Olmutinib Reverses Thioacetamide-Induced Cell Cycle Gene Alterations in Mice Liver and Kidney Tissues, While Wheat Germ Treatment Exhibits Limited Efficacy at Gene Level
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Model and Experimental Design
2.2. RNA Extraction
2.3. cDNA Synthesis
2.4. Gene Expression Analysis by Real-Time PCR
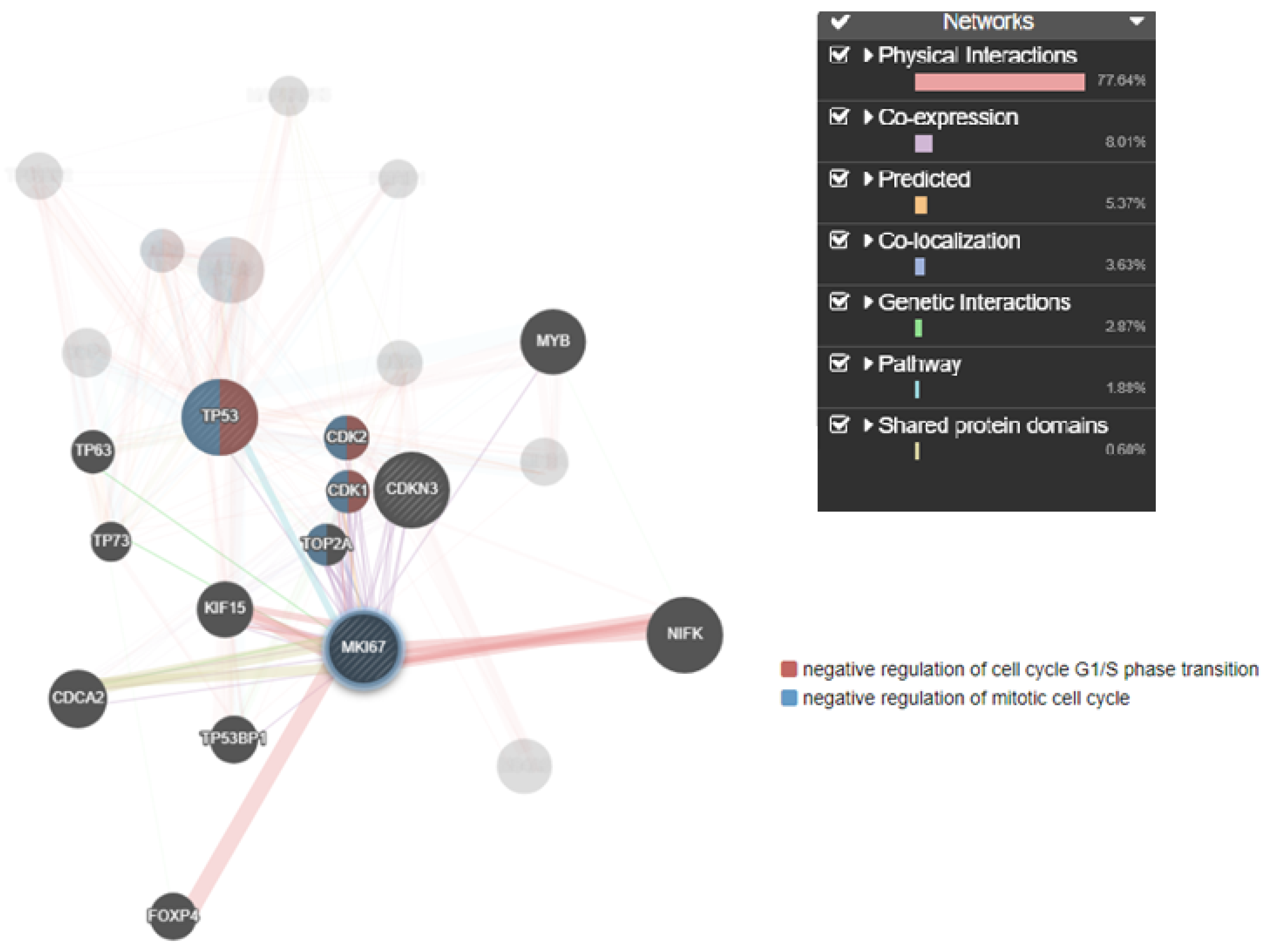
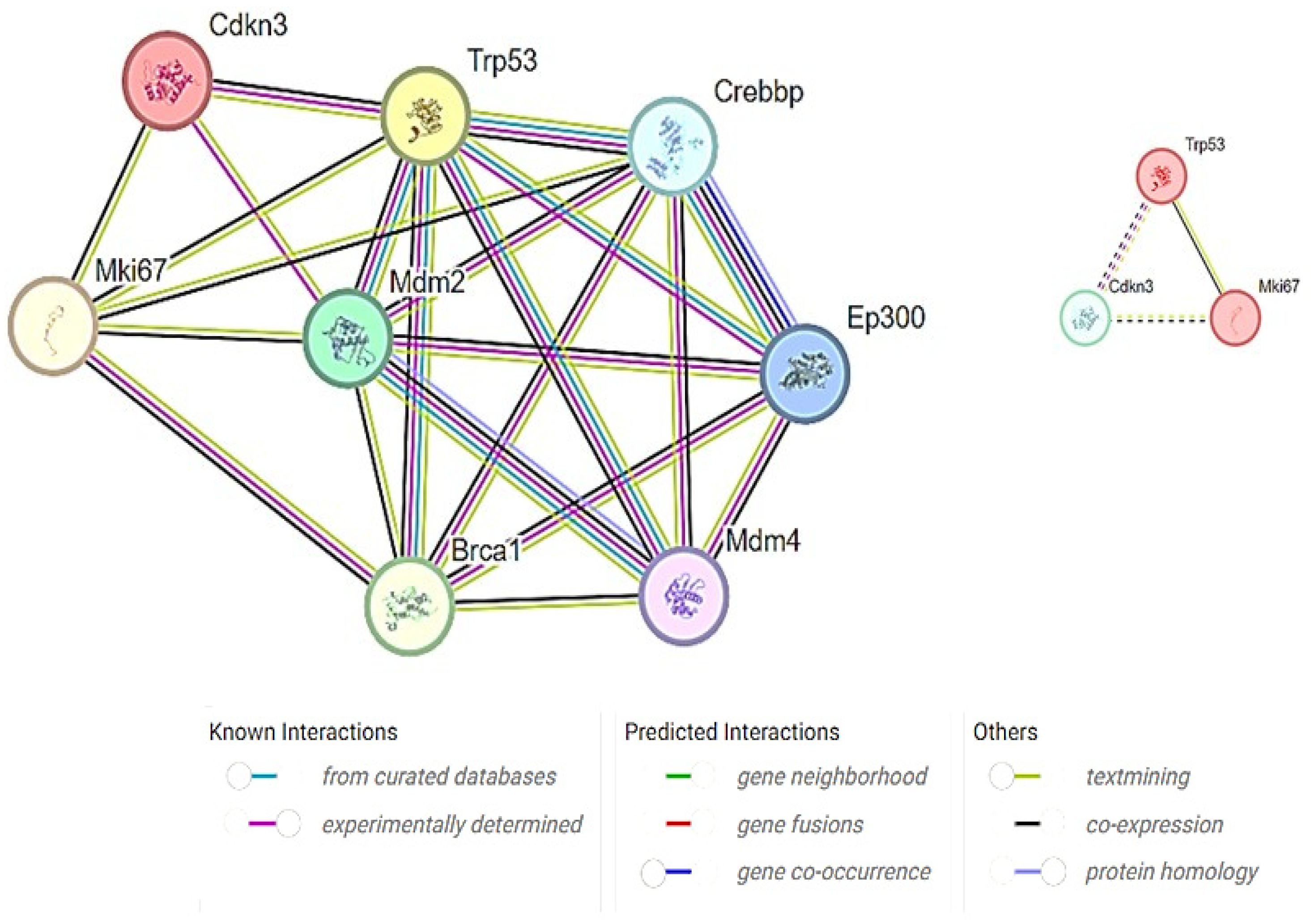
2.5. Co-Expression Network Analysis
2.6. Statistical Analysis
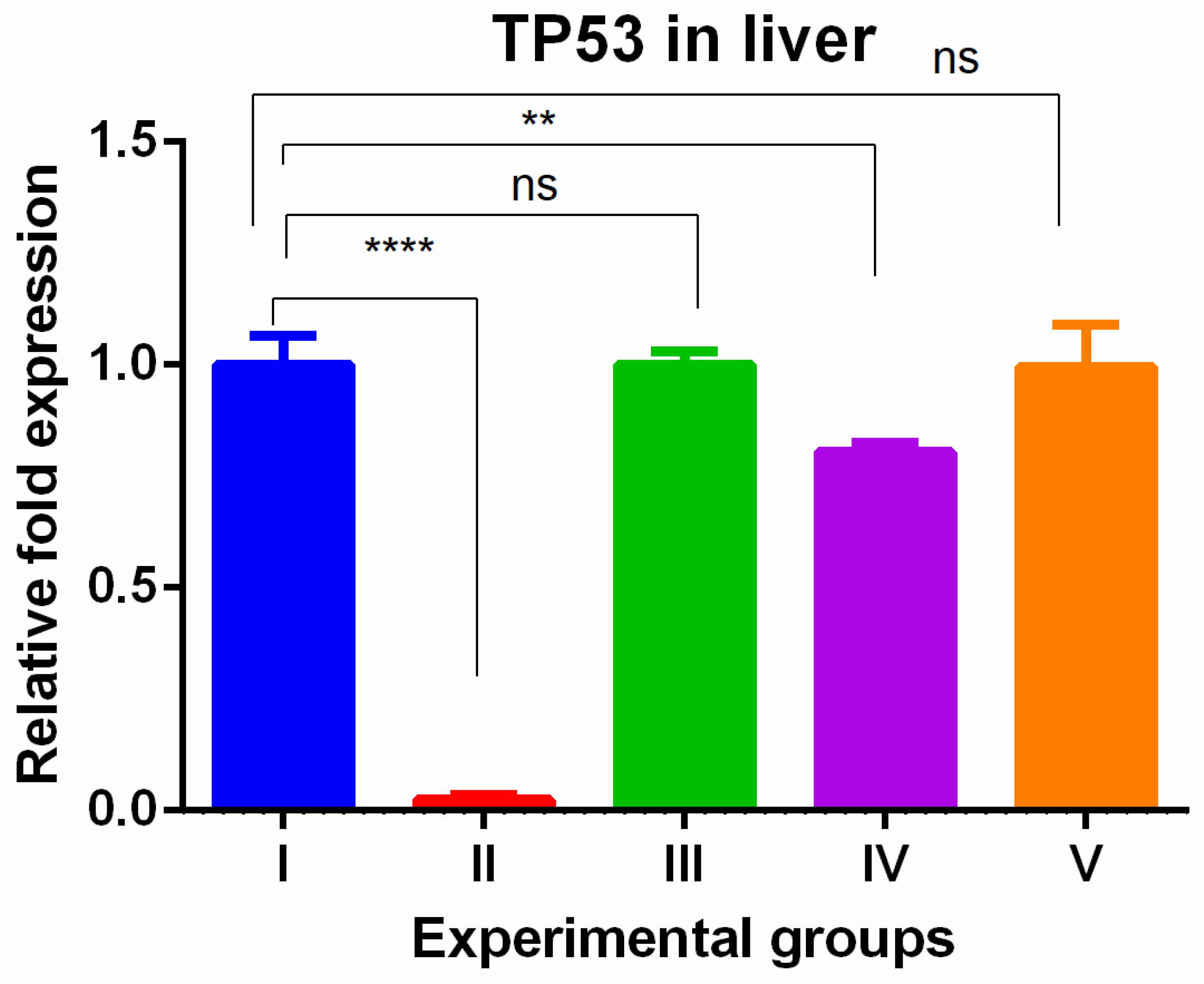
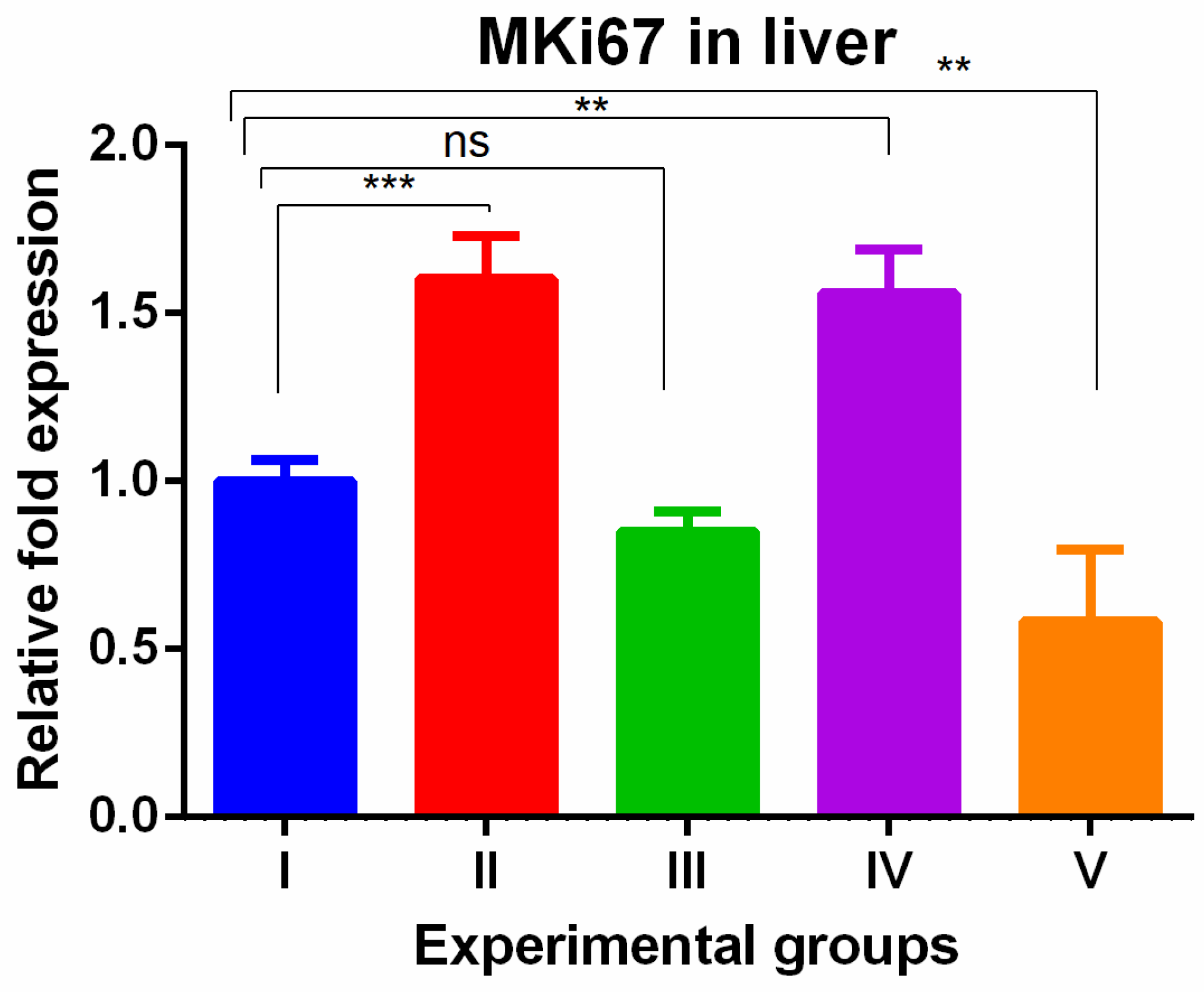
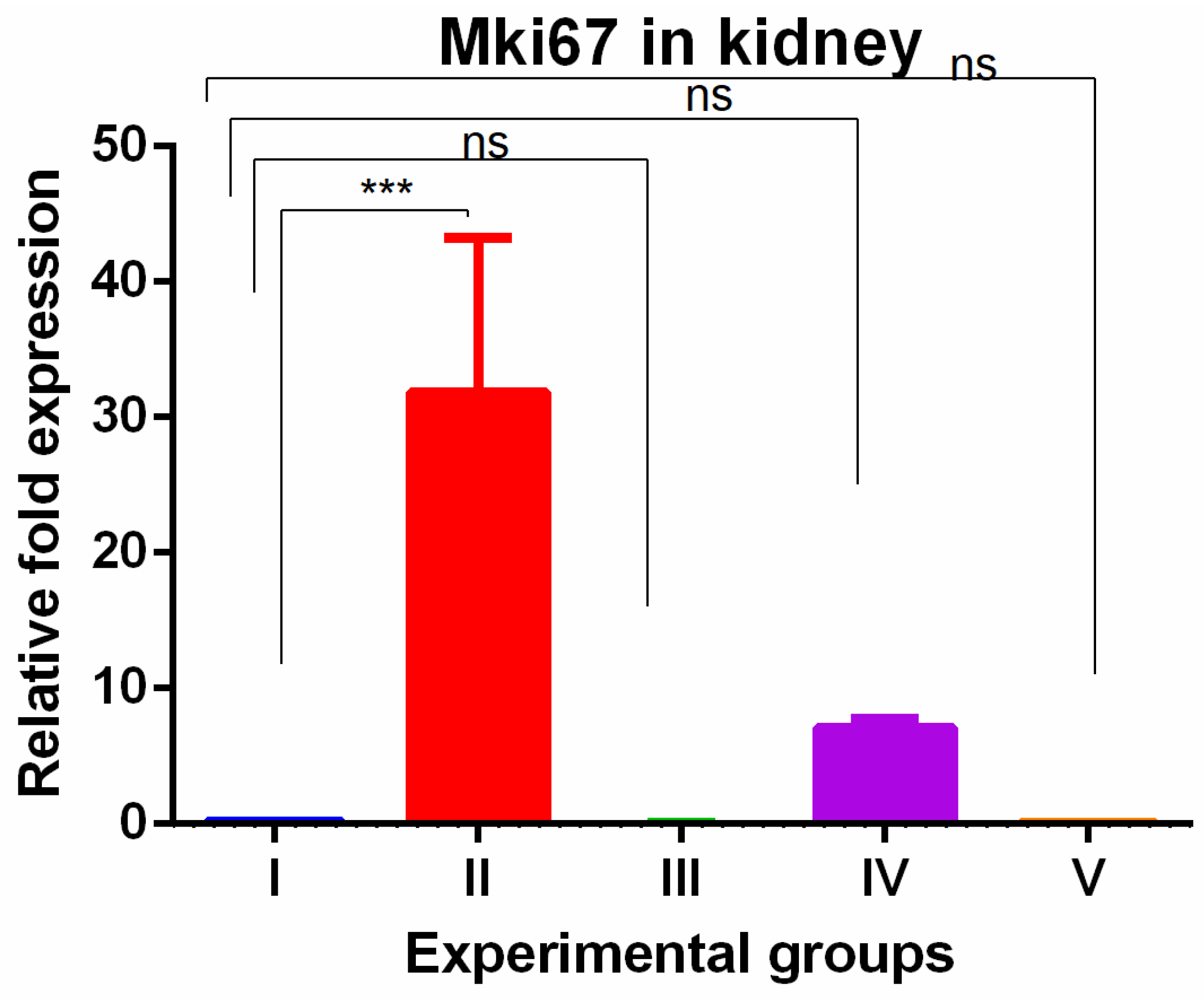
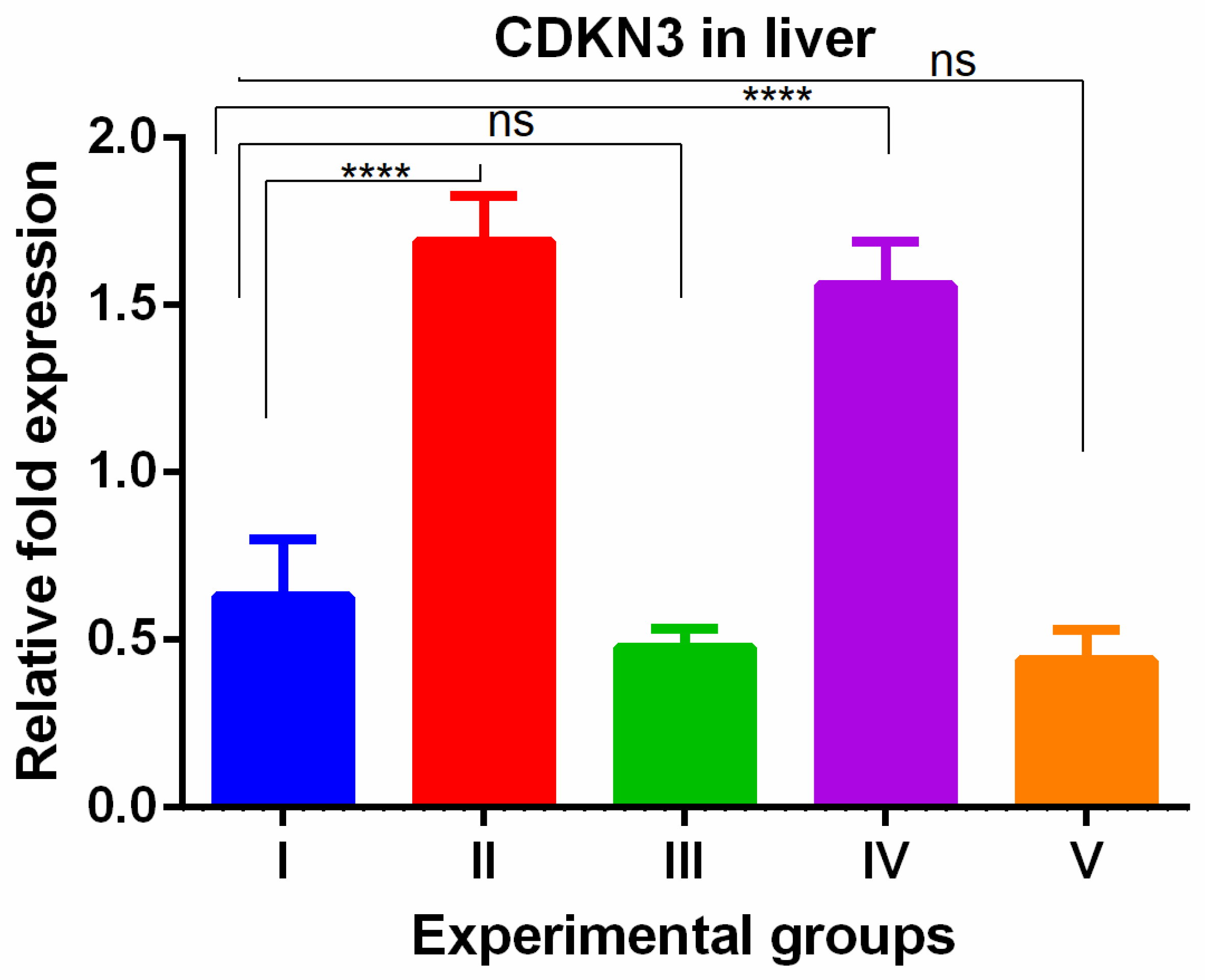
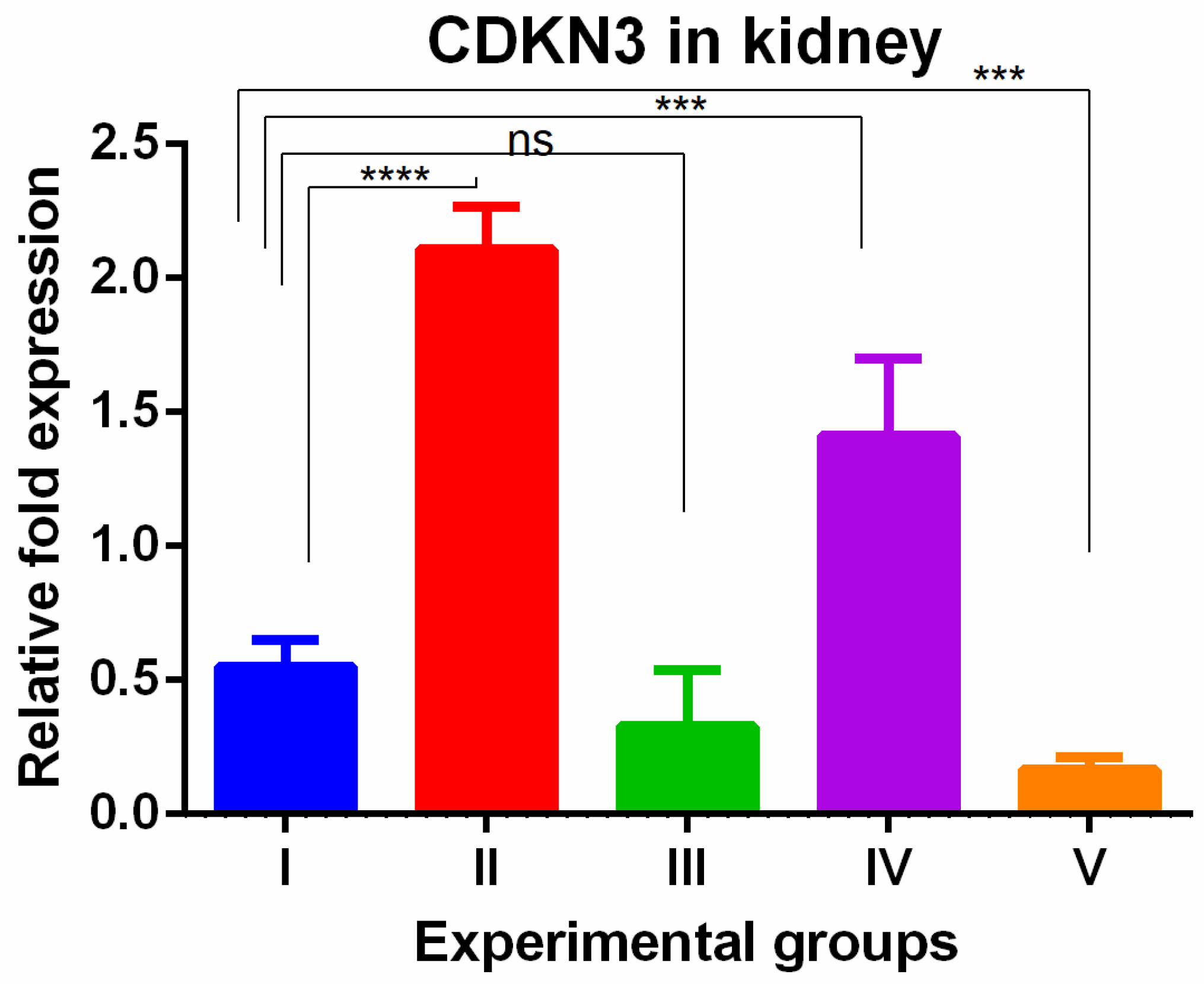
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alamery, S.; Zargar, S.; Yaseen, F.; Wani, T.A.; Siyal, A. Evaluation of the effect of wheat germ oil and olmutinib on the thioacetamide-induced liver and kidney toxicity in mice. Life 2022, 12, 900. [Google Scholar] [CrossRef] [PubMed]
- Ezhilarasan, D. Molecular mechanisms in thioacetamide-induced acute and chronic liver injury models. Environ. Toxicol. Pharmacol. 2023, 99, 104093. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Sharma, D.; Singh, A.P.; Kaur, S. Amelioration of hepatic function, oxidative stress, and histopathologic damages by Cassia fistula L. fraction in thioacetamide-induced liver toxicity. Environ. Sci. Pollut. Res. 2019, 26, 29930–29945. [Google Scholar] [CrossRef] [PubMed]
