CRISPR/Cas-Based Prenatal Screening for Aneuploidy: Challenges and Opportunities for Early Diagnosis
Abstract
1. Introduction
2. Sampling
2.1. Chorionic Villus Sampling
2.2. Amniocentesis
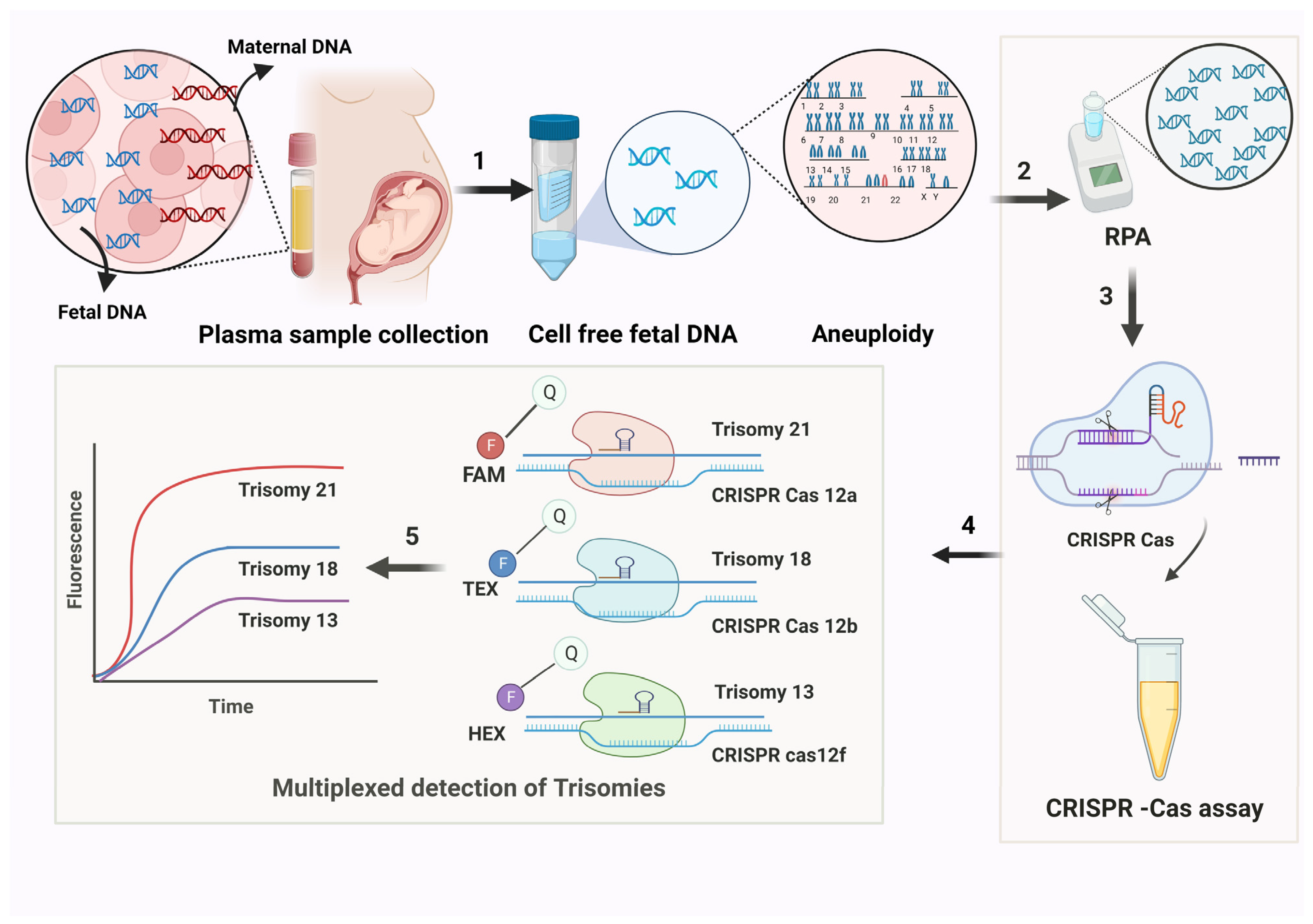
2.3. Cell-Free Fetal DNA
3. Prenatal Screening Tests
3.1. Triple, Quad, and Penta Screening
3.2. Ultrasonographic Screening
3.3. Non-Invasive Prenatal Screening (NIPT)
4. Molecular Diagnostic Methods
4.1. Karyotyping
4.2. Fluorescent In Situ Hybridization (FISH)
4.3. Real-Time Quantitative PCR (RT-qPCR)
4.4. Microarray
4.5. Gene Sequencing
4.6. CRISPR/Cas Based Diagnostics
4.7. CRISPR/Cas-Based Diagnostics of Infectious Disease
4.8. CRISPR/Cas-Based Diagnostics of Non-Infectious Disease
4.9. CRISPR/Cas-Based Diagnostics for Trisomy Detection
4.10. Advantages of CRISPR/Cas-Based Diagnostics for Trisomy
4.11. Advantages of CRISPR/Cas-Based Diagnostics in ARF and IVF Conception
4.12. Challenges of CRISPR/Cas-Based Diagnostics for Trisomy
5. Conclusions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- McFeely, R.A. Chromosome abnormalities. Vet. Clin. N. Am. Food Anim. Pract. 1993, 9, 11–22. [Google Scholar] [CrossRef]
- Nussbaum, R.L.; McInnes, R.R.; Willard, H.F. Principles of clinical cytogenetics and genome analysis. In Thompson & Thompson Genetics in Medicine, 8th ed.; Elsevier: Philadelphia, PA, USA, 2016; Available online: https://shop.elsevier.com/books/thompson-and-thompson-genetics-in-medicine/nussbaum/978-1-4377-0696-3 (accessed on 28 February 2025).
- Mennuti, M.E.; Kuller, J.A.; Dugoff, L. (Eds.) Cytogenetics: Part 1, General Concepts and Aneuploid Conditions. In Perinatal Genetics; Elsevier Asia Bookstore: Norton, VA, USA, 2019; ISBN 9780323530941. Available online: https://www.asia.elsevierhealth.com/perinatal-genetics-9780323530941.html (accessed on 28 February 2025).
