 77, e13–e16. [Google Scholar] [CrossRef] [PubMed]
- Pascoal, L.B.; Rodrigues, P.B.; Genaro, L.M.; Gomes, A.B.D.S.P.; Toledo-Teixeira, D.A.; Parise, P.L.; Bispo-Dos-Santos, K.; Simeoni, C.L.; Guimarães, P.V.; Buscaratti, L.I.; et al. Microbiota-derived short-chain fatty acids do not interfere with SARS-CoV-2 infection of human colonic samples. Gut Microbes 2021, 13, 1–9. [Google Scholar] [CrossRef]
- Hirayama, M.; Nishiwaki, H.; Hamaguchi, T.; Ito, M.; Ueyama, J.; Maeda, T.; Kashihara, K.; Tsuboi, Y.; Ohno, K. Intestinal Collinsella may mitigate infection and exacerbation of COVID-19 by producing ursodeoxycholate. PLoS ONE 2021, 16, e0260451. [Google Scholar] [CrossRef] [PubMed]
- Brevini, T.; Maes, M.; Webb, G.J.; John, B.V.; Fuchs, C.D.; Buescher, G.; Wang, L.; Griffiths, C.; Brown, M.L.; Scott, W.E., 3rd; et al. FXR inhibition may protect from SARS-CoV-2 infection by reducing ACE2. Nature 2023, 615, 134–142. [Google Scholar] [CrossRef] [PubMed]
- McAllister, M.J.; Kirkwood, K.; Chuah, S.C.; Thompson, E.J.; Cartwright, J.A.; Russell, C.D.; Dorward, D.A.; Lucas, C.D.; Ho, G.T. Intestinal Protein Characterisation of SARS-CoV-2 Entry Molecules ACE2 and TMPRSS2 in Inflammatory Bowel Disease (IBD) and Fatal COVID-19 Infection. Inflammation 2022, 45, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Jablaoui, A.; Kriaa, A.; Mkaouar, H.; Akermi, N.; Soussou, S.; Wysocka, M.; Wołoszyn, D.; Amouri, A.; Gargouri, A.; Maguin, E.; et al. Fecal Serine Protease Profiling in Inflammatory Bowel Diseases. Front. Cell Infect. Microbiol. 2020, 10, 21. [Google Scholar] [CrossRef]
- Potdar, A.A.; Dube, S.; Naito, T.; Li, K.; Botwin, G.; Haritunians, T.; Li, D.; Casero, D.; Yang, S.; Bilsborough, J.; et al. Altered Intestinal ACE2 Levels Are Associated with Inflammation, Severe Disease, and Response to Anti-Cytokine Therapy in Inflammatory Bowel Disease. Gastroenterology 2021, 160, 809–822. [Google Scholar] [CrossRef]
- Potdar, A.A.; Dube, S.; Naito, T.; Botwin, G.; Haritunians, T.; Li, D.; Yang, S.; Bilsborough, J.; Denson, L.A.; Daly, M.; et al. Reduced expression of COVID-19 host receptor, ACE2 is associated with small bowel inflammation, more severe disease, and response to anti-TNF therapy in Crohn’s disease. medRxiv 2020. [Google Scholar]
- Nowak, J.K.; Lindstrøm, J.C.; Kalla, R.; Ricanek, P.; Halfvarson, J.; Satsangi, J. Age, Inflammation, and Disease Location Are Critical Determinants of Intestinal Expression of SARS-CoV-2 Receptor ACE2 and TMPRSS2 in Inflammatory Bowel Disease. Gastroenterology 2020, 159, 1151–1154. [Google Scholar] [CrossRef]
- Garg, M.; Burrell, L.M.; Velkoska, E.; Griggs, K.; Angus, P.W.; Gibson, P.R.; Lubel, J.S. Upregulation of circulating components of the alternative renin-angiotensin system in inflammatory bowel disease: A pilot study. J. Renin Angiotensin Aldosterone Syst. 2015, 16, 559–569. [Google Scholar] [CrossRef]
- Krishnamurthy, S.; Lockey, R.F.; Kolliputi, N. Soluble ACE2 as a potential therapy for COVID-19. Am. J. Physiol. Cell Physiol. 2021, 320, C279–C281. [Google Scholar] [CrossRef] [PubMed]
