BRAF V600E Mutation of Non-Small Cell Lung Cancer in Korean Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients, Tissue Specimens, and DNA
2.2. PNA-Mediated Clamping PCR (PNA Clamping PCR)
2.3. Real-Time PCR
2.4. Immunohistochemistry
2.5. Direct Sanger Sequencing
3. Results
3.1. Clinicopathologic Characteristics of Resected Non-Small Cell Lung Cancers
3.2. BRAF V600 PNA Clamping and BRAF V600E Real-Time PCR
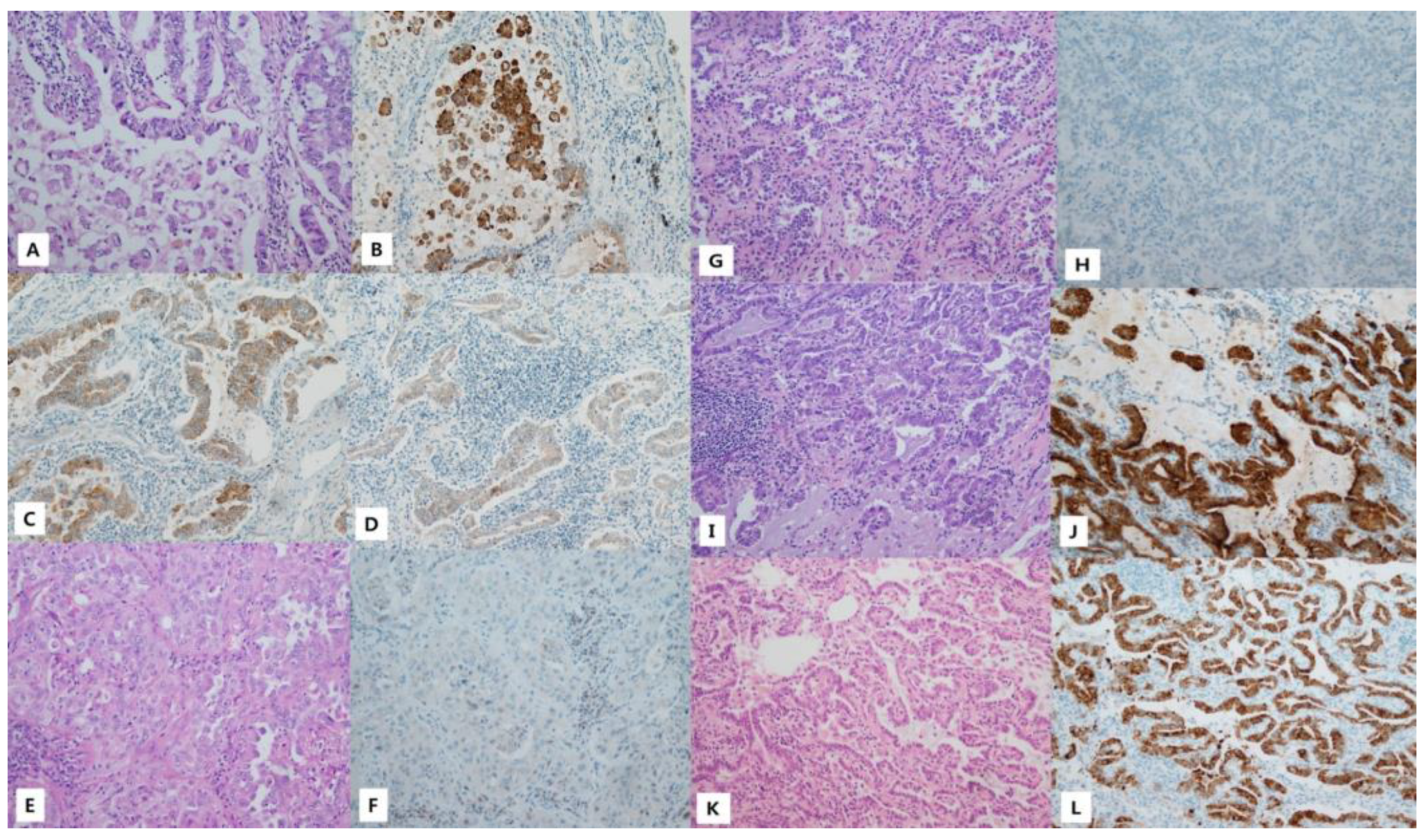
3.3. Immunohistochemistry for VE1
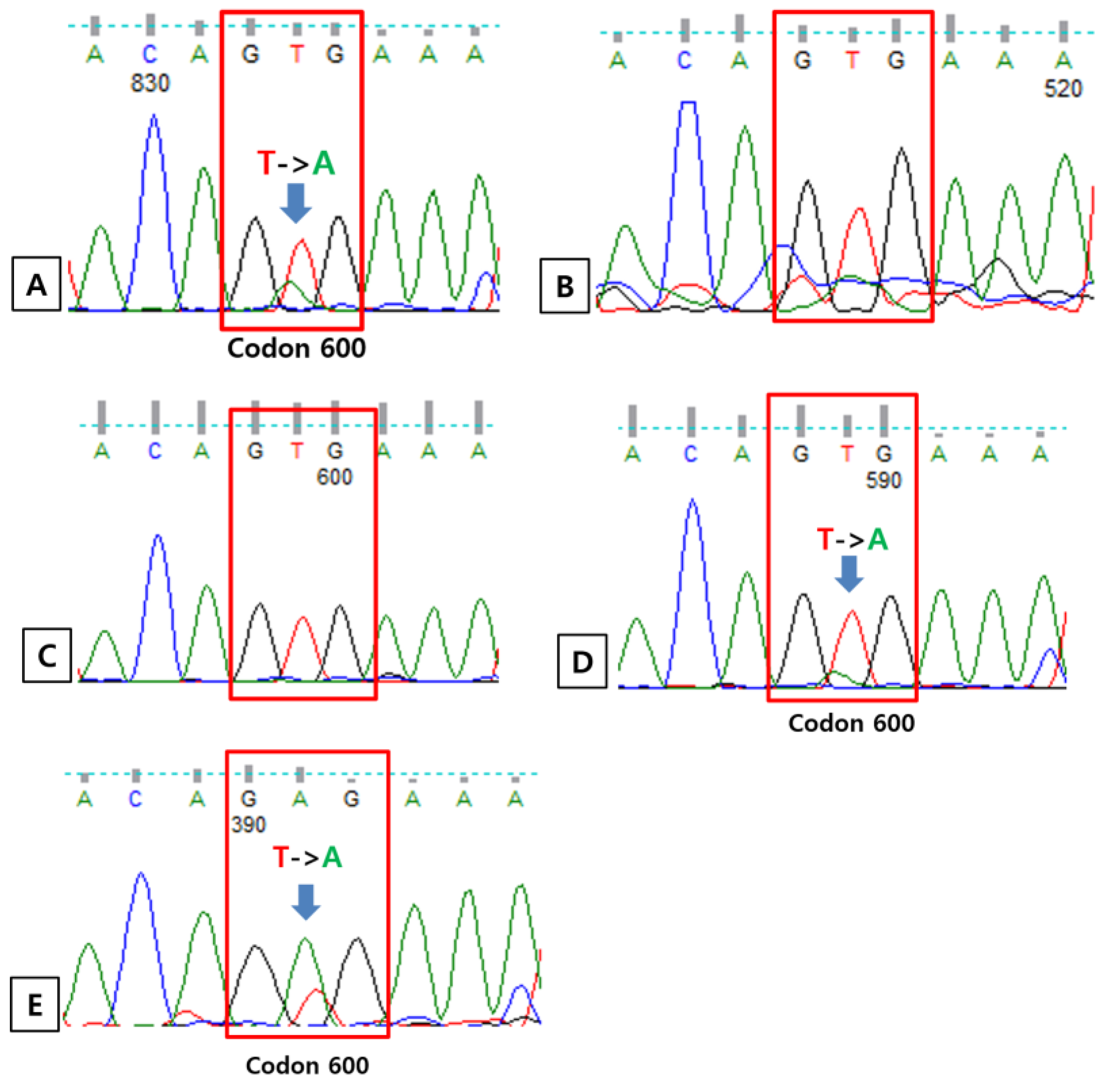
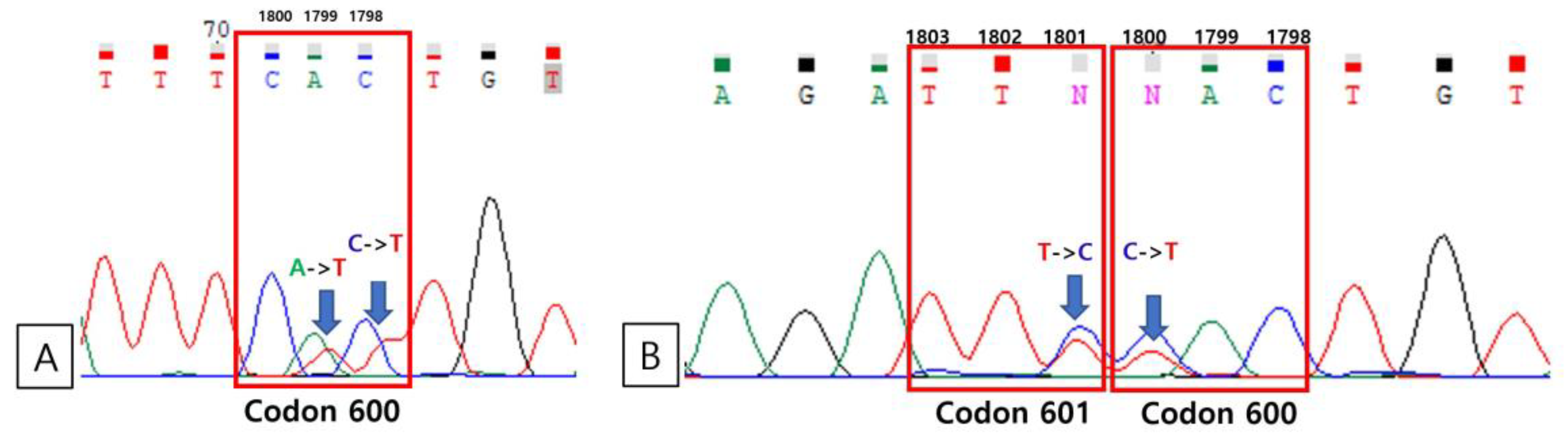
3.4. Direct Sequencing
3.5. Clinicopathologic Aspects of BRAF Mutation in Lung Cancers
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yan, N.; Guo, S.; Zhang, H.; Zhang, Z.; Shen, S.; Li, X. BRAF-Mutated Non-Small Cell Lung Cancer: Current Treatment Status and Future Perspective. Front. Oncol. 2022, 12, 863043. [Google Scholar] [CrossRef] [PubMed]
- Raman, M.; Chen, W.; Cobb, M.H. Differential regulation and properties of MAPKs. Oncogene 2007, 26, 3100–3112. [Google Scholar] [CrossRef] [PubMed]
- Forbes, S.A.; Bindal, N.; Bamford, S.; Cole, C.; Kok, C.Y.; Beare, D.; Jia, M.; Shepherd, R.; Leung, K.; Menzies, A.; et al. COSMIC: Mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011, 39, D945–D950. [Google Scholar] [CrossRef]
- Tabbò, F.; Pisano, C.; Mazieres, J.; Mezquita, L.; Nadal, E.; Planchard, D.; Pradines, A.; Santamaria, D.; Swalduz, A.; Ambrogio, C.; et al. How far we have come targeting BRAF-mutant non-small cell lung cancer (NSCLC). Cancer Treat. Rev. 2022, 103, 102335. [Google Scholar] [CrossRef] [PubMed]
- Leonetti, A.; Facchinetti, F.; Rossi, G.; Minari, R.; Conti, A.; Friboulet, L.; Tiseo, M.; Planchard, D. BRAF in non-small cell lung cancer (NSCLC): Pickaxing another brick in the wall. Cancer Treat. Rev. 2018, 66, 82–94. [Google Scholar] [CrossRef]
