Comprehensive Analysis of NKX3.2 in Liver Hepatocellular Carcinoma by Bigdata
Abstract
:1. Introduction
2. Materials and Methods
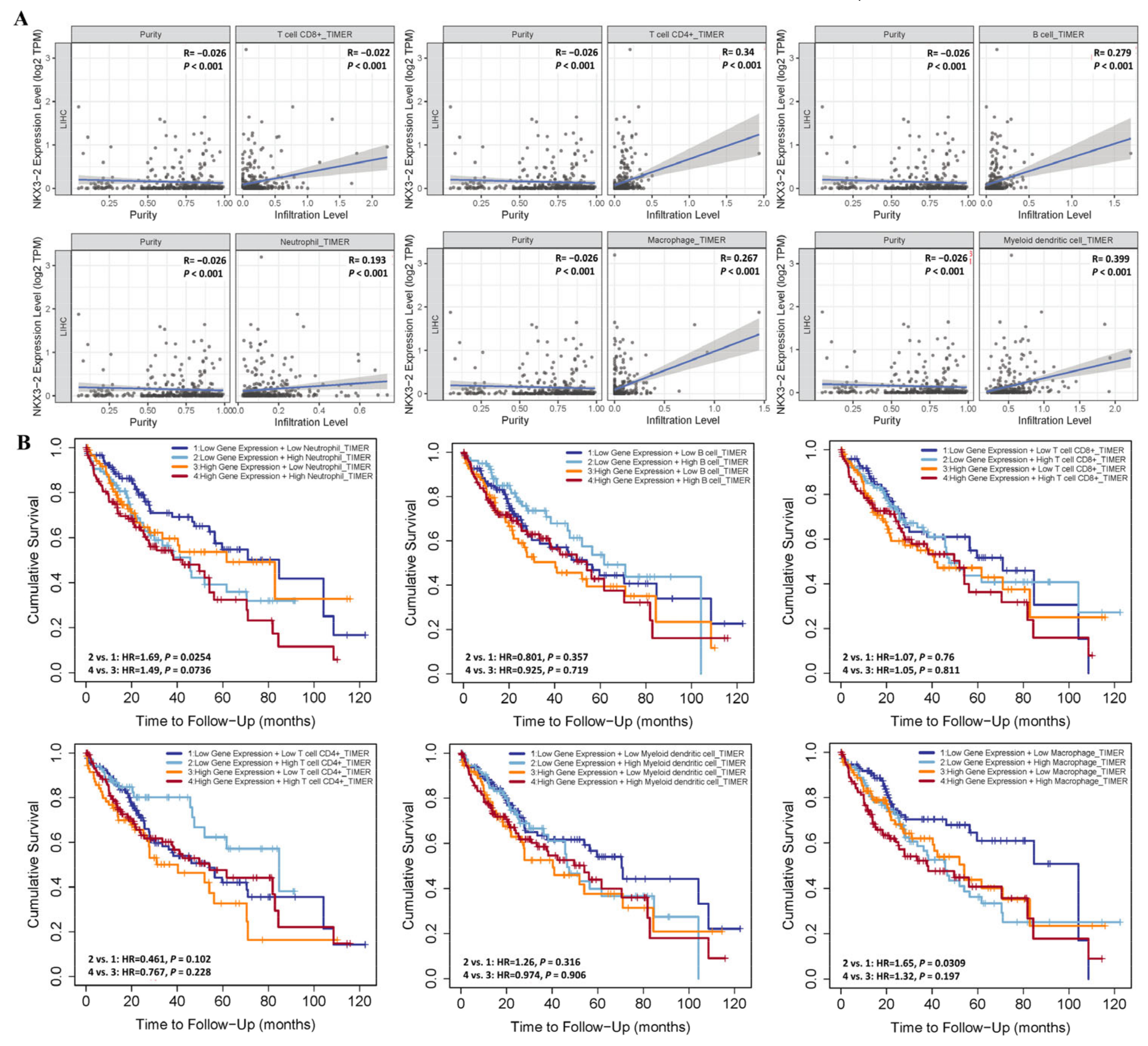
2.1. TIMER Database Analysis
2.2. UALCAN Database Analysis
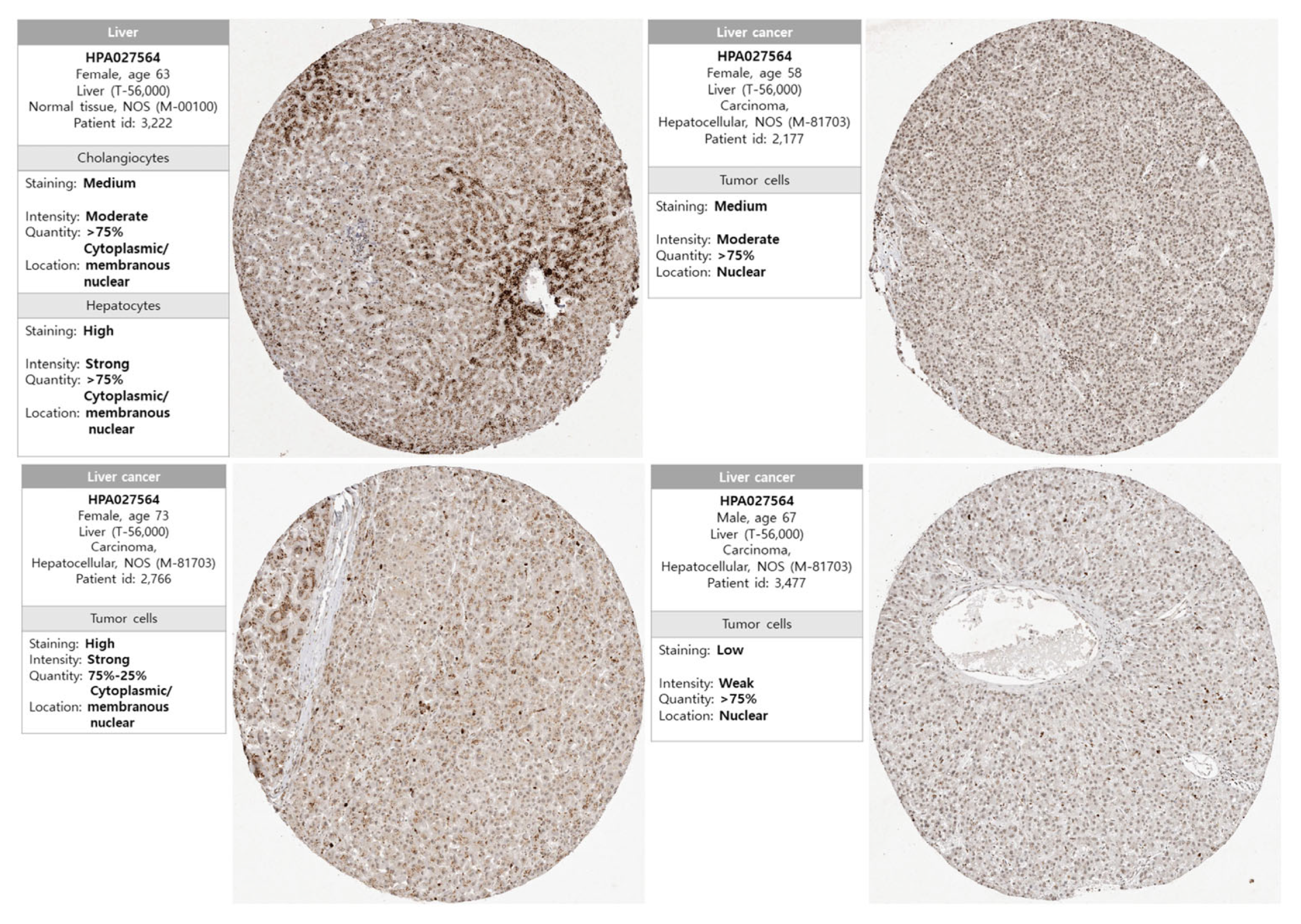
2.3. Immunohistochemistry (IHC) Staining Analysis
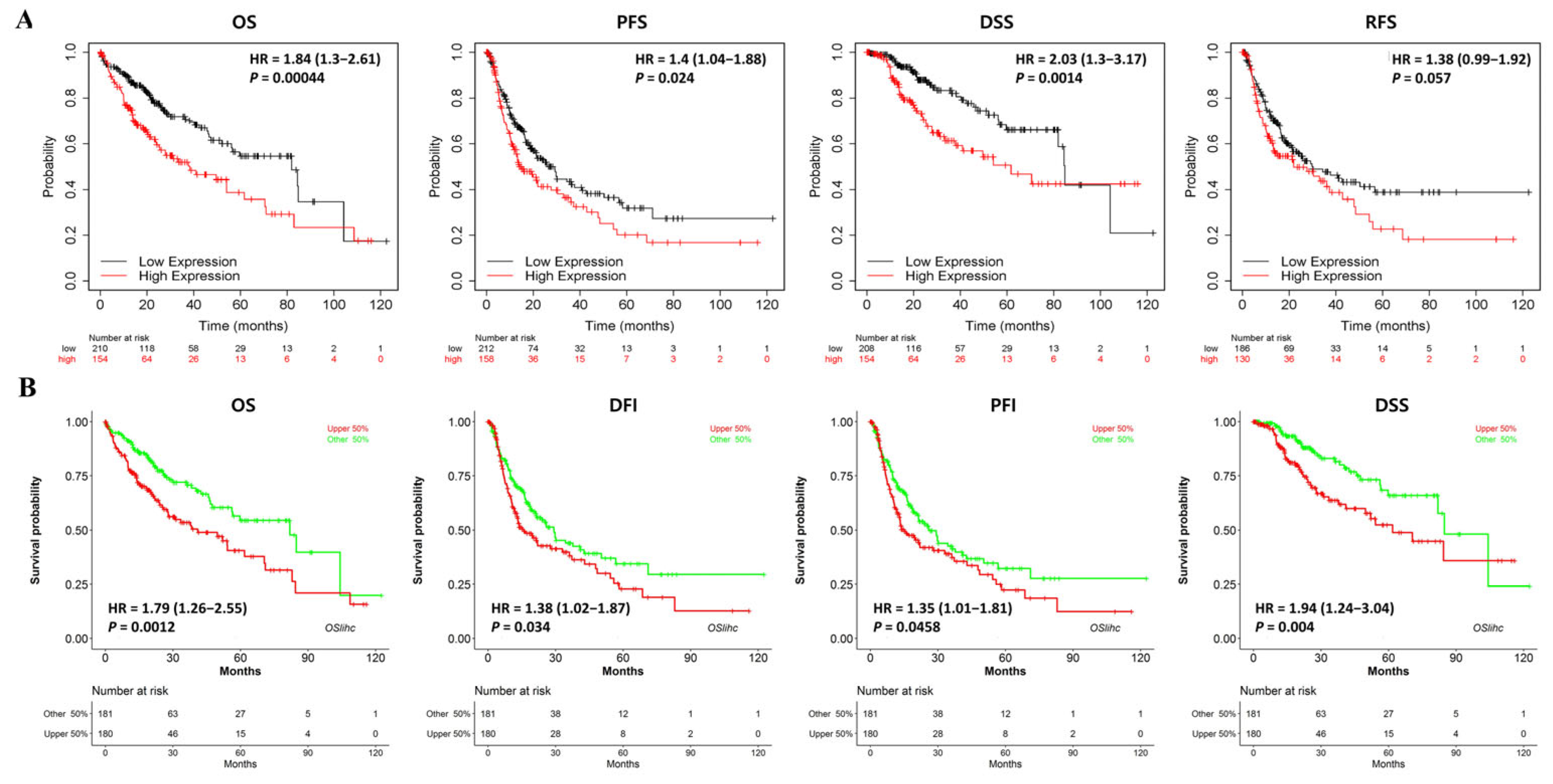
2.4. KM Database Analysis
2.5. OSlihc Database Analysis
2.6. LinkedOmics Database Analysis
2.7. GEPIA2 Database Analysis
2.8. Statistical Analysis
3. Results
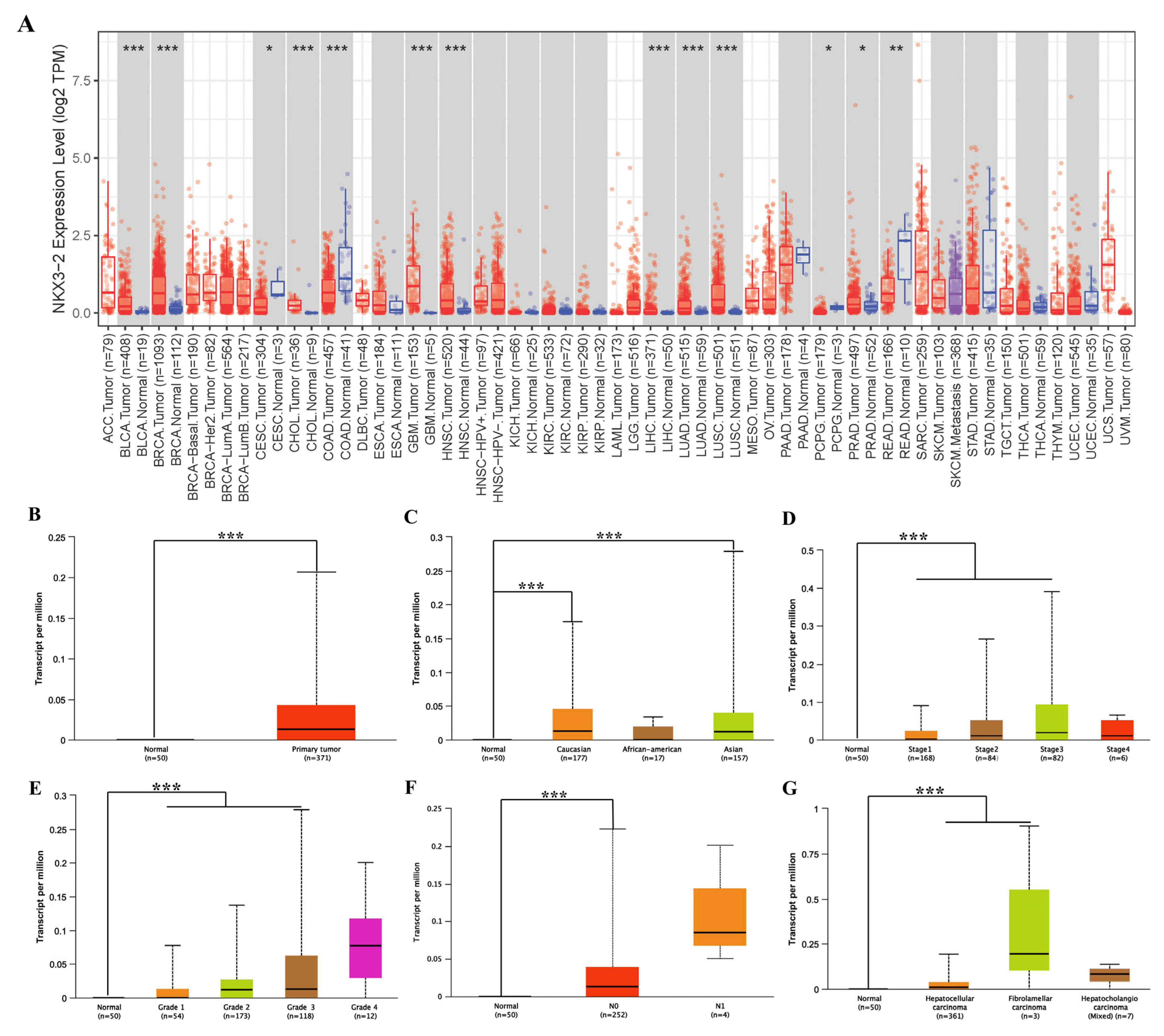
3.1. Assessment of NKX3.2 Expression in Different Cancer and Normal Tissues
3.2. Protein Expression of NKX3.2 in LICH
3.3. Multifaceted Prognostic Value of NKX3.2 in LIHC
3.4. Association of NKX3.2 Expression with Immune Cell Infiltration in LIHC
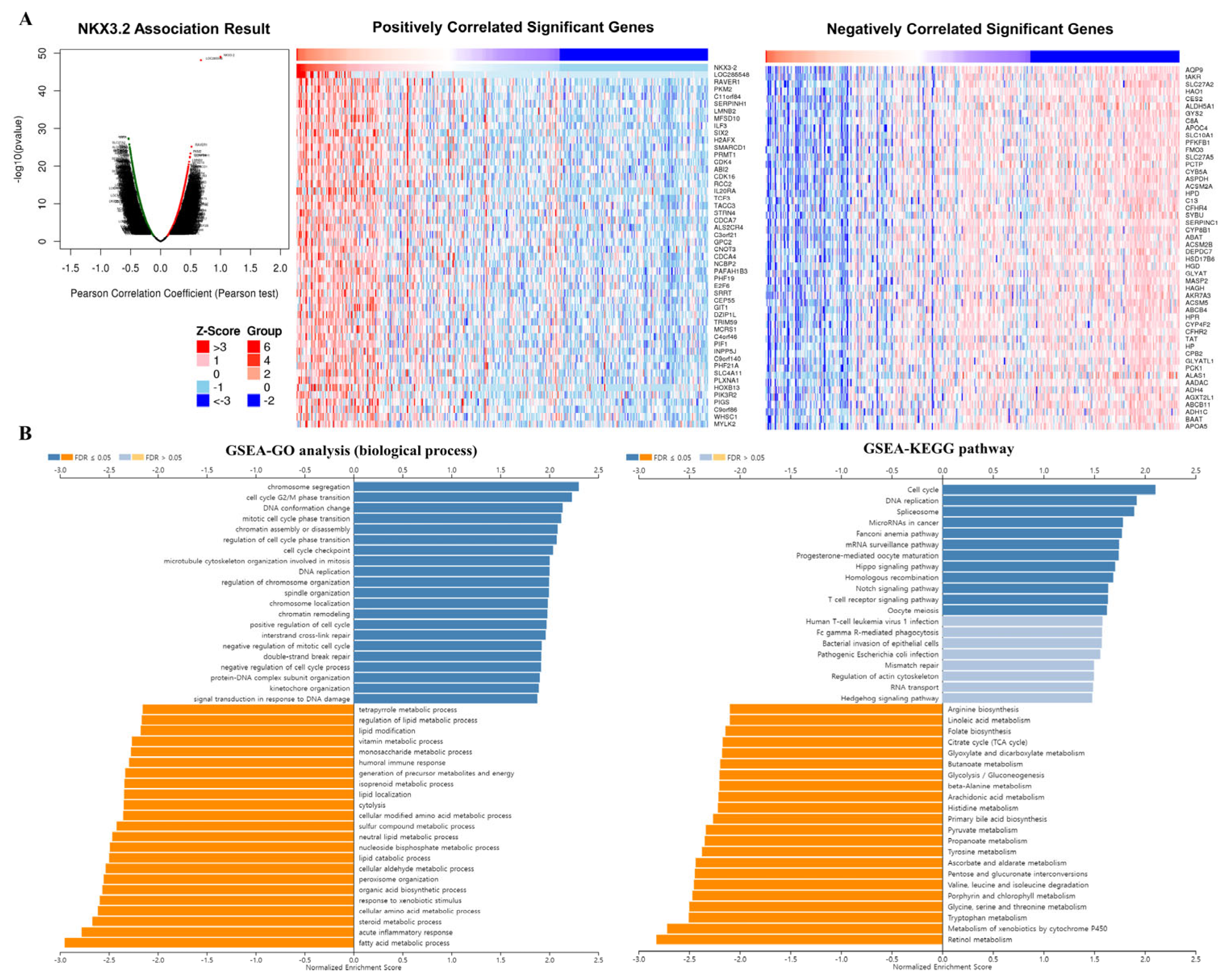
3.5. Co-Expression Analysis of the NKX3.2 Gene