- Bangma, A.; Voskuil, M.D.; Weersma, R.K. TNFα-Antagonist Use and Mucosal Inflammation Are Associated with Increased Intestinal Expression of SARS-CoV-2 Host Protease TMPRSS2 in Patients with Inflammatory Bowel Disease. Gastroenterology 2021, 160, 2621–2622. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Lu, Y.; Peng, G.; Li, J.; Li, W.; Li, M.; Wang, H.; Liu, L.; Zhao, Q. Furin inhibits epithelial cell injury and alleviates experimental colitis by activating the Nrf2-Gpx4 signaling pathway. Dig. Liver Dis. 2021, 53, 1276–1285. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, N.S.F.; Martins, C.A.; Quaresma, A.B.; Hino, A.A.F.; Steinwurz, F.; Ungaro, R.C.; Kotze, P.G. COVID-19 outcomes in patients with inflammatory bowel diseases in Latin America: Results from SECURE-IBD registry. J. Gastroenterol. Hepatol. 2021, 36, 3033–3040. [Google Scholar] [CrossRef]
- Derikx, L.A.A.P.; Lantinga, M.A.; de Jong, D.J.; van Dop, W.A.; Creemers, R.H.; Römkens, T.E.H.; Jansen, J.M.; Mahmmod, N.; West, R.L.; Tan, A.C.I.T.L.; et al. Clinical Outcomes of COVID-19 in Patients with Inflammatory Bowel Disease: A Nationwide Cohort Study. J. Crohns Colitis 2021, 15, 529–539. [Google Scholar] [CrossRef]
- Attauabi, M.; Poulsen, A.; Theede, K.; Pedersen, N.; Larsen, L.; Jess, T.; Rosager Hansen, M.; Verner-Andersen, M.K.; Haderslev, K.V.; Berg Lødrup, A.; et al. Prevalence and Outcomes of COVID-19 Among Patients with Inflammatory Bowel Disease-A Danish Prospective Population-based Cohort Study. J. Crohns Colitis 2021, 15, 540–550. [Google Scholar] [CrossRef]
- Alrashed, F.; Battat, R.; Abdullah, I.; Charabaty, A.; Shehab, M. Impact of medical therapies for inflammatory bowel disease on the severity of COVID-19: A systematic review and meta-analysis. BMJ Open Gastroenterol. 2021, 8, e000774. [Google Scholar] [CrossRef]
- Xu, J.; Chu, M.; Zhong, F.; Tan, X.; Tang, G.; Mai, J.; Lai, N.; Guan, C.; Liang, Y.; Liao, G. Digestive symptoms of COVID-19 and expression of ACE2 in digestive tract organs. Cell Death Discov. 2020, 6, 76. [Google Scholar] [CrossRef]
- Chiang, A.W.T.; Duong, L.D.; Shoda, T.; Nhu, Q.M.; Ruffner, M.; Hara, T.; Aaron, B.; Joplin, E.; Manresa, M.C.; Abonia, J.P.; et al. Type 2 Immunity and Age Modify Gene Expression of Coronavirus-induced Disease 2019 Receptors in Eosinophilic Gastrointestinal Disorders. J. Pediatr. Gastroenterol. Nutr. 2021, 72, 718–722. [Google Scholar] [CrossRef]
- Li, J.; Tian, A.; Yang, D.; Zhang, M.; Chen, L.; Wen, J.; Chen, P. Celiac Disease and the Susceptibility of COVID-19 and the Risk of Severe COVID-19: A Mendelian Randomization Study. Clin. Transl. Gastroenterol. 2022, 13, e00480. [Google Scholar] [CrossRef]
- Zevit, N.; Chehade, M.; Leung, J.; Marderfeld, L.; Dellon, E.S. Eosinophilic Esophagitis Patients Are Not at Increased Risk of Severe COVID-19: A Report from a Global Registry. J. Allergy Clin. Immunol. Pract. 2022, 10, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Santacroce, G.; Lenti, M.V.; Aronico, N.; Miceli, E.; Lovati, E.; Lucotti, P.C.; Coppola, L.; Gentile, A.; Latorre, M.A.; Di Terlizzi, F.; et al. Impact of COVID-19 in immunosuppressive drug-naïve autoimmune disorders: Autoimmune gastritis, celiac disease, type 1 diabetes, and autoimmune thyroid disease. Pediatr. Allergy Immunol. 2022, 33, 105–107. [Google Scholar] [CrossRef] [PubMed]
- Treskova-Schwarzbach, M.; Haas, L.; Reda, S.; Pilic, A.; Borodova, A.; Karimi, K.; Koch, J.; Nygren, T.; Scholz, S.; Schönfeld, V.; et al. Pre-existing health conditions and severe COVID-19 outcomes: An umbrella review approach and meta-analysis of global evidence. BMC Med. 2021, 19, 212. [Google Scholar] [CrossRef] [PubMed]