- Levine, M.S.; Holland, A.J. The impact of mitotic errors on cell proliferation and tumorigenesis. Genes Dev. 2018, 32, 620–638. [Google Scholar] [CrossRef] [PubMed]
- ACOG. Practice Bulletin No. 163: Screening for Fetal Aneuploidy. Obstet. Gynecol. 2016, 127, e123–e137. [Google Scholar] [CrossRef] [PubMed]
- Sherman, S.L.; Allen, E.G.; Bean, L.H.; Freeman, S.B. Epidemiology of Down syndrome. Ment. Retard. Dev. Disabil. Res. 2007, 13, 221–227. [Google Scholar] [CrossRef]
- Genetic Alliance; The New York-Mid-Atlantic Consortium for Genetic and Newborn ScreeningServices. Understanding Genetics: A New York, Mid-Atlantic Guide for Patients and Health Professionals. In Genetic Alliance Monographs and Guides; Genetic Alliance: Washington, DC, USA, 2009. Available online: http://www.ncbi.nlm.nih.gov/books/NBK115563/ (accessed on 28 February 2025).
- Anderson, C.L.; Brown, C.E.L. Fetal Chromosomal Abnormalities: Antenatal Screening and Diagnosis. Am. Fam. Physician 2009, 79, 117–123. [Google Scholar]
- Hixson, L.; Goel, S.; Schuber, P.; Faltas, V.; Lee, J.; Narayakkadan, A.; Leung, H.; Osborne, J. An Overview on Prenatal Screening for Chromosomal Aberrations. J. Lab. Autom. 2015, 20, 562–573. [Google Scholar] [CrossRef]
- Wilson, R.D. Safety and fetal outcome of early and midtrimester amniocentesis. Lancet 1998, 351, 1435–1436. [Google Scholar] [CrossRef]
- Kypri, E.; Ioannides, M.; Achilleos, A.; Koumbaris, G.; Patsalis, P.; Stumm, M. Non-invasive prenatal screening tests—Update 2022. J. Lab. Med. 2022, 46, 311–320. [Google Scholar] [CrossRef]
- Jain, M.; Balatsky, A.V.; Revina, D.B.; Samokhodskaya, L.M. Direct comparison of QIAamp DSP Virus Kit and QIAamp Circulating Nucleic Acid Kit regarding cell-free fetal DNA isolation from maternal peripheral blood. Mol. Cell. Probes 2019, 43, 13–19. [Google Scholar] [CrossRef]
- Chan, K.A.; Ding, C.; Gerovassili, A.; Yeung, S.W.; Chiu, R.W.; Leung, T.N.; Lau, T.K.; Chim, S.S.; Chung, G.T.; Nicolaides, K.H.; et al. Hypermethylated RASSF1A in Maternal Plasma: A Universal Fetal DNA Marker that Improves the Reliability of Noninvasive Prenatal Diagnosis. Clin. Chem. 2006, 52, 2211–2218. [Google Scholar] [CrossRef]
- Yang, Q.; Du, Z.; Song, Y.; Gao, S.; Yu, S.; Zhu, H.; Ren, M.; Zhang, G. Size-selective separation and overall-amplification of cell-free fetal DNA fragments using PCR-based enrichment. Sci. Rep. 2017, 7, 40936. [Google Scholar] [CrossRef]
- Liu, L.; Guo, Z.; Huang, Z.; Zhuang, J.; Yang, W. Size-selective separation of DNA fragments by using lysine-functionalized silica particles. Sci. Rep. 2016, 6, 22029. [Google Scholar] [CrossRef]
- Baer, R.J.; Norton, M.E.; Shaw, G.M.; Flessel, M.C.; Goldman, S.; Currier, R.J. Risk of selected structural abnormalities in infants after increased nuchal translucency measurement. Am. J. Obstet. Gynecol. 2014, 211, 675.e1–675.e19. [Google Scholar] [CrossRef] [PubMed]
- Nicolaides, K.H.; Azar, G.; Byrne, D.; Mansur, C.; Marks, K. Fetal nuchal translucency: Ultrasound screening for chromosomal defects in first trimester of pregnancy. BMJ 1992, 304, 867–869. [Google Scholar] [CrossRef]
- Driscoll, D.A.; Gross, S.J. Screening for fetal aneuploidy and neural tube defects. Genet. Med. 2009, 11, 818–821. [Google Scholar] [CrossRef]
- Palomaki, G.E.; Neveux, L.M.; Knight, G.J.; Haddow, J.E.; Pandian, R. Maternal serum invasive trophoblast antigen (hyperglycosylated hCG) as a screening marker for Down syndrome during the second trimester. Clin. Chem. 2004, 50, 1804–1808. [Google Scholar] [CrossRef]
- Conde-Agudelo, A.; Kafury-Goeta, A.C. Triple-marker test as screening for Down syndrome: A meta-analysis. Obstet. Gynecol. Surv. 1998, 53, 369–376. [Google Scholar] [CrossRef]
- Carlson, L.M.; Vora, N.L. Prenatal Diagnosis: Screening and Diagnostic Tools. Obstet. Gynecol. Clin. N. Am. 2017, 44, 245–256. [Google Scholar] [CrossRef]
- Lau, T.K.; Zhu, X.; Kwok, Y.K.Y.; Leung, T.Y.; Choy, K.W. Recent Advances in the Noninvasive Prenatal Testing for Chromosomal Abnormalities Using Maternal Plasma DNA. J. Fetal Med. 2023, 7, 17–23. [Google Scholar] [CrossRef]
- Dar, P.; Shani, H.; Evans, M.I. Cell-free DNA: Comparison of Technologies. Clin. Lab. Med. 2016, 36, 199–211. [Google Scholar] [CrossRef]
- Gil, M.M.; Quezada, M.S.; Revello, R.; Akolekar, R.; Nicolaides, K.H. Analysis of cell-free DNA in maternal blood in screening for fetal aneuploidies: Updated meta-analysis. Ultrasound Obstet. Gynecol. 2015, 45, 249–266. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, D.W.; Hanson, J. Sharpening the tools: A summary of a National Institutes of Health workshop on new technologies for detection of fetal cells in maternal blood for early prenatal diagnosis. J. Matern. Fetal Neonatal Med. 2006, 19, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Chitty, L.S.; Lo, Y.M.D. Noninvasive Prenatal Screening for Genetic Diseases Using Massively Parallel Sequencing of Maternal Plasma DNA. Cold Spring Harb. Perspect. Med. 2015, 5, a023085. [Google Scholar] [CrossRef]