3.6. Prognosis Analysis of the NKX3.2-Related Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Z.; Zhang, G.; Wu, J.; Jia, M. Adjuvant therapy for hepatocellular carcinoma: Current situation and prospect. Drug Discov. Ther. 2013, 7, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Alqahtani, A.; Khan, Z.; Alloghbi, A.; Ahmed, T.S.S.; Ashraf, M.; Hammouda, D.M. Hepatocellular Carcinoma: Molecular Mechanisms and Targeted Therapies. Medicina 2019, 55, 526. [Google Scholar] [CrossRef] [PubMed]
- Mulero-Sánchez, A.; Ramirez, C.F.A.; du Chatinier, A.; Wang, H.; Koomen, S.J.I.; Song, J.; de Groot, M.H.P.; Lieftink, C.; Bosma, A.; Burylo, A.; et al. Rational combination of SHP2 and mTOR inhibition for the treatment of hepatocellular carcinoma. Mol. Oncol. 2023, 17, 964–980. [Google Scholar] [CrossRef]
- Fang, X.; Yan, Q.; Liu, S.; Guan, X.Y. Cancer Stem Cells in Hepatocellular Carcinoma: Intrinsic and Extrinsic Molecular Mechanisms in Stemness Regulation. Int. J. Mol. Sci. 2022, 23, 12327. [Google Scholar] [CrossRef] [PubMed]