- Zargar, S. Protective effect of Trigonella foenum-graecum on thioacetamide induced hepatotoxicity in rats. Saudi J. Biol. Sci. 2014, 21, 139–145. [Google Scholar] [PubMed]
- Schyman, P.; Printz, R.L.; Estes, S.K.; Boyd, K.L.; Shiota, M.; Wallqvist, A. Identification of the toxicity pathways associated with thioacetamide-induced injuries in rat liver and kidney. Front. Pharmacol. 2018, 9, 1272. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhao, X.; Gu, X.; Chen, Y.; Yu, G. The preventive role of hydrogen-rich water in thioacetamide-induced cholangiofibrosis in rat assessed by automated histological classification. Front. Pharmacol. 2021, 12, 632045. [Google Scholar] [PubMed]
- Türkmen, N.B.; Hande, Y.; Taşlidere, A.; Şahin, Y.; Çiftçi, O. The ameliorate effects of nerolidol on thioacetamide-induced oxidative damage in heart and kidney tissue. Turk. J. Pharm. Sci. 2022, 19, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ionescu, A.; Altman, T.; Perlson, E. Looking for answers far away from the soma—The (un) known axonal functions of TDP-43, and their contribution to early NMJ disruption in ALS. Mol. Neurodegener. 2023, 18, 35. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Tan, L.; Tian, C.; Wang, W.; Zhang, H.; Zhu, Z.; Li, Y. Research progress on rodent models and its mechanisms of liver injury. Life Sci. 2023, 337, 122343. [Google Scholar]