- Benn, P. Non-Invasive Prenatal Testing Using Cell Free DNA in Maternal Plasma: Recent Developments and Future Prospects. J. Clin. Med. 2014, 3, 537–565. [Google Scholar] [CrossRef]
- Norton, M.E.; Baer, R.J.; Wapner, R.J.; Kuppermann, M.; Jelliffe-Pawlowski, L.L.; Currier, R.J. Cell-free DNA vs sequential screening for the detection of fetal chromosomal abnormalities. Am. J. Obstet. Gynecol. 2016, 214, 727.e1–727.e6. [Google Scholar] [CrossRef]
- Fruhman, G.; Van den Veyver, I.B. Applications of array comparative genomic hybridization in obstetrics. Obstet. Gynecol. Clin. N. Am. 2010, 37, 71–85. [Google Scholar] [CrossRef]
- Hahn, S.; Jackson, L.G.; Kolla, V.; Mahyuddin, A.P.; Choolani, M. Noninvasive prenatal diagnosis of fetal aneuploidies and Mendelian disorders: New innovative strategies. Expert Rev. Mol. Diagn. 2009, 9, 613–621. [Google Scholar] [CrossRef]
- Philip, J.; Silver, R.K.; Wilson, R.D.; Thom, E.A.; Zachary, J.M.; Mohide, P.; Mahoney, M.J.; Simpson, J.L.; Platt, L.D.; Pergament, E.; et al. Late first-trimester invasive prenatal diagnosis: Results of an international randomized trial. Obstet. Gynecol. 2004, 103, 1164–1173. [Google Scholar] [CrossRef]
- Bridge, J.A. Advantages and limitations of cytogenetic, molecular cytogenetic, and molecular diagnostic testing in mesenchymal neoplasms. J. Orthop. Sci. 2008, 13, 273–282. [Google Scholar] [CrossRef]
- Caine, A.; Maltby, A.E.; Parkin, C.A.; Waters, J.J.; Crolla, J.A.; UK Association of Clinical Cytogeneticists (ACC). Prenatal detection of Down’s syndrome by rapid aneuploidy testing for chromosomes 13, 18, and 21 by FISH or PCR without a full karyotype: A cytogenetic risk assessment. Lancet Lond. Engl. 2005, 366, 123–128. [Google Scholar] [CrossRef]
- Hu, L.; Ru, K.; Zhang, L.; Huang, Y.; Zhu, X.; Liu, H.; Zetterberg, A.; Cheng, T.; Miao, W. Fluorescence in situ hybridization (FISH): An increasingly demanded tool for biomarker research and personalized medicine. Biomark. Res. 2014, 2, 3. [Google Scholar] [CrossRef]
- Lalonde, E.; Rentas, S.; Lin, F.; Dulik, M.C.; Skraban, C.M.; Spinner, N.B. Genomic Diagnosis for Pediatric Disorders: Revolution and Evolution. Front. Pediatr. 2020, 8, 373. [Google Scholar] [CrossRef]
- Tepperberg, J.; Pettenati, M.J.; Rao, P.N.; Lese, C.M.; Rita, D.; Wyandt, H.; Gersen, S.; White, B.; Schoonmaker, M.M. Prenatal diagnosis using interphase fluorescence in situ hybridization (FISH): 2-year multi-center retrospective study and review of the literature. Prenat. Diagn. 2001, 21, 293–301. [Google Scholar] [CrossRef]
- Savola, S.; Nardi, F.; Scotlandi, K.; Picci, P.; Knuutila, S. Microdeletions in 9p21.3 induce false negative results in CDKN2A FISH analysis of Ewing sarcoma. Cytogenet. Genome Res. 2007, 119, 21–26. [Google Scholar] [CrossRef]
- Salomon, L.J.; Sotiriadis, A.; Wulff, C.B.; Odibo, A.; Akolekar, R. Risk of miscarriage following amniocentesis or chorionic villus sampling: Systematic review of literature and updated meta-analysis. Ultrasound Obstet. Gynecol. 2019, 54, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Kubista, M.; Andrade, J.M.; Bengtsson, M.; Forootan, A.; Jonák, J.; Lind, K.; Sindelka, R.; Sjöback, R.; Sjögreen, B.; Strömbom, L.; et al. The real-time polymerase chain reaction. Mol. Asp. Med. 2006, 27, 95–125. [Google Scholar] [CrossRef]
- Guo, L.; Accorsi, A.; He, S.; Guerrero-Hernández, C.; Sivagnanam, S.; McKinney, S.; Gibson, M.; Alvarado, A.S. An adaptable chromosome preparation methodology for use in invertebrate research organisms. BMC Biol. 2018, 16, 25. [Google Scholar] [CrossRef]
- Lou, J.; Sun, M.; Zhao, Y.; Ji, Z.; Liu, F.; Li, D.; Xu, W.; Lin, Y.; Liu, Y. Rapid and simultaneous detection of common aneuploidies by quadruplex real-time polymerase chain reaction combined with melting curve analysis. PLoS ONE 2017, 12, e0171886. [Google Scholar] [CrossRef]
- Murphy, J.; Bustin, S.A. Reliability of real-time reverse-transcription PCR in clinical diagnostics: Gold standard or substandard? Expert Rev. Mol. Diagn. 2009, 9, 187–197. [Google Scholar] [CrossRef]
- Traeger-Synodinos, J. Real-time PCR for prenatal and preimplantation genetic diagnosis of monogenic diseases. Mol. Asp. Med. 2006, 27, 176–191. [Google Scholar] [CrossRef]