- Waldmann, L.; Leyhr, J.; Zhang, H.; Öhman-Mägi, C.; Allalou, A.; Haitina, T. The broad role of Nkx3.2 in the development of the zebrafish axial skeleton. PLoS ONE 2021, 16, e0255953. [Google Scholar] [CrossRef]
- Lettice, L.; Hecksher-Sørensen, J.; Hill, R. The role of Bapx1 (Nkx3.2) in the development and evolution of the axial skeleton. J. Anat. 2001, 199, 181–187. [Google Scholar] [CrossRef]
- Kim, J.A.; Im, S.; Cantley, L.C.; Kim, D.W. Suppression of Nkx3.2 by phosphatidylinositol-3-kinase signaling regulates cartilage development by modulating chondrocyte hypertrophy. Cell. Signal. 2015, 27, 2389–2400. [Google Scholar] [CrossRef]
- Rainbow, R.S.; Won, H.K.; Zeng, L. The role of Nkx3.2 in chondrogenesis. Front. Biol. 2014, 9, 376–381. [Google Scholar] [CrossRef]
- Simon, M.; Campos-Xavier, A.B.; Mittaz-Crettol, L.; Valadares, E.R.; Carvalho, D.; Speck-Martins, C.E.; Nampoothiri, S.; Alanay, Y.; Mihci, E.; van Bever, Y.; et al. Severe neurologic manifestations from cervical spine instability in spondylo-megaepiphyseal-metaphyseal dysplasia. Am. J. Med. Genet. Part C Semin. Med. Genet. 