- Salem, A.A.; Ismail, A.F. Protective impact of Spirulina platensis against γ-irradiation and thioacetamide-induced nephrotoxicity in rats mediated by regulation of micro-RNA 1 and micro-RNA 146a. Toxicol. Res. 2021, 10, 453–466. [Google Scholar] [CrossRef]
- Patel, K.; Murray, M.G.; Whelan, K.A. Roles for GADD45 in development and cancer. Gadd45 Stress Sens. Genes 2022, 1360, 23–39. [Google Scholar]
- Sun, W.; Lei, Y.; Jiang, Z.; Wang, K.; Liu, H.; Xu, T. BPA and low-Se exacerbate apoptosis and mitophagy in chicken pancreatic cells by regulating the PTEN/PI3K/AKT/mTOR pathway. J. Adv. Res. 2024, in press. [Google Scholar] [CrossRef]
- Scholzen, T.; Gerdes, J. The Ki-67 protein: From the known and the unknown. J. Cell. Physiol. 2000, 182, 311–322. [Google Scholar] [CrossRef]
- Sequist, L.V.; Soria, J.-C.; Goldman, J.W.; Wakelee, H.A.; Gadgeel, S.M.; Varga, A.; Papadimitrakopoulou, V.; Solomon, B.J.; Oxnard, G.R.; Dziadziuszko, R. Rociletinib in EGFR-mutated non–small-cell lung cancer. N. Engl. J. Med. 2015, 372, 1700–1709. [Google Scholar] [CrossRef]
- Zhang, W.; Fan, Y.-F.; Cai, C.-Y.; Wang, J.-Q.; Teng, Q.-X.; Lei, Z.-N.; Zeng, L.; Gupta, P.; Chen, Z.-S. Olmutinib (BI1482694/HM61713), a novel epidermal growth factor receptor tyrosine kinase inhibitor, reverses ABCG2-mediated multidrug resistance in cancer cells. Front. Pharmacol. 2018, 9, 1097. [Google Scholar] [CrossRef]
