Evaluation of the Copy Number Variants and Single-Nucleotide Polymorphisms of ABCA3 in Newborns with Respiratory Distress Syndrome—A Pilot Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Molecular Analysis
2.3. Data Analysis
2.3.1. Descriptive Analysis
2.3.2. Inferential Analysis
3. Results
3.1. Clinical Characteristics of the Patients
3.2. CNV Analysis in RDS Patients
3.3. Genotype and Allele Frequency Analysis of ABCA3 SNPs and Their Effects on RDS Susceptibility
3.4. Haplotype Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alfoldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, R.A.; Belmont, J.W.; Hardenbol, P.; Willis, T.D.; Yu, F.L.; Yang, H.M.; Ch’ang, L.Y.; Huang, W.; Liu, B.; Shen, Y.; et al. The International HapMap Project. Nature 2003, 426, 789–796. [Google Scholar] [CrossRef]
- Shen, C.L.; Zhang, Q.; Meyer Hudson, J.; Cole, F.S.; Wambach, J.A. Genetic Factors Contribute to Risk for Neonatal Respiratory Distress Syndrome among Moderately Preterm, Late Preterm, and Term Infants. J. Pediatr. 2016, 172, 69–74.e2. [Google Scholar] [CrossRef] [PubMed]
- Feuk, L.; Carson, A.R.; Scherer, S.W. Structural variation in the human genome. Nat. Rev. Genet. 2006, 7, 85–97. [Google Scholar] [CrossRef]
- Genomes Project, C.; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Henkel, J.; Saif, R.; Jagannathan, V.; Schmocker, C.; Zeindler, F.; Bangerter, E.; Herren, U.; Posantzis, D.; Bulut, Z.; Ammann, P.; et al. Selection signatures in goats reveal copy number variants underlying breed-defining coat color phenotypes. PLoS Genet. 2019, 15, e1008536. [Google Scholar] [CrossRef] [PubMed]
- Elder, P.J.D.; Ramsden, D.B.; Burnett, D.; Weickert, M.O.; Barber, T.M. Human amylase gene copy number variation as a determinant of metabolic state. Expert Rev. Endocrinol. Metab. 2018, 13, 193–205. [Google Scholar] [CrossRef]
- Gonzalez, E.; Kulkarni, H.; Bolivar, H.; Mangano, A.; Sanchez, R.; Catano, G.; Nibbs, R.J.; Freedman, B.I.; Quinones, M.P.; Bamshad, M.J.; et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 2005, 307, 1434–1440. [Google Scholar] [CrossRef]
- Poole, A.C.; Goodrich, J.K.; Youngblut, N.D.; Luque, G.G.; Ruaud, A.; Sutter, J.L.; Waters, J.L.; Shi, Q.; El-Hadidi, M.; Johnson, L.M.; et al. Human Salivary Amylase Gene Copy Number Impacts Oral and Gut Microbiomes. Cell Host Microbe 2019, 25, 553–564.e7. [Google Scholar] [CrossRef]
- Auwerx, C.; Jõeloo, M.; Sadler, M.C.; Tesio, N.; Ojavee, S.; Clark, C.J.; Mägi, R.; Estonian Biobank Research Team; Reymond, A.; Kutalik, Z. Rare copy-number variants as modulators of common disease susceptibility. Genome Med. 2024, 16, 5. [Google Scholar] [CrossRef]
- Redon, R.; Ishikawa, S.; Fitch, K.R.; Feuk, L.; Perry, G.H.; Andrews, T.D.; Fiegler, H.; Shapero, M.H.; Carson, A.R.; Chen, W.; et al. Global variation in copy number in the human genome. Nature 2006, 444, 444–454. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Ventura, M.; She, X.; Khaitovich, P.; Graves, T.; Osoegawa, K.; Church, D.; DeJong, P.; Wilson, R.K.; Paabo, S.; et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature 2005, 437, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Werdyani, S.; Carey, M.; Parfrey, P.; Yilmaz, Y.E.; Savas, S. A comprehensive analysis of SNPs and CNVs identifies novel markers associated with disease outcomes in colorectal cancer. Mol. Oncol. 2021, 15, 3329–3347. [Google Scholar] [CrossRef] [PubMed]