- Golla, R.; Vuyyuru, S.; Kante, B.; Kumar, P.; Thomas, D.M.; Makharia, G.; Kedia, S.; Ahuja, V. Long-term Gastrointestinal Sequelae Following COVID-19: A Prospective Follow-up Cohort Study. Clin. Gastroenterol. Hepatol. 2023, 21, 789–796. [Google Scholar] [CrossRef]
- Schmulson, M.; Ghoshal, U.C.; Barbara, G. Managing the inevitable surge of post–COVID-19 functional gastrointestinal disorders. Am. J. Gastroenterol. 2021, 116, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Spiller, R.C. Estimating the importance of infection in IBS. Am. J. Gastroenterol. 2003, 98, 238–241. [Google Scholar] [CrossRef]
- Porter, C.K.; Gormley, R.; Tribble, D.R.; Cash, B.D.; Riddle, M.S. The Incidence and gastrointestinal infectious risk of functional gastrointestinal disorders in a healthy US adult population. Am. J. Gastroenterol. 2011, 106, 130–138. [Google Scholar] [CrossRef]
- Neal, K.R.; Barker, L.; Spiller, R.C. Prognosis in post infective irritable bowel syndrome: A 6 year follow up study. Gut 2002, 51, 410–413. [Google Scholar] [CrossRef]
- Liu, Q.; Mak, J.W.Y.; Su, Q.; Yeoh, Y.K.; Lui, G.C.; Ng, S.S.S.; Zhang, F.; Li, A.Y.L.; Lu, W.; Hui, D.S.; et al. Gut microbiota dynamics in a prospective cohort of patients with post-acute COVID-19 syndrome. Gut 2022, 71, 544–552. [Google Scholar] [CrossRef]
- Prasad, R.; Patton, M.J.; Floyd, J.L.; Fortmann, S.; DuPont, M.; Harbour, A.; Wright, J.; Lamendella, R.; Stevens, B.R.; Oudit, G.Y.; et al. Plasma Microbiome in COVID-19 Subjects: An Indicator of Gut Barrier Defects and Dysbiosis. Int. J. Mol. Sci. 2022, 23, 9141. [Google Scholar] [CrossRef]
- Okuyucu, M.; Yalcin Kehribar, D.; Çapraz, M.; Çapraz, A.; Arslan, M.; Çelik, Z.B.; Usta, B.; Birinci, A.; Ozgen, M. The Relationship Between COVID-19 Disease Severity and Zonulin Levels. Cureus 2022, 14, e28255. [Google Scholar] [CrossRef]
- Osman, I.O.; Garrec, C.; de Souza, G.A.P.; Zarubica, A.; Belhaouari, D.B.; Baudoin, J.P.; Lepidi, H.; Mege, J.L.; Malissen, B.; Scola, B.; et al. Control of CDH1/E-Cadherin Gene Expression and Release of a Soluble Form of E-Cadherin in SARS-CoV-2 Infected Caco-2 Intestinal Cells: Physiopathological Consequences for the Intestinal Forms of COVID-19. Front. Cell Infect. Microbiol. 2022, 12, 798767. [Google Scholar] [CrossRef]
- Natarajan, A.; Zlitni, S.; Brooks, E.F.; Vance, S.E.; Dahlen, A.; Hedlin, H.; Park, R.M.; Han, A.; Schmidtke, D.T.; Verma, R.; et al. Gastrointestinal symptoms and fecal shedding of SARS-CoV-2 RNA suggest prolonged gastrointestinal infection. Med 2022, 3, 371–387.e9. [Google Scholar] [CrossRef]
- Zollner, A.; Koch, R.; Jukic, A.; Pfister, A.; Meyer, M.; Rössler, A.; Kimpel, J.; Adolph, T.E.; Tilg, H. Post-acute COVID19 is characterized by gut viral antigen persistence in inflammatory bowel diseases. Gastroenterology 2022, 163, 495–506. [Google Scholar] [CrossRef]
- Chalkias, S.; Harper, C.; Vrbicky, K.; Walsh, S.R.; Essink, B.; Brosz, A.; McGhee, N.; Tomassini, J.E.; Chen, X.; Chang, Y.; et al. A Bivalent Omicron-Containing Booster Vaccine against COVID-19. N. Engl. J. Med. 2022, 387, 1279–1291. [Google Scholar] [CrossRef]
- Willan, J.; Agarwal, G.; Bienz, N. Mortality and burden of post-COVID-19 syndrome have reduced with time across SARS-CoV-2 variants in haematology patients. Br. J. Haematol. 2023, 201, 640–644. [Google Scholar] [CrossRef]