- Kumari, A.; Kaur, S.; Sharma, N.; Kaur, J.; Krishania, M.; Tiwari, V.; Garg, M. Effect of processing on the phytochemicals and quality attributes of vermicelli developed from colored wheat. J. Cereal Sci. 2022, 108, 103560. [Google Scholar] [CrossRef]
- Arshad, M.U.; Anjum, F.M.; Zahoor, T. Nutritional assessment of cookies supplemented with defatted wheat germ. Food Chem. 2007, 102, 123–128. [Google Scholar]
- Zargar, S.; Wani, T.A.; Rizwan Ahamad, S. An insight into wheat germ oil nutrition, identification of its bioactive constituents and computer-aided multidimensional data analysis of its potential anti-inflammatory effect via molecular connections. Life 2023, 13, 526. [Google Scholar] [CrossRef]
- Uxa, S.; Castillo-Binder, P.; Kohler, R.; Stangner, K.; Müller, G.A.; Engeland, K. Ki-67 gene expression. Cell Death Differ. 2021, 28, 3357–3370. [Google Scholar] [CrossRef]
- Babamohamadi, M.; Babaei, E.; Ahmed Salih, B.; Babamohammadi, M.; Jalal Azeez, H.; Othman, G. Recent findings on the role of wild-type and mutant p53 in cancer development and therapy. Front. Mol. Biosci. 2022, 9, 903075. [Google Scholar] [CrossRef]
- Wang, J.; Che, W.; Wang, W.; Su, G.; Zhen, T.; Jiang, Z. CDKN3 promotes tumor progression and confers cisplatin resistance via RAD51 in esophageal cancer. Cancer Manag. Res. 2019, 11, 3253–3264. [Google Scholar] [CrossRef]