- Riudavets, M.; Cascetta, P.; Planchard, D. Targeting BRAF-mutant non-small cell lung cancer: Current status and future directions. Lung Cancer 2022, 169, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Owsley, J.; Stein, M.K.; Porter, J.; In, G.K.; Salem, M.; O’Day, S.; Elliott, A.; Poorman, K.; Gibney, G.; VanderWalde, A. Prevalence of class I-III BRAF mutations among 114,662 cancer patients in a large genomic database. Exp. Biol. Med. (Maywood) 2021, 246, 31–39. [Google Scholar] [CrossRef]
- Mazieres, J.; Cropet, C.; Montané, L.; Barlesi, F.; Souquet, P.J.; Quantin, X.; Dubos-Arvis, C.; Otto, J.; Favier, L.; Avrillon, V.; et al. Vemurafenib in non-small-cell lung cancer patients with BRAF. Ann. Oncol. 2020, 31, 289–294. [Google Scholar] [CrossRef]
- Ilie, M.; Long, E.; Hofman, V.; Dadone, B.; Marquette, C.H.; Mouroux, J.; Vignaud, J.M.; Begueret, H.; Merlio, J.P.; Capper, D.; et al. Diagnostic value of immunohistochemistry for the detection of the BRAFV600E mutation in primary lung adenocarcinoma Caucasian patients. Ann. Oncol. 2013, 24, 742–748. [Google Scholar] [CrossRef]
- Kang, S.H.; Pyo, J.Y.; Yang, S.W.; Hong, S.W. Detection of BRAF V600E mutation with thyroid tissue using pyrosequencing: Comparison with PNA-clamping and real-time PCR. Am. J. Clin. Pathol. 2013, 139, 759–764. [Google Scholar] [CrossRef]
- Luk, P.P.; Yu, B.; Ng, C.C.; Mercorella, B.; Selinger, C.; Lum, T.; Kao, S.; O’Toole, S.A.; Cooper, W.A. BRAF mutations in non-small cell lung cancer. Transl. Lung Cancer Res. 2015, 4, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Paik, P.K.; Arcila, M.E.; Fara, M.; Sima, C.S.; Miller, V.A.; Kris, M.G.; Ladanyi, M.; Riely, G.J. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J. Clin. Oncol. 2011, 29, 2046–2051. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, A.; Felicioni, L.; Malatesta, S.; Grazia Sciarrotta, M.; Guetti, L.; Chella, A.; Viola, P.; Pullara, C.; Mucilli, F.; Buttitta, F. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J. Clin. Oncol. 2011, 29, 3574–3579. [Google Scholar] [CrossRef] [PubMed]