- Grace, M.R.; Hardisty, E.; Dotters-Katz, S.K.; Vora, N.L.; Kuller, J.A. Cell-Free DNA Screening: Complexities and Challenges of Clinical Implementation. Obstet. Gynecol. Surv. 2016, 71, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Fan, Z.; Long, J.; Weng, X.; Tang, W.; Pang, W. Rapid prenatal diagnosis of aneuploidy for chromosomes 21, 18, 13, X, and Y using segmental duplication quantitative fluorescent PCR (SD-QF-PCR). Gene 2017, 627, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Li, L.; Sun, L.; Fu, K.; Long, J.; Weng, X.; Ye, X.; Liu, X.; Wang, B.; Yan, S.; et al. Rapid Diagnosis of Aneuploidy Using Segmental Duplication Quantitative Fluorescent PCR. PLoS ONE 2014, 9, e88932. [Google Scholar] [CrossRef]
- Atef, S.H.; Hafez, S.; Helmy, S.; Helmy, N. QF-PCR as a Rapid Technique for Routine Prenatal Diagnosis of Fetal Aneuploidies. Pediatr. Res. 2011, 70, 412. [Google Scholar] [CrossRef]
- Committee on Practice Bulletins—Obstetrics; Committee on Genetics; Society for Maternal–Fetal Medicine. Practice Bulletin No. 162: Prenatal Diagnostic Testing for Genetic Disorders. Obstet. Gynecol. 2016, 127, e108–e122. [Google Scholar] [CrossRef]
- Wapner, R.J.; Martin, C.L.; Levy, B.; Ballif, B.C.; Eng, C.M.; Zachary, J.M.; Savage, M.; Platt, L.D.; Saltzman, D.; Grobman, W.A.; et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N. Engl. J. Med. 2012, 367, 2175–2184. [Google Scholar] [CrossRef]
- Liu, X.; Liu, S.; Wang, H.; Hu, T. Potentials and challenges of chromosomal microarray analysis in prenatal diagnosis. Front. Genet. 2022, 13, 938183. [Google Scholar] [CrossRef]
- Tabor, A.; Alfirevic, Z. Update on procedure-related risks for prenatal diagnosis techniques. Fetal Diagn. Ther. 2010, 27, 1–7. [Google Scholar] [CrossRef]
- Van den Veyver, I.B.; Eng, C.M. Genome-Wide Sequencing for Prenatal Detection of Fetal Single-Gene Disorders. Cold Spring Harb. Perspect. Med. 2015, 5, a023077. [Google Scholar] [CrossRef]
- Lepri, F.R.; Scavelli, R.; Digilio, M.C.; Gnazzo, M.; Grotta, S.; Dentici, M.L.; Pisaneschi, E.; Sirleto, P.; Capolino, R.; Baban, A.; et al. Diagnosis of Noonan syndrome and related disorders using target next generation sequencing. BMC Med. Genet. 2014, 15, 14. [Google Scholar] [CrossRef]
- Filges, I.; Friedman, J.M. Exome sequencing for gene discovery in lethal fetal disorders—Harnessing the value of extreme phenotypes. Prenat. Diagn. 2015, 35, 1005–1009. [Google Scholar] [CrossRef] [PubMed]
- Schouten, J.; Van Vught, P.; Galjaard, R.-J. Multiplex Ligation-Dependent Probe Amplification (MLPA) for Prenatal Diagnosis of Common Aneuploidies. In Prenatal Diagnosis; Levy, B., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; Volume 1885, pp. 161–170. [Google Scholar] [CrossRef]
- Blyth, U.; Craciunas, L.; Hudson, G.; Choudhary, M. Maternal germline factors associated with aneuploid pregnancy loss: A systematic review. Hum. Reprod. Update 2021, 27, 866–884. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Marraffini, L.A. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol. Cell 2014, 54, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Caplan, A.L.; Parent, B.; Shen, M.; Plunkett, C. No time to waste—The ethical challenges created by CRISPR. EMBO Rep. 2015, 16, 1421–1426. [Google Scholar] [CrossRef]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Hilton, I.B.; D’ippolito, A.M.; Vockley, C.M.; Thakore, P.I.; Crawford, G.E.; Reddy, T.E.; Gersbach, C.A. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015, 33, 510–517. [Google Scholar] [CrossRef]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef]
- Chen, B.; Gilbert, L.A.; Cimini, B.A.; Schnitzbauer, J.; Zhang, W.; Li, G.W.; Park, J.; Blackburn, E.H.; Weissman, J.S.; Qi, L.S.; et al. Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell 2013, 155, 1479–1491. [Google Scholar] [CrossRef]
- Sheth, R.U.; Yim, S.S.; Wu, F.L.; Wang, H.H. Multiplex recording of cellular events over time on CRISPR biological tape. Science 2017, 358, 1457–1461. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Peng, Y.; Hua, K.; Deng, Y.; Bellizzi, M.; Gupta, D.R.; Mahmud, N.U.; Urashima, A.S.; Paul, S.L.; Peterson, G.; et al. Rapid Detection of Wheat Blast Pathogen Magnaporthe oryzae Triticum Pathotype Using Genome-Specific Primers and Cas12a-mediated Technology. Engineering 2021, 7, 1326–1335. [Google Scholar] [CrossRef]
- Sánchez, E.; Ali, Z.; Islam, T.; Mahfouz, M. A CRISPR-based lateral flow assay for plant genotyping and pathogen diagnostics. Plant Biotechnol. J. 2022, 20, 2418–2429. [Google Scholar] [CrossRef]
- Yuan, J.; Wang, L.; Huang, L.; He, K.; Wang, H.; Xu, X.; Su, B.; Wang, J. CRISPR-Cas12a-Mediated Hue-Recognition Lateral Flow Assay for Point-of-Need Detection of Salmonella. Anal. Chem. 2024, 96, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Jolany Vangah, S.; Katalani, C.; Boone, H.A.; Hajizade, A.; Sijercic, A.; Ahmadian, G. CRISPR-Based Diagnosis of Infectious and Noninfectious Diseases. Biol. Proced. Online 2020, 22, 22. [Google Scholar] [CrossRef]
- Rahimi, S.; Balusamy, S.R.; Perumalsamy, H.; Ståhlberg, A.; Mijakovic, I. CRISPR-Cas target recognition for sensing viral and cancer biomarkers. Nucleic Acids Res. 2024, 52, 10040–10067. [Google Scholar] [CrossRef]