- Andrés-Sánchez, N.; Fisher, D.; Krasinska, L. Physiological functions and roles in cancer of the proliferation marker Ki-67. J. Cell Sci. 2022, 135, jcs258932. [Google Scholar]
- Liu, L.-M.; Huang, Q.; Chen, Q.-N.; Liu, Z.-Y.; Ge, B.-J. A critical role of CDKN3 in gastric cancer and regulates tumor cell proliferation. Int. J. Clin. Exp. Med. 2016, 9, 172–178. [Google Scholar]
- Wong, E.Y.L.; Wong, S.C.C.; Chan, C.M.L.; Lam, E.K.Y.; Ho, L.Y.; Lau, C.P.Y.; Au, T.C.C.; Chan, A.K.C.; Tsang, C.M.; Tsao, S.W. TP53-induced glycolysis and apoptosis regulator promotes proliferation and invasiveness of nasopharyngeal carcinoma cells. Oncol. Lett. 2015, 9, 569–574. [Google Scholar]
- Brisco, P.; Sankbeil, J.; Kephart, D. RNA purification: A rapid and versatile protocol for the isolation of total RNA. Promega Notes 1997, 64. [Google Scholar]
- Thermo Scientific. SuperScript™ First-Strand Synthesis System for RT-PCR; Thermo Scientific: Waltham, MA, USA, 2003. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar]
- Alamery, S.; AlAjmi, A.; Wani, T.A.; Zargar, S. In Silico and In Vitro Exploration of Poziotinib and Olmutinib Synergy in Lung Cancer: Role of hsa-miR-7-5p in Regulating Apoptotic Pathway Marker Genes. Medicina 2023, 59, 1923. [Google Scholar] [CrossRef]