- Tissot, C.; Couraud, S.; Tanguy, R.; Bringuier, P.P.; Girard, N.; Souquet, P.J. Clinical characteristics and outcome of patients with lung cancer harboring BRAF mutations. Lung Cancer 2016, 91, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Kinno, T.; Tsuta, K.; Shiraishi, K.; Mizukami, T.; Suzuki, M.; Yoshida, A.; Suzuki, K.; Asamura, H.; Furuta, K.; Kohno, T.; et al. Clinicopathological features of nonsmall cell lung carcinomas with BRAF mutations. Ann. Oncol. 2014, 25, 138–142. [Google Scholar] [CrossRef]
- Kim, H.C.; Kang, Y.R.; Ji, W.; Kim, Y.J.; Yoon, S.; Lee, J.C.; Choi, C.M. Frequency and clinical features of BRAF mutations among patients with stage III/IV lung adenocarcinoma without EGFR/ALK aberrations. Onco Targets Ther. 2019, 12, 6045–6052. [Google Scholar] [CrossRef]
- Brustugun, O.T.; Khattak, A.M.; Trømborg, A.K.; Beigi, M.; Beiske, K.; Lund-Iversen, M.; Helland, Å. BRAF-mutations in non-small cell lung cancer. Lung Cancer 2014, 84, 36–38. [Google Scholar] [CrossRef]
- Kalemkerian, G.P.; Narula, N.; Kennedy, E.B.; Biermann, W.A.; Donington, J.; Leighl, N.B.; Lew, M.; Pantelas, J.; Ramalingam, S.S.; Reck, M.; et al. Molecular Testing Guideline for the Selection of Patients With Lung Cancer for Treatment With Targeted Tyrosine Kinase Inhibitors: American Society of Clinical Oncology Endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Clinical Practice Guideline Update. J. Clin. Oncol. 2018, 36, 911–919. [Google Scholar] [CrossRef]
- Zengarini, C.; Mussi, M.; Veronesi, G.; Alessandrini, A.; Lambertini, M.; Dika, E. BRAF V600K vs. BRAF V600E: A comparison of clinical and dermoscopic characteristics and response to immunotherapies and targeted therapies. Clin. Exp. Dermatol. 2022, 47, 1131–1136. [Google Scholar] [CrossRef]
- Gow, C.H.; Hsieh, M.S.; Lin, Y.T.; Liu, Y.N.; Shih, J.Y. Validation of Immunohistochemistry for the Detection of. Cancers 2019, 11, 866. [Google Scholar] [CrossRef]