- Escaramis, G.; Docampo, E.; Rabionet, R. A decade of structural variants: Description, history and methods to detect structural variation. Brief Funct. Genom. 2015, 14, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Wambach, J.A.; Wegner, D.J.; Depass, K.; Heins, H.; Druley, T.E.; Mitra, R.D.; An, P.; Zhang, Q.; Nogee, L.M.; Cole, F.S.; et al. Single ABCA3 mutations increase risk for neonatal respiratory distress syndrome. Pediatrics 2012, 130, e1575–e1582. [Google Scholar] [CrossRef]
- Jouza, M.; Jimramovsky, T.; Sloukova, E.; Pecl, J.; Seehofnerova, A.; Jezova, M.; Urik, M.; Kunovsky, L.; Slaba, K.; Stourac, P.; et al. A Newly Observed Mutation of the ABCA3 Gene Causing Lethal Respiratory Failure of a Full-Term Newborn: A Case Report. Front. Genet. 2020, 11, 568303. [Google Scholar] [CrossRef] [PubMed]
- Anciuc-Crauciuc, M.; Cucerea, M.C.; Tripon, F.; Crauciuc, G.-A.; Bănescu, C.V. Descriptive and Functional Genomics in Neonatal Respiratory Distress Syndrome: From Lung Development to Targeted Therapies. Int. J. Mol. Sci. 2024, 25, 649. [Google Scholar] [CrossRef]
- Amatya, S.; Ye, M.; Yang, L.; Gandhi, C.K.; Wu, R.; Nagourney, B.; Floros, J. Single Nucleotide Polymorphisms Interactions of the Surfactant Protein Genes Associated With Respiratory Distress Syndrome Susceptibility in Preterm Infants. Front. Pediatr. 2021, 9, 682160. [Google Scholar] [CrossRef]
- Kala, P.; Have, T.; Nielsen, H.; Dunn, M.; Floros, J. Association of Pulmonary Surfactant Protein A (SP-A) Gene and Respiratory Distress Syndrome: Interaction with SP-B. Pediatr. Res. 1998, 43, 169–177. [Google Scholar] [CrossRef]
- Lahti, M.; Marttila, R.; Hallman, M. Surfactant protein C gene variation in the Finnish population—Association with perinatal respiratory disease. Eur. J. Hum. Genet. 2004, 12, 312–320. [Google Scholar] [CrossRef]
- Sorensen, G.L.; Dahl, M.; Tan, Q.; Bendixen, C.; Holmskov, U.; Husby, S. Surfactant protein-D-encoding gene variant polymorphisms are linked to respiratory outcome in premature infants. J. Pediatr. 2014, 165, 683–689. [Google Scholar] [CrossRef] [PubMed]
- Hilgendorff, A.; Heidinger, K.; Bohnert, A.; Kleinsteiber, A.; Konig, I.R.; Ziegler, A.; Lindner, U.; Frey, G.; Merz, C.; Lettgen, B.; et al. Association of polymorphisms in the human surfactant protein-D (SFTPD) gene and postnatal pulmonary adaptation in the preterm infant. Acta Paediatr. 2009, 98, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Wert, S.E.; Whitsett, J.A.; Nogee, L.M. Genetic disorders of surfactant dysfunction. Pediatr. Dev. Pathol. 2009, 12, 253–274. [Google Scholar] [CrossRef] [PubMed]
- Nogee, L.M. Genetic causes of surfactant protein abnormalities. Curr. Opin. Pediatr. 2019, 31, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Wambach, J.A.; Casey, A.M.; Fishman, M.P.; Wegner, D.J.; Wert, S.E.; Cole, F.S.; Hamvas, A.; Nogee, L.M. Genotype-phenotype correlations for infants and children with ABCA3 deficiency. Am. J. Respir. Crit. Care Med. 2014, 189, 1538–1543. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wu, Y.D.; Xu, X.F.; Du, L.Z. Polymorphism analysis of the ABCA3 gene: Association with neonatal respiratory distress syndrome in preterm infants. Chin. Med. J. 2012, 125, 1594–1598. [Google Scholar] [PubMed]
- Karjalainen, M.K.; Haataja, R.; Hallman, M. Haplotype analysis of ABCA3: Association with respiratory distress in very premature infants. Ann. Med. 2008, 40, 56–65. [Google Scholar] [CrossRef]