- Donehower, L.A.; Soussi, T.; Korkut, A.; Liu, Y.; Schultz, A.; Cardenas, M.; Li, X.; Babur, O.; Hsu, T.-K.; Lichtarge, O. Integrated analysis of TP53 gene and pathway alterations in the cancer genome atlas. Cell Rep. 2019, 28, 1370–1384.e1375. [Google Scholar]
- Qin, K.; Hou, H.; Liang, Y.; Zhang, X. Prognostic value of TP53 concurrent mutations for EGFR-TKIs and ALK-TKIs based targeted therapy in advanced non-small cell lung cancer: A meta-analysis. BMC Cancer 2020, 20, 328. [Google Scholar] [CrossRef]
- Gu, W.; Hu, M.; Xu, L.; Ren, Y.; Mei, J.; Wang, W.; Wang, C. The Ki-67 proliferation index-related nomogram to predict the response of first-line tyrosine kinase inhibitors or chemotherapy in non-small cell lung cancer patients with epidermal growth factor receptor-mutant status. Front. Med. 2021, 8, 728575. [Google Scholar] [CrossRef]
- Das, S.; Dey, M.K.; Devireddy, R.; Gartia, M.R. Biomarkers in Cancer Detection, Diagnosis, and Prognosis. Sensors 2023, 24, 37. [Google Scholar] [CrossRef]
- Yu, C.; Cao, H.; He, X.; Sun, P.; Feng, Y.; Chen, L.; Gong, H. Cyclin-dependent kinase inhibitor 3 (CDKN3) plays a critical role in prostate cancer via regulating cell cycle and DNA replication signaling. Biomed. Pharmacother. 2017, 96, 1109–1118. [Google Scholar] [CrossRef]
- Dai, W.; Miao, H.; Fang, S.; Fang, T.; Chen, N.; Li, M. CDKN3 expression is negatively associated with pathological tumor stage and CDKN3 inhibition promotes cell survival in hepatocellular carcinoma. Mol. Med. Rep. 2016, 14, 1509–1514. [Google Scholar] [CrossRef]