2012, 160C, 230–237. [Google Scholar] [CrossRef]
- Kim, J.; Park, J.H.; Lee, J.H. Analysis of the Cancer Genome Atlas Data to Determine the Prognostic Value of GABPB1L and TERT in Glioblastoma. Keimyung Med. J. 2021, 40, 73–76. [Google Scholar] [CrossRef]
- Kim, J.; Jung, S.J.; Lee, J.H. Clinical and Prognostic Values of DNMT3B Expression in Hepatocellular Carcinoma. Keimyung Med. J. 2022, 41, 13–16. [Google Scholar] [CrossRef]
- Gil Lee, J.; Kang, C.M.; Park, J.S.; Kim, K.S.; Yoon, D.S.; Choi, J.S.; Lee, W.J.; Kim, B.R. The actual five-year survival rate of hepatocellular carcinoma patients after curative resection. Yonsei Med. J. 2006, 47, 105–112. [Google Scholar]
- Matsumura, H.; Nirei, K.; Nakamura, H.; Higuchi, T.; Arakawa, Y.; Ogawa, M.; Tanaka, N.; Moriyama, M. Histopathology of type C liver disease for determining hepatocellular carcinoma risk factors. World J. Gastroenterol. 2013, 19, 4887–4896. [Google Scholar] [CrossRef] [PubMed]