- Wambach, J.A.; Wegner, D.J.; Heins, H.B.; Druley, T.E.; Mitra, R.D.; Hamvas, A.; Cole, F.S. Synonymous ABCA3 variants do not increase risk for neonatal respiratory distress syndrome. J. Pediatr. 2014, 164, 1316–1321.e3. [Google Scholar] [CrossRef]
- Chen, Y.J.; Wambach, J.A.; DePass, K.; Wegner, D.J.; Chen, S.K.; Zhang, Q.Y.; Heins, H.; Cole, F.S.; Hamvas, A. Population-based frequency of surfactant dysfunction mutations in a native Chinese cohort. World J. Pediatr. 2016, 12, 190–195. [Google Scholar] [CrossRef]
- Silverman, W.A.; Andersen, D.H. A controlled clinical trial of effects of water mist on obstructive respiratory signs, death rate and necropsy findings among premature infants. Pediatrics 1956, 17, 1–10. [Google Scholar]
- Cucerea, M.; Moscalu, M.; Moldovan, E.; Santa, R.; Gall, Z.; Suciu, L.M.; Simon, M. Early Surfactant Therapy for Respiratory Distress Syndrome in Very Preterm Infants. Healthcare 2023, 11, 439. [Google Scholar] [CrossRef] [PubMed]
- National Center of Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/snp (accessed on 27 December 2023).
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Sweet, D.G.; Carnielli, V.P.; Greisen, G.; Hallman, M.; Klebermass-Schrehof, K.; Ozek, E.; Te Pas, A.; Plavka, R.; Roehr, C.C.; Saugstad, O.D.; et al. European Consensus Guidelines on the Management of Respiratory Distress Syndrome: 2022 Update. Neonatology 2023, 120, 3–23. [Google Scholar] [CrossRef] [PubMed]
- Linkage Disequilibrium Calculator. Available online: https://www.ensembl.org/Homo_sapiens/Tools/LD (accessed on 27 December 2023).
- Whitsett, J.A.; Wert, S.E.; Weaver, T.E. Alveolar surfactant homeostasis and the pathogenesis of pulmonary disease. Annu. Rev. Med. 2010, 61, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Nkadi, P.O.; Merritt, T.A.; Pillers, D.A. An overview of pulmonary surfactant in the neonate: Genetics, metabolism, and the role of surfactant in health and disease. Mol. Genet. Metab. 2009, 97, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Jobe, A.H. Lung Development and maturation. In Neonatal-Perinatal Medicine; Martin, R.J., Fanaroff, A.A., Walsh, M.C., Eds.; Elsevier Mosby: St. Louis, MO, USA, 2010; Volume 2, p. 1075. [Google Scholar]
- Naderi, H.M.; Murray, J.C.; Dagle, J.M. Single mutations in ABCA3 increase the risk for neonatal respiratory distress syndrome in late preterm infants (gestational age 34-36 weeks). Am. J. Med. Genet. A 2014, 164A, 2676–2678. [Google Scholar] [CrossRef] [PubMed]
- Benton, M.L.; Abraham, A.; LaBella, A.L.; Abbot, P.; Rokas, A.; Capra, J.A. The influence of evolutionary history on human health and disease. Nat. Rev. Genet. 2021, 22, 269–283. [Google Scholar] [CrossRef] [PubMed]
- McCombie, W.R.; McPherson, J.D.; Mardis, E.R. Next-Generation Sequencing Technologies. Cold Spring Harb. Perspect. Med. 2019, 9, a036798. [Google Scholar] [CrossRef] [PubMed]
- Stenson, P.D.; Mort, M.; Ball, E.V.; Chapman, M.; Evans, K.; Azevedo, L.; Hayden, M.; Hey-wood, S.; Millar, D.S.; Phillips, A.D.; et al. The Human Gene Mutation Database (HGMD®): Optimizing its use in a clinical diagnostic or research setting. Hum. Genet. 2020, 139, 1197–1207. [Google Scholar] [CrossRef]
- The Human Gene Mutation Database (HGMD®). Available online: https://www.hgmd.cf.ac.uk (accessed on 3 February 2024).