- Hwang, I.; Choi, Y.L.; Lee, H.; Hwang, S.; Lee, B.; Yang, H.; Chelakkot, C.; Han, J. Selection Strategies and Practical Application of BRAF V600E-Mutated Non-Small Cell Lung Carcinoma. Cancer Res. Treat. 2022, 54, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Capper, D.; Preusser, M.; Habel, A.; Sahm, F.; Ackermann, U.; Schindler, G.; Pusch, S.; Mechtersheimer, G.; Zentgraf, H.; von Deimling, A. Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol. 2011, 122, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Hofman, V.; Benzaquen, J.; Heeke, S.; Lassalle, S.; Poudenx, M.; Long, E.; Lantéri, E.; Bordone, O.; Lespinet, V.; Tanga, V.; et al. Real-world assessment of the BRAF status in non-squamous cell lung carcinoma using VE1 immunohistochemistry: A single laboratory experience (LPCE, Nice, France). Lung Cancer 2020, 145, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Choi, Y.L.; Shim, H.S.; Lee, G.K.; Ha, S.Y.; Group, K.C.P.S. Usefulness of BRAF VE1 immunohistochemistry in non-small cell lung cancers: A multi-institutional study by 15 pathologists in Korea. J. Pathol. Transl. Med. 2022, 56, 334–341. [Google Scholar] [CrossRef] [PubMed]
| Characteristics | Number (%) |
|---|---|
| Age (years) | 66.84 ± 8.76 |
| Sex | |
| Male | 238 (63.0) |
| Female | 140 (37.0) |
| Smoking status | |
| Never-smoker | 168 (44.4) |
| Current smoker | 98 (25.9) |
| Ex-smoker | 112 (29.6) |
| Pack-years among ever-smokers (years) * | 34.68 ± 19.81 |
| Tumor size (cm) | 3.37 ± 1.55 |
| Histologic type | |
| ADC | 255 (67.5) |
| SqCC | 91 (24.1) |
| SC | 15 (4.0) |
| LCNEC | 9 (2.4) |
| ADSqCC | 5 (1.3) |
| Other | 3 (0.8) |
| Differentiation | |
| WD | 19 (5.0) |
| MD | 269 (71.2) |
| PD | 90 (23.8) |
| Stage | |
| Early (I–II) | 305 (80.7) |
| Advanced (III–IV) | 73 (19.3) |
| EGFR mutation | |
| Absent | 258 (68.3) |
| Present | 120 (31.7) |
| ALK translocation | |
| Absent | 367 (97.1) |
| Present | 11 (2.9) |