- Chen, X.; Xia, Z.; Wan, Y.; Huang, P. Identification of hub genes and candidate drugs in hepatocellular carcinoma by integrated bioinformatics analysis. Medicine 2021, 100, e27117. [Google Scholar] [CrossRef]
- Montojo, J.; Zuberi, K.; Rodriguez, H.; Kazi, F.; Wright, G.; Donaldson, S.L.; Morris, Q.; Bader, G.D. GeneMANIA Cytoscape plugin: Fast gene function predictions on the desktop. Bioinformatics 2010, 26, 2927–2928. [Google Scholar] [CrossRef]
- Annenkov, A. Receptor tyrosine kinase (RTK) signalling in the control of neural stem and progenitor cell (NSPC) development. Mol. Neurobiol. 2014, 49, 440–471. [Google Scholar] [CrossRef]
- Kaewjanthong, P.; Sooksai, S.; Sasano, H.; Hutvagner, G.; Bajan, S.; McGowan, E.; Boonyaratanakornkit, V. Cell-penetrating peptides containing the progesterone receptor polyproline domain inhibits EGF signaling and cell proliferation in lung cancer cells. PLoS ONE 2022, 17, e0264717. [Google Scholar] [CrossRef]

| Name of Gene | Primers | Primer Name | |
|---|---|---|---|
| MKi67 | Forward | AAGAAGAGCCCACAGCACAGAGAA | M_MKi67-E15F |
| Reverse | AAGAAGAGCCCACAGCACAGAGAA | M_MKi67-E16R | |
| CDKN3 | Forward | TTCTGCCATTCTCACCGTGTCCTT | M_CDKN3-E4F |
| Reverse | TGCGATAACAAGCTCCGTCCATCT | M_CDKN3-E6R | |
| TP53 | Forward | AACAATGGCCCGAGTCTAATGGGA | M_TP53-E2F |