- Li, L.; Li, S.; Wu, N.; Wu, J.; Wang, G.; Zhao, G.; Wang, J. HOLMESv2: A CRISPR-Cas12b-Assisted Platform for Nucleic Acid Detection and DNA Methylation Quantitation. ACS Synth. Biol. 2019, 8, 2228–2237. [Google Scholar] [CrossRef]
- Kostyusheva, A.; Brezgin, S.; Babin, Y.; Vasilyeva, I.; Glebe, D.; Kostyushev, D.; Chulanov, V. CRISPR-Cas systems for diagnosing infectious diseases. Methods 2022, 203, 431–446. [Google Scholar] [CrossRef]
- Zhang, X.; He, X.; Zhang, Y.; Chen, L.; Pan, Z.; Huang, Y.; Li, H. A new method for the detection of Mycobacterium tuberculosis based on the CRISPR/Cas system. BMC Infect. Dis. 2023, 23, 680. [Google Scholar] [CrossRef]
- Qian, B.; Liao, K.; Zeng, D.; Peng, W.; Wu, X.; Li, J.; Bo, Z.; Hu, Y.; Nan, W.; Wen, Y.; et al. Clustered Regularly Interspaced Short Palindromic Repeat/Cas12a Mediated Multiplexable and Portable Detection Platform for GII Genotype Porcine Epidemic Diarrhoea Virus Rapid Diagnosis. Front. Microbiol. 2022, 13, 920801. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Li, L.; Zhao, Y.; Liu, Y.; Liu, C.; Wang, Q.; Dong, Y.; Wang, S.; Chi, T.; Song, F.; et al. Clinical Validation of Two Recombinase-Based Isothermal Amplification Assays (RPA/RAA) for the Rapid Detection of African Swine Fever Virus. Front. Microbiol. 2020, 11, 1696. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef]
- Zhong, M.; Liu, J.; Wu, J.; Li, J.; Luo, N.; Zhu, C.; Liu, R.; Xia, Q.; Ju, H. Proximity sequence enhanced CRISPR-Cas12a connected through hybridization chain reaction for sensitive biosensing of dengue virus. Sens. Actuators B Chem. 2022, 366, 132011. [Google Scholar] [CrossRef]
- Song, D.; Han, X.; Xu, W.; Liu, J.; Zhuo, Y.; Zhu, A.; Long, F. Target nucleic acid amplification-free detection of Escherichia coli O157:H7 by CRISPR/Cas12a and hybridization chain reaction based on an evanescent wave fluorescence biosensor. Sens. Actuators B Chem. 2022, 376, 133005. [Google Scholar] [CrossRef]
- Lin, S.; Lin, Y.; Wu, J.; Li, G.; Wu, X.; Luo, N.; Li, W.; Zhu, C.; Liu, R.; Xu, Q.; et al. A ratiometric fluorescent biosensor for rapid detection of Burkholderia pseudomallei by dual CRISPR/Cas12a trans-cleavage assisted signal enhancement. Sens. Actuators B Chem. 2022, 379, 133204. [Google Scholar] [CrossRef]
- Huang, S.; Wang, X.; Chen, X.; Liu, X.; Xu, Q.; Zhang, L.; Huang, G.; Wu, J. Rapid and sensitive detection of Pseudomonas aeruginosa by isothermal amplification combined with Cas12a-mediated detection. Sci. Rep. 2023, 13, 19199. [Google Scholar] [CrossRef]
- Li, C.; Wu, Y.; Chen, Y.; Xu, C.; Yao, H.; Yu, W.; Wang, Z.; Du, X. Violet phosphorene nanosheets coupled with CRISPR/Cas12a in a biosensor with a low background signal for onsite detection of tigecycline-resistant hypervirulent Klebsiella pneumoniae. Sens. Actuators B Chem. 2023, 395, 134509. [Google Scholar] [CrossRef]
- Xu, D.; Zeng, H.; Wu, W.; Liu, H.; Wang, J. Isothermal Amplification and CRISPR/Cas12a-System-Based Assay for Rapid, Sensitive and Visual Detection of Staphylococcus aureus. Foods 2023, 12, 4432. [Google Scholar] [CrossRef]
- Yu, L.; Lan, H.; Zhang, Y.; Yi, H.; Shu, W.; Cui, K.; He, W.; Chen, M.; Huang, Q.; Li, L.; et al. A novel CRISPR/Cas12a biosensor for sensitive detection of Helicobacter pylori from clinical patients. Sens. Actuators B Chem. 2024, 412, 135818. [Google Scholar] [CrossRef]
- Hao, J.; Xie, L.; Yang, T.; Huo, Z.; Liu, G.; Liu, Y.; Xiong, W.; Zeng, Z. Naked-eye on-site detection platform for Pasteurella multocida based on the CRISPR-Cas12a system coupled with recombinase polymerase amplification. Talanta 2023, 255, 124220. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, P.; Nanaware, N.S.; Behera, S.P.; Kulkarni, S.; Deval, H.; Kumar, R.; Dwivedi, G.R.; Kant, R.; Singh, R. CRISPR/Cas12a-Based Detection Platform for Early and Rapid Diagnosis of Scrub Typhus. Biosensors 2023, 13, 12. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Lin, Z.; Huang, X.; Lu, J.; Zhou, Y.; Zheng, L.; Lou, Y. Rapid and Sensitive Detection of Vibrio vulnificus Using CRISPR/Cas12a Combined With a Recombinase-Aided Amplification Assay. Front. Microbiol. 2021, 12, 767315. [Google Scholar] [CrossRef] [PubMed]
- Jirawannaporn, S.; Limothai, U.; Tachaboon, S.; Dinhuzen, J.; Kiatamornrak, P.; Chaisuriyong, W.; Srisawat, N. The combination of RPA-CRISPR/Cas12a and Leptospira IgM RDT enhances the early detection of leptospirosis. PLOS Neglected Trop. Dis. 2023, 17, e0011596. [Google Scholar] [CrossRef]
- Singh, M.; Misra, C.S.; Bindal, G.; Rangu, S.S.; Rath, D. CRISPR-Cas12a assisted specific detection of mpox virus. J. Med. Virol. 2023, 95, e28974. [Google Scholar] [CrossRef]