- Alhourani, E.; Rincic, M.; Othman, M.A.; Pohle, B.; Schlie, C.; Glaser, A.; Liehr, T. Comprehensive chronic lymphocytic leukemia diagnostics by combined multiplex ligation dependent probe amplification (MLPA) and interphase fluorescence in situ hybridization (iFISH). Mol. Cytogenet. 2014, 7, 79. [Google Scholar] [CrossRef]
- Fuka, G.; Farias-Vieira, T.M.; Hummel, L.; Blunck, C.B.; Santoro, J.C.; Terra-Granado, E.; Barbosa, T.C.; Emerenciano, M.; Pombo-de-Oliveira, M.S. Evaluation of multiplex ligation dependent probe amplification (MLPA) for identification of acute lymphoblastic leukemia with an intrachromosomal amplification of chromosome 21 (iAMP21) in a Brazilian population. Mol. Cytogenet. 2015, 8, 35. [Google Scholar] [CrossRef] [PubMed]
- Balla, B.; Tripon, F.; Candea, M.; Banescu, C. Copy Number Variations and Gene Mutations Identified by Multiplex Ligation-Dependent Probe Amplification in Romanian Chronic Lymphocytic Leukemia Patients. J. Pers. Med. 2023, 13, 1239. [Google Scholar] [CrossRef] [PubMed]
- Kerkhof, J.; Schenkel, L.C.; Reilly, J.; McRobbie, S.; Aref-Eshghi, E.; Stuart, A.; Rupar, C.A.; Adams, P.; Hegele, R.A.; Lin, H.; et al. Clinical Validation of Copy Number Variant Detection from Targeted Next-Generation Sequencing Panels. J. Mol. Diagn. 2017, 19, 905–920. [Google Scholar] [CrossRef] [PubMed]
- Talevich, E.; Shain, A.H.; Botton, T.; Bastian, B.C. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput. Biol. 2016, 12, e1004873. [Google Scholar] [CrossRef]
- Kolb, S.J.; Kissel, J.T. Spinal Muscular Atrophy. Neurol. Clin. 2015, 33, 831–846. [Google Scholar] [CrossRef] [PubMed]
- Perne, A.; Zhang, X.; Lehmann, L.; Groth, M.; Stuber, F.; Book, M. Comparison of multiplex ligation-dependent probe amplification and real-time PCR accuracy for gene copy number quantification using the beta-defensin locus. Biotechniques 2009, 47, 1023–1028. [Google Scholar] [CrossRef] [PubMed]