| Reverse | ACAGATGTTGCCTGATGTCTGGGT | M_TP53-E4R | |
| GAPDH | Forward | ACCACAGTCCATGCCATCAC | M_GAPDH-F |
| Reverse | ACCACAGTCCATGCCATCAC | M_GAPDH-R | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zargar, S.; Wani, T.A.; Alamery, S.; Yaseen, F. Olmutinib Reverses Thioacetamide-Induced Cell Cycle Gene Alterations in Mice Liver and Kidney Tissues, While Wheat Germ Treatment Exhibits Limited Efficacy at Gene Level. Medicina 2024, 60, 639. https://doi.org/10.3390/medicina60040639
Zargar S, Wani TA, Alamery S, Yaseen F. Olmutinib Reverses Thioacetamide-Induced Cell Cycle Gene Alterations in Mice Liver and Kidney Tissues, While Wheat Germ Treatment Exhibits Limited Efficacy at Gene Level. Medicina. 2024; 60(4):639. https://doi.org/10.3390/medicina60040639
Chicago/Turabian StyleZargar, Seema, Tanveer A. Wani, Salman Alamery, and Fatimah Yaseen. 2024. "Olmutinib Reverses Thioacetamide-Induced Cell Cycle Gene Alterations in Mice Liver and Kidney Tissues, While Wheat Germ Treatment Exhibits Limited Efficacy at Gene Level" Medicina 60, no. 4: 639. https://doi.org/10.3390/medicina60040639
APA StyleZargar, S., Wani, T. A., Alamery, S., & Yaseen, F. (2024). Olmutinib Reverses Thioacetamide-Induced Cell Cycle Gene Alterations in Mice Liver and Kidney Tissues, While Wheat Germ Treatment Exhibits Limited Efficacy at Gene Level. Medicina, 60(4), 639. https://doi.org/10.3390/medicina60040639