| Ethnicity Korean | 378 (100.0) |
| Case Number | BRAF V600 PNA Clamping | BRAF V600E Real-Time PCR | IHC for VE1 | Direct Sanger Sequencing | Direct Sanger Sequencing of the Clamping PCR Product |
|---|---|---|---|---|---|
| 142 | + | + | + | V600E | Not done |
| 270 | + | – | – | WT | V600K |
| 324 | + | – | – | WT | V600V, K601E |
| 348 | + | + | + | V600E | Not done |
| 358 | + | + | + | V600E | Not done |
| Case Number | DNA Loading (ng) | Cycle Threshold (CT) | ΔCT-2 | ΔCT-1 | |
|---|---|---|---|---|---|
| Non-PNA | V600 | V600 | V600 | ||
| Clamping Control | 24.38 | 36.33 | 11.95 | −1.33 | |
| Positive Control | 24.03 | 30 | 5.97 | 5 | |
| 142 | 10 | 28.59 | 31.7 | 3.11 | 3.3 |
| 270 | 10 | 26.91 | 32.44 | 5.53 | 2.56 |
| 324 | 10 | 25.98 | 31.82 | 5.84 | 3.18 |
| 348 | 10 | 26.02 | 30.3 | 4.28 | 4.7 |
| 358 | 10 | 26.21 | 31.46 | 5.26 | 3.54 |
| 142 | 25 | 27.15 | 31.13 | 3.97 | 3.87 |
| 270 | 25 | 26.31 | 32.42 | 6.1 | 2.58 |
| 324 | 25 | 25.15 | 31.89 | 6.74 | 3.11 |
| 348 | 25 | 25.22 | 29.6 | 4.38 | 5.4 |
| 358 | 25 | 25.26 | 31.02 | 5.76 | 3.98 |
| Patient Number | Internal Control CT | Sample CT | Result |
|---|---|---|---|
| 142 | 25.9 | 31.9 | + |
| 270 | 24.6 | NA | − |
| 324 | 26.6 | NA | − |
| 348 | 24.7 | 29.6 | + |
| 358 | 24.4 | 30.6 | + |
| Case Number | Proportion of Cytoplasm Positive Rate | Intensity Pattern | Intensity Scores of Tumor Cells | Result |
|---|---|---|---|---|
| 142 | 100 | Heterogeneous | 3+: 40% 2+: 50% 1+: 10% 0: 0% | + |
| 270 | 0 | Homogeneous | 3+: 0% 2+: 0% 1+: 0% 0: 100% | − |
| 324 | 0 | Homogeneous | 3+: 0% 2+: 0% 1+: 0% 0: 100% | − |
| 348 | 100 | Homogeneous | 3+: 100% 2+: 0% 1+: 0% 0: 0% | + |
| 358 | 100 | Heterogeneous | 3+:70% 2+:30% 1+: 0% 0: 0% | + |
| BRAF Mutation | BRAF V600E Mutation | BRAF Non-V600E Mutation | |
|---|---|---|---|
| N (%) | N (%) | N (%) | |
| NSCLC (n = 378) | 5 (1.3) | 3 (0.8) | 2 (0.5) |
| Adenocarcinomas (n = 255) | 5 (2.0) | 3 (1.2) | 2 (0.8) |