- Dorgaleleh, S.; Naghipoor, K.; Ahmad Barahouie, A.; Oladnabi, M. Detection of the Duplication in Exons 56-63 of Duchenne Muscular Dystrophy Patients with MLPA. Int. J. Pediatr. 2020, 8, 11617–11623. [Google Scholar] [CrossRef]
- Massalska, D.; Ozdarska, K.; Bijok, J.; Roszkowski, T.; Kucinska-Chahwan, A.; Jakiel, G.; Panek, G.M.; Zimowski, J.G. Usefulness of methylation-specific multiplex ligation-dependent probe amplification for identification of parental origin of triploidy. J. Hum. Genet. 2020, 65, 889–894. [Google Scholar] [CrossRef]
- Fu, X.; Shi, Y.; Ma, J.; Zhang, K.; Wang, G.; Li, G.; Xiao, L.; Wang, H. Advances of multiplex ligation-dependent probe amplification technology in molecular diagnostics. Biotechniques 2022, 73, 205–213. [Google Scholar] [CrossRef]
- Eid, O.M.; Eid, M.M.; Farid, M.; Abdel Kader, R.M.A.; Mahrous, R.; El-Dessouky, S.H. MLPA as a genetic assay for the prenatal diagnosis of common aneuploidy: The first Egyptian experience. J. Genet. Eng. Biotechnol. 2022, 20, 112. [Google Scholar] [CrossRef]
- Schrijver, I.; Rappahahn, K.; Pique, L.; Kharrazi, M.; Wong, L.J. Multiplex ligation-dependent probe amplification identification of whole exon and single nucleotide deletions in the CFTR gene of Hispanic individuals with cystic fibrosis. J. Mol. Diagn. 2008, 10, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Stuppia, L.; Antonucci, I.; Palka, G.; Gatta, V. Use of the MLPA assay in the molecular diagnosis of gene copy number alterations in human genetic diseases. Int. J. Mol. Sci. 2012, 13, 3245–3276. [Google Scholar] [CrossRef] [PubMed]