- Low, S.J.; O’Neill, M.T.; Kerry, W.J.; Krysiak, M.; Papadakis, G.; Whitehead, L.W.; Savic, I.; Prestedge, J.; Williams, L.; Cooney, J.P.; et al. Rapid detection of monkeypox virus using a CRISPR-Cas12a mediated assay: A laboratory validation and evaluation study. Lancet Microbe 2023, 4, e800–e810. [Google Scholar] [CrossRef]
- Tu, Q.; Cao, X.; Ling, C.; Xiang, L.; Yang, P.; Huang, S. Point-of-care detection of Neisseria gonorrhoeae based on RPA-CRISPR/Cas12a. AMB Express 2023, 13, 50. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, S.; Ma, Y.; Li, Y.; Deng, G.; Shi, J.; Wang, X. Rapid detection of avian influenza virus based on CRISPR-Cas12a. Virol. J. 2023, 20, 261. [Google Scholar] [CrossRef]
- Ali, Z.; Sánchez, E.; Tehseen, M.; Mahas, A.; Marsic, T.; Aman, R.; Rao, G.S.; Alhamlan, F.S.; Alsanea, M.; Alqahtani, A.A.; et al. Bio-SCAN: A CRISPR/dCas9-Based Lateral Flow Assay for Rapid, Specific, and Sensitive Detection of SARS-CoV-2. ACS Synth. Biol. 2022, 11, 406–419. [Google Scholar] [CrossRef]
- Awan, M.J.A.; Amin, I.; Mansoor, S. CRISPR-Cas12c: A noncleaving DNA binder with minimal PAM requirement. Trends Biotechnol. 2022, 40, 1141–1143. [Google Scholar] [CrossRef]
- Taglauer, E.S.; Wilkins-Haug, L.; Bianchi, D.W. Review: Cell-free fetal DNA in the maternal circulation as an indication of placental health and disease. Placenta 2014, 35, S64–S68. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-X.; Zhang, C.; Lu, S.; Tong, X.; Zhang, K.; Yin, H.; Zhang, Y. Cas12a-based one-pot SNP detection with high accuracy. Cell Insight 2023, 2, 100080. [Google Scholar] [CrossRef] [PubMed]
- Kohabir, K.A.V.; Nooi, L.O.; Brink, A.; Brakenhoff, R.H.; Sistermans, E.A.; Wolthuis, R.M.F. In Vitro CRISPR-Cas12a-Based Detection of Cancer-Associated TP53 Hotspot Mutations Beyond the crRNA Seed Region. CRISPR J. 2023, 6, 127–139. [Google Scholar] [CrossRef]
- Yang, W.; Tao, D.; Xu, B.; Zheng, Y.; Zhao, S. Detecting Melanocortin 1 Receptor Gene’s SNPs by CRISPR/enAsCas12a. Genes 2023, 14, 394. [Google Scholar] [CrossRef]
- Feng, Z.; Kong, D.; Jin, W.; He, K.; Zhao, J.; Liu, B.; Xu, H.; Yu, X.; Feng, S. Rapid detection of isocitrate dehydrogenase 1 mutation status in glioma based on Crispr-Cas12a. Sci. Rep. 2023, 13, 5748. [Google Scholar] [CrossRef]
- Xu, W.; Peng, J.; Guo, C.; Chai, Y.; Zhou, H.; Wang, J.; Li, X. Rapid and ultra-sensitive early detection of cervical cancer using CRISPR/Cas12-based assay based on methylated SEPT9. Sens. Actuators B Chem. 2023, 379, 133231. [Google Scholar] [CrossRef]
- Kaminski, M.M.; Abudayyeh, O.O.; Gootenberg, J.S.; Zhang, F.; Collins, J.J. CRISPR-based diagnostics. Nat. Biomed. Eng. 2021, 5, 643–656. [Google Scholar] [CrossRef]
- Brokowski, C.; Adli, M. CRISPR ethics: Moral considerations for applications of a powerful tool. J. Mol. Biol. 2019, 431, 88–101. [Google Scholar] [CrossRef]
- Chirinskaite, A.V.; Zelinsky, A.A.; Sopova, J.V.; Leonova, E.I. Development of the Cas12a-based microdeletion and microinsertion detection system. Ecol. Genet. 2023, 21, 20–21. [Google Scholar] [CrossRef]
- Zuo, E.; Huo, X.; Yao, X.; Hu, X.; Sun, Y.; Yin, J.; He, B.; Wang, X.; Shi, L.; Ping, J.; et al. CRISPR/Cas9-mediated targeted chromosome elimination. Genome Biol. 2017, 18, 224. [Google Scholar] [CrossRef]
- Cullot, G.; Boutin, J.; Toutain, J.; Prat, F.; Pennamen, P.; Rooryck, C.; Teichmann, M.; Rousseau, E.; Lamrissi-Garcia, I.; Guyonnet-Duperat, V.; et al. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat. Commun. 2019, 10, 1136. [Google Scholar] [CrossRef] [PubMed]
- Tai, D.J.C.; Ragavendran, A.; Manavalan, P.; Stortchevoi, A.; Seabra, C.M.; Erdin, S.; Collins, R.L.; Blumenthal, I.; Chen, X.; Shen, Y.; et al. Engineering microdeletions and microduplications by targeting segmental duplications with CRISPR. Nat. Neurosci. 2016, 19, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, C.A.; Brandes, N.; Bueno, R.; Trinidad, M.; Mazumder, T.; Yu, B.; Hwang, B.; Chang, C.; Liu, J.; Sun, Y.; et al. Mitigation of chromosome loss in clinical CRISPR-Cas9-engineered T cells. Cell 2023, 186, 4567–4582.e20. [Google Scholar] [CrossRef] [PubMed]
- Mandrioli, D.; Belpoggi, F.; Silbergeld, E.K.; Perry, M.J. Aneuploidy: A common and early evidence-based biomarker for carcinogens and reproductive toxicants. Environ. Health 2016, 15, 97. [Google Scholar] [CrossRef]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019, 14, 2986–3012. [Google Scholar] [CrossRef]
- D’Hulst, C.; Parvanova, I.; Tomoiaga, D.; Sapar, M.L.; Feinstein, P. Fast quantitative real-time PCR-based screening for common chromosomal aneuploidies in mouse embryonic stem cells. Stem Cell Rep. 2013, 1, 350–359. [Google Scholar] [CrossRef]
- Hong, S.; Lee, S.M.; Oh, S.; Kim, S.Y.; Jung, Y.M.; Kim, S.M.; Lee, S.M.; Oh, S.; Kim, S.Y.; Jung, Y.M.; et al. Simple and rapid detection of common fetal aneuploidies using peptide nucleic acid probe-based real-time polymerase chain reaction. Sci. Rep. 2022, 12, 150. [Google Scholar] [CrossRef]