- Cortellini, A.; Tabernero, J.; Mukherjee, U.; Salazar, R.; Sureda, A.; Maluquer, C.; Ferrante, D.; Bower, M.; Sharkey, R.; Mirallas, O.; et al. SARS-CoV-2 omicron (B.1.1.529)-related COVID-19 sequelae in vaccinated and unvaccinated patients with cancer: Results from the OnCovid registry. Lancet Oncol. 2023, 24, 335–346. [Google Scholar] [CrossRef]
- Kahlert, C.R.; Strahm, C.; Güsewell, S.; Cusini, A.; Brucher, A.; Goppel, S.; Möller, E.; Möller, J.C.; Ortner, M.; Ruetti, M.; et al. Post-acute sequelae after SARS-CoV-2 infection by viral variant and vaccination status: A multicenter cross-sectional study. Clin. Infect. Dis. 2023, 77, 194–202. [Google Scholar] [CrossRef]
- Bolesławska, I.; Kowalówka, M.; Bolesławska-Król, N.; Przysławski, J. Ketogenic Diet and Ketone Bodies as Clinical Support for the Treatment of SARS-CoV-2-Review of the Evidence. Viruses 2023, 15, 1262. [Google Scholar] [CrossRef]
- Sharma, S.; Di Castelnuovo, A.; Costanzo, S.; Persichillo, M.; Panzera, T.; Ruggiero, E.; De Curtis, A.; Storto, M.; Cavallo, P.; Gianfagna, F.; et al. Habitual adherence to a traditional Mediterranean diet and risk of SARS-CoV-2 infection and Coronavirus disease 2019 (COVID-19): A longitudinal analysis. Int. J. Food Sci. Nutr. 2023, 74, 382–394. [Google Scholar] [CrossRef]
- Tomkinson, S.; Triscott, C.; Schenk, E.; Foey, A. The Potential of Probiotics as Ingestible Adjuvants and Immune Modulators for Antiviral Immunity and Management of SARS-CoV-2 Infection and COVID-19. Pathogens 2023, 12, 928. [Google Scholar] [CrossRef] [PubMed]
- Mardi, A.; Kamran, A.; Pourfarzi, F.; Zare, M.; Hajipour, A.; Doaei, S.; Abediasl, N.; Hackett, D. Potential of macronutrients and probiotics to boost immunity in patients with SARS-CoV-2: A narrative review. Front. Nutr. 2023, 10, 1161894. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Somasundaram, I.; Das, D.; Jain Manoj, S.; Banu, H.; Mitta Suresh, P.; Paul, S.; Bisgin, A.; Zhang, H.; Sun, X.F.; et al. Functional Foods: A Promising Strategy for Restoring Gut Microbiota Diversity Impacted by SARS-CoV-2 Variants. Nutrients 2023, 15, 2631. [Google Scholar] [CrossRef]
- Cheong, K.L.; Yu, B.; Teng, B.; Veeraperumal, S.; Xu, B.; Zhong, S.; Tan, K. Post-COVID-19 syndrome management: Utilizing the potential of dietary polysaccharides. Biomed. Pharmacother. 2023, 166, 115320. [Google Scholar] [CrossRef]
- Cheong, K.L.; Chen, S.; Teng, B.; Veeraperumal, S.; Zhong, S.; Tan, K. Oligosaccharides as Potential Regulators of Gut Microbiota and Intestinal Health in Post-COVID-19 Management. Pharmaceuticals 2023, 16, 860. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vernia, F.; Ashktorab, H.; Cesaro, N.; Monaco, S.; Faenza, S.; Sgamma, E.; Viscido, A.; Latella, G. COVID-19 and Gastrointestinal Tract: From Pathophysiology to Clinical Manifestations. Medicina 2023, 59, 1709. https://doi.org/10.3390/medicina59101709
Vernia F, Ashktorab H, Cesaro N, Monaco S, Faenza S, Sgamma E, Viscido A, Latella G. COVID-19 and Gastrointestinal Tract: From Pathophysiology to Clinical Manifestations. Medicina. 2023; 59(10):1709. https://doi.org/10.3390/medicina59101709
Chicago/Turabian StyleVernia, Filippo, Hassan Ashktorab, Nicola Cesaro, Sabrina Monaco, Susanna Faenza, Emanuele Sgamma, Angelo Viscido, and Giovanni Latella. 2023. "COVID-19 and Gastrointestinal Tract: From Pathophysiology to Clinical Manifestations" Medicina 59, no. 10: 1709. https://doi.org/10.3390/medicina59101709