- Flamein, F.; Riffault, L.; Muselet-Charlier, C.; Pernelle, J.; Feldmann, D.; Jonard, L.; Durand-Schneider, A.M.; Coulomb, A.; Maurice, M.; Nogee, L.M.; et al. Molecular and cellular characteristics of ABCA3 mutations associated with diffuse parenchymal lung diseases in children. Hum. Mol. Genet. 2012, 21, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Garmany, T.H.; Wambach, J.A.; Heins, H.B.; Watkins-Torry, J.M.; Wegner, D.J.; Bennet, K.; An, P.; Land, G.; Saugstad, O.D.; Henderson, H.; et al. Population and disease-based prevalence of the common mutations associated with surfactant deficiency. Pediatr. Res. 2008, 63, 645–649. [Google Scholar] [CrossRef]
- Bozkurt, H.B.; Sahin, Y. Whole exome sequencing identifies a novel variant in ABCA3 in an individual with fatal congenital surfactant protein deficiency. Turk. J. Pediatr. 2021, 63, 703–707. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Fan, J.; Zhang, Y.; Huang, L.; Shi, Y. ABCA3 gene mutations shape the clinical profiles of severe unexplained respiratory distress syndrome in late preterm and term infants. Transl. Pediatr. 2021, 10, 350–358. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, Z.; Lin, Y.; Wang, R.; Xu, J.; He, Y.; Zhang, F.; Wu, L.; Chen, D. A novel synonymous ABCA3 variant identified in a Chinese family with lethal neonatal respiratory failure. BMC Med. Genom. 2021, 14, 256. [Google Scholar] [CrossRef]
- Mei, H.; Zhang, Y.; Zhang, Y.; Song, D.; Huo, M.; Du, Q.; Wang, X. A study on the correlation of ABCA3 gene mutation and neonatal respiratory distress syndrome. Chin. J. Neonatol. 2018, 6, 415–418. [Google Scholar]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009, 4, 1073–1081. [Google Scholar] [CrossRef]
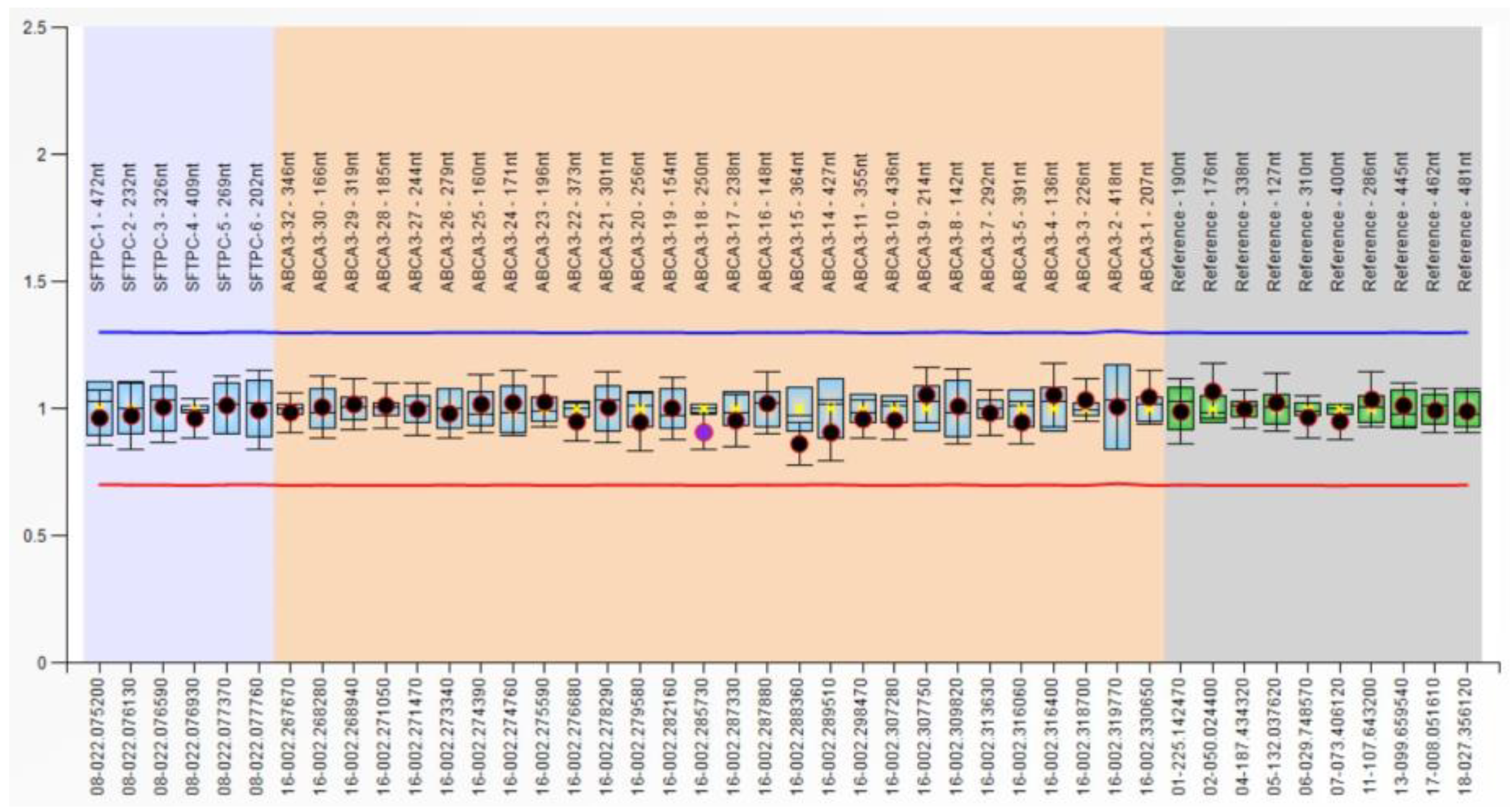
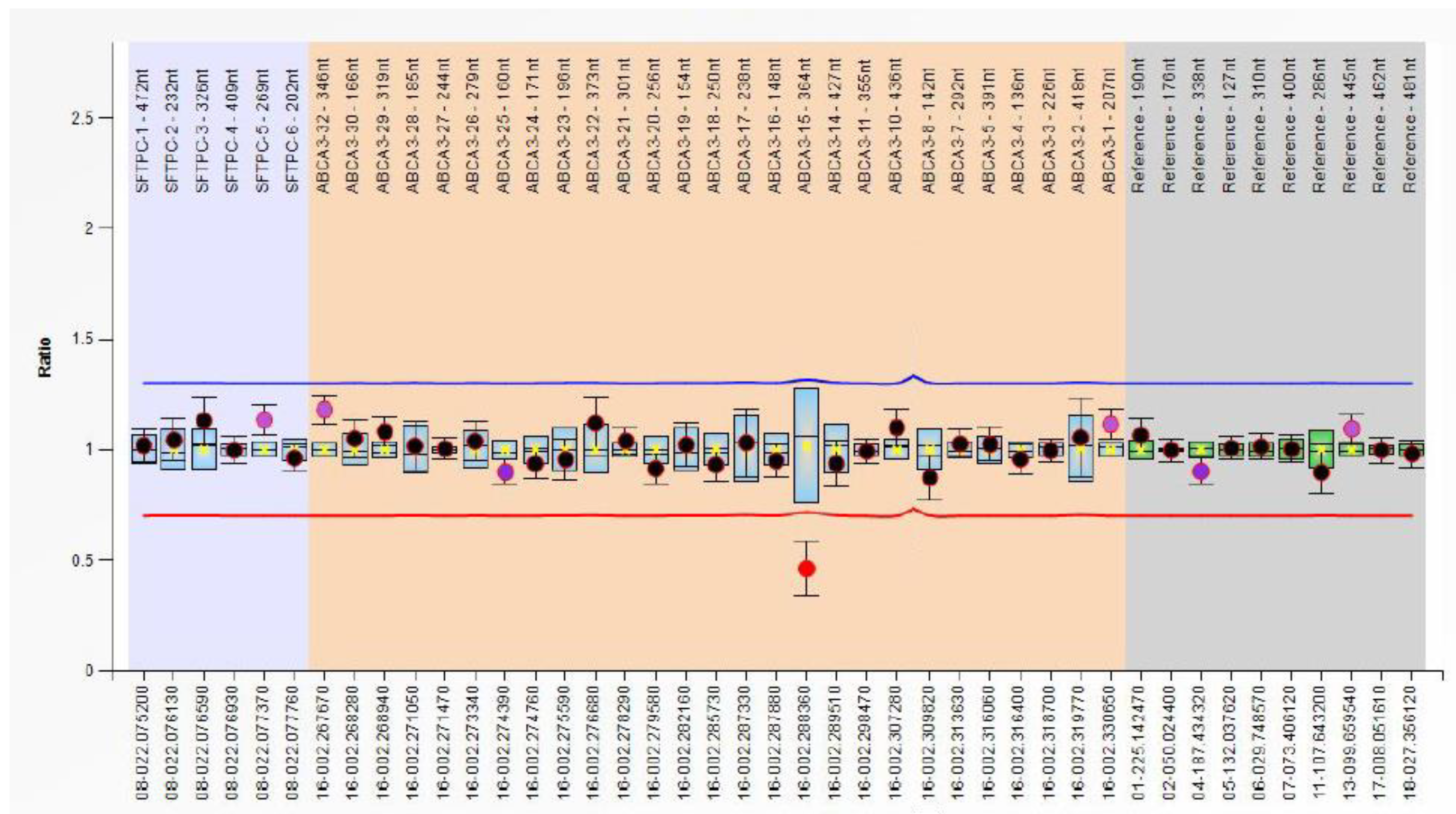
| Variables | RDS Patients | Controls (without RDS) | p-Values |
|---|---|---|---|
| Gestational age (weeks): mean ± SD | 31 ± 3.72 | 32 ± 2.80 | <0.015 * |
| Gender: n (%) | |||
| Female | 26 (34.2) | 80 (37.4) | 0.678 ‡ |
| Male | 50 (65.5) | 134 (62.6) | |
| Birth weight (g): mean ± SD | 1473.10 ± 504.51 | 1620.20 ± 310.5 | <0.003 * |