| NSCLC lacking EGFR mutation and ALK translocation (n = 246) | 5 (2.0) | 3 (1.2) | 2 (0.8) |
| Adenocarcinomas lacking EGFR mutation and ALK translocation (n = 129) | 5 (3.8) | 3 (2.3) | 2 (1.5) |
| Case No. | BRAF Mutation | Age (Years) | Sex | Smoking Status | Pack Years | Predominant Histological Subtype | Present of Micropapillary Component | pT Size (cm) | pN Stage | pM Stage | Stage |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 142 | V600E | 72 | Male | Current | 50 | Solid | + | 6.5 | 2 | 0 | IIIA |
| 270 | V600K | 83 | Male | Current | 15 | Acinar | - | 3.6 | 0 | 0 | IB |
| 324 | V600V, K601E | 67 | Female | Never | 0 | Acinar | - | 1.6 | 0 | 0 | IA |
| 348 | V600E | 51 | Female | Never | 0 | Acinar | + | 2.2 | 0 | 0 | IA |
| 358 | V600E | 52 | Male | Ex-smoker | 7.5 | Acinar | + | 1.8 | 0 | 0 | IA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahn, H.Y.; Lee, C.H.; Lee, M.K.; Eom, J.S.; Jeong, Y.J.; Kim, Y.D.; Cho, J.S.; Lee, J.; Lee, S.J.; Shin, D.H.; et al. BRAF V600E Mutation of Non-Small Cell Lung Cancer in Korean Patients. Medicina 2023, 59, 1085. https://doi.org/10.3390/medicina59061085
Ahn HY, Lee CH, Lee MK, Eom JS, Jeong YJ, Kim YD, Cho JS, Lee J, Lee SJ, Shin DH, et al. BRAF V600E Mutation of Non-Small Cell Lung Cancer in Korean Patients. Medicina. 2023; 59(6):1085. https://doi.org/10.3390/medicina59061085
Chicago/Turabian StyleAhn, Hyo Yeong, Chang Hun Lee, Min Ki Lee, Jung Seop Eom, Yeon Joo Jeong, Yeong Dae Kim, Jeong Su Cho, Jonggeun Lee, So Jeong Lee, Dong Hoon Shin, and et al. 2023. "BRAF V600E Mutation of Non-Small Cell Lung Cancer in Korean Patients" Medicina 59, no. 6: 1085. https://doi.org/10.3390/medicina59061085
APA StyleAhn, H. Y., Lee, C. H., Lee, M. K., Eom, J. S., Jeong, Y. J., Kim, Y. D., Cho, J. S., Lee, J., Lee, S. J., Shin, D. H., & Kim, A. (2023). BRAF V600E Mutation of Non-Small Cell Lung Cancer in Korean Patients. Medicina, 59(6), 1085. https://doi.org/10.3390/medicina59061085