- Bianchi, D.W.; Chudova, D.; Sehnert, A.J.; Bhatt, S.; Murray, K.; Prosen, T.L.; Garber, J.E.; Wilkins-Haug, L.; Vora, N.L.; Warsof, S.; et al. Noninvasive Prenatal Testing and Incidental Detection of Occult Maternal Malignancies. JAMA 2015, 314, 162–169. [Google Scholar] [CrossRef]
- Chen, C.P.; Chern, S.R.; Wang, L.K.; Chen, S.W.; Wu, F.T.; Town, D.D.; Wang, W. Detection of paternal origin of fetal trisomy 18 in a pregnancy conceived by assisted reproductive technology and in vitro fertilization. Taiwan. J. Obstet. Gynecol. 2020, 59, 607–609. [Google Scholar] [CrossRef]
- Boroujen, P.T.; Nemati, M.; Naderipour, F. Two case reports of rare Down’s syndrome during in vitro fertilization and embryo transfer (IVF-ET). Medicine 2025, 104, e37872. [Google Scholar] [CrossRef]
- Jafri, S.K.; Harman, K.E. Neurocognitive abilities in individuals with Down syndrome-a narrative review. Turk. J. Pediatr. 2020, 62, 897–905. [Google Scholar] [CrossRef]
- Gullo, G.; Scaglione, M.; Cucinella, G.; Perino, A.; Chiantera, V.; D’Anna, R.; Laganà, A.S.; Buzzaccarini, S. Impact of assisted reproduction techniques on the neuro-psycho-motor outcome of newborns: A critical appraisal. J. Obstet. Gynaecol. 2022, 42, 2583–2587. [Google Scholar] [CrossRef] [PubMed]
- Zaami, S.; Gullo, G.; Varone, M.C.; Umani Ronchi, F.; Vergallo, G.M. From the maternal uterus to the “uterus device”? Ethical and scientific considerations on partial ectogenesis. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 7354–7362. Available online: https://www.europeanreview.org/wp/wp-content/uploads/7354-7362.pdf (accessed on 22 March 2025). [PubMed]
- Zhang, X.-H.; Tee, L.Y.; Wang, X.-G.; Huang, Q.-S.; Yang, S.-H. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol. Ther. Nucleic Acids 2015, 4, e264. [Google Scholar] [CrossRef]
- Liu, M.-S.; Gong, S.; Yu, H.-H.; Jung, K.; Johnson, K.A.; Taylor, D.W. Engineered CRISPR/Cas9 enzymes improve discrimination by slowing DNA cleavage to allow release of off-target DNA. Nat. Commun. 2020, 11, 3576. [Google Scholar] [CrossRef]
- Tsai, S.Q.; Wyvekens, N.; Khayter, C.; Foden, J.A.; Thapar, V.; Reyon, D.; Goodwin, M.J.; Aryee, M.J.; Joung, J.K. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014, 32, 569–576. [Google Scholar] [CrossRef]
- Pan, A.; Kraschel, K.L. CRISPR diagnostics: Underappreciated uses in perinatology. Semin. Perinatol. 2018, 42, 525–530. [Google Scholar] [CrossRef]

| Protein Family | Cas9 | Cas12 | Cas13 | Cas14 |
|---|---|---|---|---|
| Bacteria | Streptococcus pyogenes | Lachnospiraceae | Leptotrichia shahii | DPANN (Archea) |
| Nuclease domine | RUVC&HNH | RUVC | RUVC | RUVC |
| Guide RNA length | 17–150 nt | 41–44 nt | 50–64 nt | −140 nt |
| PAM Sequence | NGG | TTTN | Protospacer flanking site | TTTA |
| Cleavage site | 3–4 nt upstream of PAM | −18 bases 3 of PAM | 3′ end of the target RNA | 3′ end of the target region |
| Target nucleic acid | DNA | DNA | RNA | DNA |
| Termini of cleavage | Blunt end | Overhang ends | Sticky end | Sticky end |
| S.No | Type of Cas | Title | Infectious Disease | Reference |
|---|---|---|---|---|
| 1 | Cas12a | A new method for the detection of Mycobacterium tuberculosis based on the CRISPR/Cas system | Tuberculosis | Zhang et al. [74] |
| 2 | Cas12a | Clustered regularly interspaced short palindromic Repeat/Cas12a mediated multiplexable and portable detection platform for GII genotype Porcine Epidemic Diarrhoea Virus Rapid diagnosis | Genotype Porcine Epidemic Diarrhea Virus | Qian et al. [75] |
| 3 | Cas12a | Clinical Validation of Two Recombinase-Based Isothermal Amplification Assays (RPA/RAA) for the Rapid Detection of African Swine Fever Virus | African Swine Fever Virus | Fan et al. [76] |
| 4 | Cas12a | CRISPR-Cas12-based detection of SARS-CoV-2 | COVID-19 | Beroughton et al. [77] |
| 5 | Cas12a | Proximity sequence enhanced CRISPR-Cas12a connected through hybridization chain reaction for sensitive biosensing of dengue virus | Dengue | Zhong et al. [78] |
| 6 | Cas12a | Target nucleic acid amplification-free detection of Escherichia coli O157:H7 by CRISPR/Cas12a and hybridization chain reaction based on an evanescent wave fluorescence biosensor | E. coli | Song et al. [79] |
| 7 | Cas12a | A ratiometric fluorescent biosensor for rapid detection of Burkholderia pseudomallei by dual CRISPR/Cas12a trans-cleavage assisted signal enhancement | Burkholderia pseudomallei | Lin et al. [80] |
| 8 | Cas12a | Rapid and sensitive detection of Pseudomonas aeruginosa by isothermal amplification combined with Cas12a-mediated detection | Pseudomonas aeruginosa | Huang et al. [81] |