| Preterm labor, n (%) | 37 (48.6) | 122 (57) | 0.228 ‡ |
| Singleton pregnancy, n (%) | 70 (92.1) | 202 (94.4) | 0.579 ‡ |
| Antenatal care, n (%) | 39 (51.3) | 139 (64.9) | 0.040 ‡ |
| Mother’s age, mean ± SD | 28.7 ± 3.65 | 29.5 ± 3.35 | 0.081 * |
| Antenatal steroids, n (%) | 37 (48.6) | 12 (5.6) | <0.001 ‡ |
| PROM > 18 h, n (%) | 23 (30.2) | 32 (14.9) | <0.001 ‡ |
| Chorioamnionitis, n (%) | 10 (13.1) | 16 (7.5) | 0.160 ‡ |
| Delivery mode, n (%) | |||
| Spontaneous | 26 (34.2) | 152 (71) | <0.001 ‡ |
| C-section | 50 (65.8) | 54 (11.6) | <0.001 ‡ |
| Delivery in a tertiary center, n (%) | 60 (78.9) | 209 (94.8) | <0.001 ‡ |
| Apgar Score, mean | |||
| 1 min | 6 | 7 | <0.001 * |
| 5 min | 7 | 8 | 0.079 * |
| Surfactant use, n (%) | 66 (86.8) | - | - |
| LISA | 19 (28.7) | - | - |
| INSURE | 19 (28.7) | - | - |
| Standard | 28 (42.4) | - | - |
| Need for subsequent surfactant doses, n (%) | 7 (9.2) | - | - |
| Duration of non-invasive ventilation, hours (mean) | 222.48 ± 180.2 | 25 ± 18.9 | <0.001 * |
| Duration of MV, hours (mean) | 210.3 ± 50.2 | - | - |
| Chronic lung disease, n (%) | 15 (19.7) | - | - |
| PDA: n (%) | 10 (13.1) | 4 (1.86) | <0.001 § |
| Pulmonary hemorrhage, n (%) | 4 (5.2) | - | - |
| NICU days, mean ± SD | 27.4 ± 4.9 | 7 ± 4.9 | <0.001 * |
| Deaths, n (%) | 7 (9.2) | - | - |
| ABCA3 SNPs | Gene Models | Genotypes | Controls n (%) | RDS Cases n (%) | OR Crude (95%CI) * | p-Value |
|---|---|---|---|---|---|---|
| rs170447 (c.1741+33T>C) | Codominant | TT | 44 (20.5) | 15 (19.7) | Reference | - |
| TC | 139 (65) | 49 (64.5) | 1.03 (0.528–2.02) | 1.00 | ||
| CC | 31 (20.57) | 12 (15.8) | 1.13 (0.46–2.75) | 0.82 | ||
| Dominant | TT | 44 (19.75) | 15 (19.7) | Reference | - | |
| TC + CC | 170 (79.43) | 61 (80.3) | 1.05 (0.54–2.02) | 1.00 | ||
| rs323043 (c.1755G>C) | Codominant | GG | 161 (75.2) | 51 (67.1) | Reference | - |
| GC | 40 (18.7) | 21 (26.9) | 1.657 (0.89–3.06) | 0.137 | ||