- Pollack, H.J.; Kwon, S.C.; Wang, S.H.; Wyatt, L.C.; Trinh-Shevrin, C.; AAHBP Coalition. Chronic hepatitis B and liver cancer risks among Asian immigrants in New York City: Results from a large, community-based screening, evaluation, and treatment program. Cancer Epidemiol. Biomark. Prev. 2014, 23, 2229–2239. [Google Scholar] [CrossRef]
- Rizzo, G.E.M.; Cabibbo, G.; Craxì, A. Hepatitis B Virus-Associated Hepatocellular Carcinoma. Viruses 2022, 14, 986. [Google Scholar] [CrossRef]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Lalazar, G.; Simon, S.M. Fibrolamellar Carcinoma: Recent Advances and Unresolved Questions on the Molecular Mechanisms. Semin. Liver Dis. 2018, 38, 51–59. [Google Scholar] [CrossRef]
- Alshareefy, Y.; Shen, C.Y.; Prekash, R.J. Exploring the molecular pathogenesis, diagnosis and treatment of fibrolamellar hepatocellular carcinoma: A state of art review of the current literature. Pathol. Res. Pract. 2023, 248, 154655. [Google Scholar] [CrossRef]
- Son, B.; Lee, S.; Youn, H.; Kim, E.; Kim, W.; Youn, B. The role of tumor microenvironment in therapeutic resistance. Oncotarget 2017, 8, 3933–3945. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Yuan, L.; Ma, B.; Wang, G.; Tian, Y. Tumour microenvironment (TME) characterization identified prognosis and immunotherapy response in muscle-invasive bladder cancer (MIBC). Cancer Immunol. Immunother. 2021, 70, 1–18. [Google Scholar] [CrossRef]
- Xu, F.; Jin, T.; Zhu, Y.; Dai, C. Immune checkpoint therapy in liver cancer. J. Exp. Clin. Cancer Res. 2018, 37, 110. [Google Scholar] [CrossRef]
- Zhu, X.D.; Tang, Z.Y.; Sun, H.C. Targeting angiogenesis for liver cancer: Past, present, and future. Genes Dis. 2020, 7, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Le, Y.; Kong, H.; Gao, X.; Zhu, J. Prognostic and Immunological Significance of FUNDC1 in Hepatocellular Carcinoma: A Study on TCGA Mining. Comput. Math. Methods Med. 2022, 2022, 8371885. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.; Dong, L.; Cheng, L. Neutrophils in cancer carcinogenesis and metastasis. J. Hematol. Oncol. 2021, 14, 173. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bae, A.-N.; Kim, J.; Park, J.-H.; Lee, J.-H.; Choi, E. Comprehensive Analysis of NKX3.2 in Liver Hepatocellular Carcinoma by Bigdata. Medicina 2023, 59, 1782. https://doi.org/10.3390/medicina59101782
Bae A-N, Kim J, Park J-H, Lee J-H, Choi E. Comprehensive Analysis of NKX3.2 in Liver Hepatocellular Carcinoma by Bigdata. Medicina. 2023; 59(10):1782. https://doi.org/10.3390/medicina59101782
Chicago/Turabian StyleBae, An-Na, Jongwan Kim, Jong-Ho Park, Jae-Ho Lee, and Euncheol Choi. 2023. "Comprehensive Analysis of NKX3.2 in Liver Hepatocellular Carcinoma by Bigdata" Medicina 59, no. 10: 1782. https://doi.org/10.3390/medicina59101782