| 9 | Cas12a | Violet phosphorene nanosheets coupled with CRISPR/Cas12a in a biosensor with a low background signal for onsite detection of tigecycline-resistant hypervirulent Klebsiella pneumoniae | Klebsiella pneumoniae | Li et al. [82] |
| 10 | Cas12a | Isothermal Amplification and CRISPR/Cas12a-System-Based Assay for Rapid, Sensitive and Visual Detection of Staphylococcus aureus | Staphylococcus aureus | Xu et al. [83] |
| 11 | Cas12a | A novel CRISPR/Cas12a biosensor for sensitive detection of Helicobacter pylori from clinical patients | Helicobacter pylori | Yu et al. [84] |
| 12 | Cas 12a | Naked-eye on-site detection platform for Pasteurella multocida based on the CRISPR-Cas12a system coupled with recombinase polymerase amplification | Pasteurella multocida | Hao et al. [85] |
| 13 | Cas 12a | CRISPR/Cas12a-Based Detection Platform for Early and Rapid Diagnosis of Scrub Typhus | Scrub Typhus | Bharadwaj et al. [86] |
| 14 | Cas 12a | Rapid and Sensitive Detection of Vibrio vulnificus Using CRISPR/Cas12a Combined with a Recombinase-Aided Amplification Assay | Vibrio vulnificus | Xiao et al. [87] |
| 15 | Cas 12a | The combination of RPA-CRISPR/Cas12a and Leptospira IgM RDT enhances the early detection of leptospirosis | Leptospirosis | Jirawannaporen et al. [88] |
| 16 | Cas 12a | CRISPR-Cas12a assisted specific detection of mpox virus | Mpox virus | Singh et al. [89] |
| 17 | Cas 12a | CRISPR-Cas12a-Mediated Hue-Recognition Lateral Flow Assay for Point-of-Need Detection of Salmonella | Salmonella | Yuan et al. [69] |
| 18 | Cas 12a | Rapid detection of monkeypox virus using a CRISPR Cas12a mediated assay: a laboratory validation and evaluation study | Monkeypox virus | Low et al. [90] |
| 19 | Cas 12a | Point-of-care detection of Neisseria gonorrhoeae based on RPA-CRISPR/Cas12a | Neisseria gonorrhoeae | Tu et al. [91] |
| 20 | Cas 12a | Rapid detection of avian influenza virus based on CRISPR-Cas12a | Avian influenza virus | Zhou et al. [92] |
| S.No | Type of Cas | Title | Genes | Reference |
|---|---|---|---|---|
| 1 | Cas12a | Cas12a-based one-pot SNP detection with high accuracy | CYP2C19 | Zhang et al. [96] |
| 2 | Cas12a | In Vitro CRISPR-Cas12a-Based Detection of Cancer-Associated TP53 Hotspot Mutations Beyond the crRNA Seed Region | Cancer-Associated TP53 Hotspot | Kohabir et al. [97] |
| 3 | Cas12a | Detecting Melanocortin 1 Receptor Gene’s SNPs by CRISPR/enAsCas12a | Melanocortin 1 | Yang et al. [98] |
| 4 | Cas12a | Rapid detection of isocitrate dehydrogenase 1 mutation status in glioma based on Crispr-Cas12a | Isocitrate dehydrogenase | Feng et al. [99] |
| 5 | Cas12a | Rapid and ultra-sensitive early detection of cervical cancer using CRISPR/Cas12-based assay based on methylated SEPT9 | SEPT9 | Xu et al. [100] |
| S.No | Types of Cas | Title | Chromosome /Locus | Reference |
|---|---|---|---|---|
| 1 | Cas12a | Development of the Cas12a-based microdeletion and micro insertion detection system | Pde6b-KO mice and Grin3A-KO mice | Chirinskaite et al. [103] |
| 2 | Cas9 | CRISPR/Cas9-mediated targeted chromosome elimination | TKNEO, XIST Transgene | Zuo et al. [104] |
| 3 | Cas9 | CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations | UROS locus to model and correct congenital erythropoietic porphyria | Cullot et al. [105] |
| 4 | Cas12a | Engineering microdeletions and microduplications by targeting segmental duplications with CRISPR | 16p11.2 and 15q13.3 | Tai et al. [106] |
| 5 | Cas12a | Mitigation of chromosome loss in clinical CRISPR-Cas9-engineered T cells | TRAC locus | Tsuchida et al. [107] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the Lithuanian University of Health Sciences. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ganesh, I.; Karthiga, I.; Murugan, M.; Rangarajalu, K.; Ballambattu, V.B.; Ravikumar, S. CRISPR/Cas-Based Prenatal Screening for Aneuploidy: Challenges and Opportunities for Early Diagnosis. Medicina 2025, 61, 610. https://doi.org/10.3390/medicina61040610
Ganesh I, Karthiga I, Murugan M, Rangarajalu K, Ballambattu VB, Ravikumar S. CRISPR/Cas-Based Prenatal Screening for Aneuploidy: Challenges and Opportunities for Early Diagnosis. Medicina. 2025; 61(4):610. https://doi.org/10.3390/medicina61040610
Chicago/Turabian StyleGanesh, Irisappan, Ilangovan Karthiga, Manoranjani Murugan, Kumar Rangarajalu, Vishnu Bhat Ballambattu, and Sambandam Ravikumar. 2025. "CRISPR/Cas-Based Prenatal Screening for Aneuploidy: Challenges and Opportunities for Early Diagnosis" Medicina 61, no. 4: 610. https://doi.org/10.3390/medicina61040610
APA StyleGanesh, I., Karthiga, I., Murugan, M., Rangarajalu, K., Ballambattu, V. B., & Ravikumar, S. (2025). CRISPR/Cas-Based Prenatal Screening for Aneuploidy: Challenges and Opportunities for Early Diagnosis. Medicina, 61(4), 610. https://doi.org/10.3390/medicina61040610