| CC | 13 (6.1) | 4 (6) | 0.97 (0.30–3.11) | 1.00 | ||
| Dominant | GG | 161 (75.2) | 51 (67.1) | Reference | - | |
| GC + CC | 53 (24.8) | 25 (32.9) | 1.48 (0.84–2.63) | 0.17 | ||
| rs13332514 (c.1059G>A) | Codominant | GG | 195 (91.1) | 67 (85.9) | Reference | - |
| GA | 14 (6.54) | 9 (14.5) | 1.87 (0.77–4.52) | 0.216 | ||
| AA | 5 (2.36) | - | 0.26 (0.01–4.82) | 0.33 | ||
| Dominant | GG | 195 (91.1) | 67 (88.1) | Reference | - | |
| GA + AA | 19 (8.9) | 9 (11.9) | 1.37 (0.59–3.19) | 0.49 |
| ABCA3 SNPs | Variant Alleles | Frequency of Variant Allele in Control Group (%) | Frequency of Variant Allele in RDS Group (%) | OR Crude (95% CI) * | p-Value |
|---|---|---|---|---|---|
| rs170447 (c.1741+33T>C) | C | 46.9 | 48 | 1.04 (0.72–1.51) | 0.85 |
| rs323043 (c.1755G>C) | C | 28.9 | 23.9 | 1.293 (0.79–2.09) | 0.308 |
| rs13332514 (c.1059G>A) | A | 5.6 | 7 | 1.05 (0.48–2.33) | 0.84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anciuc-Crauciuc, M.; Cucerea, M.C.; Crauciuc, G.-A.; Tripon, F.; Bănescu, C.V. Evaluation of the Copy Number Variants and Single-Nucleotide Polymorphisms of ABCA3 in Newborns with Respiratory Distress Syndrome—A Pilot Study. Medicina 2024, 60, 419. https://doi.org/10.3390/medicina60030419
Anciuc-Crauciuc M, Cucerea MC, Crauciuc G-A, Tripon F, Bănescu CV. Evaluation of the Copy Number Variants and Single-Nucleotide Polymorphisms of ABCA3 in Newborns with Respiratory Distress Syndrome—A Pilot Study. Medicina. 2024; 60(3):419. https://doi.org/10.3390/medicina60030419
Chicago/Turabian StyleAnciuc-Crauciuc, Mădălina, Manuela Camelia Cucerea, George-Andrei Crauciuc, Florin Tripon, and Claudia Violeta Bănescu. 2024. "Evaluation of the Copy Number Variants and Single-Nucleotide Polymorphisms of ABCA3 in Newborns with Respiratory Distress Syndrome—A Pilot Study" Medicina 60, no. 3: 419. https://doi.org/10.3390/medicina60030419





