Abstract
Omega-3 fatty acids are essential fatty acids that are not synthesised by the human body and have been linked with the prevention of chronic illnesses such as cardiovascular and neurodegenerative diseases. However, the current dietary habits of the majority of the population include lower omega-3 content compared to omega-6, which does not promote good health. To overcome this, pharmaceutical and nutraceutical companies aim to produce omega-3-fortified foods. For this purpose, various approaches have been employed to obtain omega-3 concentrates from sources such as fish and algal oil with higher amounts of eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA). Among these techniques, enzymatic enrichment using lipase enzymes has gained tremendous interest as it is low in capital cost and simple in operation. Microorganism-derived lipases are preferred as they are easily produced due to their higher growth rate, and they hold the ability to be manipulated using genetic modification. This review aims to highlight the recent studies that have been carried out using marine lipases for the enrichment of omega-3, to provide insight into future directions. Overall, the covalent bond-based lipase immobilization to various support materials appears most promising; however, greener and less expensive options need to be strengthened.
1. Introduction
Marine environments cover the majority (70%) of the total Earth’s biosphere, representing an exhaustive and wide range of diverse groups of flora and fauna that have developed a unique way to adapt to extreme environmental conditions, producing novel metabolites [1]. As such, marine ecosystems are considered a renewable resource for various commercially valuable components such as enzymes, vitamins, antibiotics, drugs, bio-emulsifiers, biosurfactants, and biofuels, which have significant applications in biotechnology and biomedical industries [2,3,4]. Similarly, the biomolecules, carotenoids, lipids, saponins, phenolics, and polysaccharides obtained from marine sources also have great biological value in functional foods and nutraceuticals [5]. In particular, over the past few decades, marine microorganisms have attracted attention towards unexplored marine enzymes and their application in food processing as they are more stable compared to those from animal and plant origins [2,5]. Various studies have identified the special function of lipase enzymes from marine fungi, bacteria, actinomycetes and other microorganisms, which have been utilised in different industries such as leather, textile, pharmaceuticals, food, biodiesel, agrochemical, and cosmetic industries due to their properties and ability to catalyse various biotechnological reactions, and the diversity of microbial, animal, and plant genes that encode lipases [6,7,8,9,10]. More recently, the use of lipases has proven effective in concentrating polyunsaturated fatty acids (PUFAs) [11], which is significant as the global demand for essential omega-3 was valued at USD 7.5 billion in 2024 and is expected to increase to USD 14.1 billion in 2029 [12] as a result of increased health awareness and associated health benefits.
Omega-3 and omega-6 fatty acids are the two major classes of PUFAs, which are named based on the position of the first double bond from the methylated end of the fatty acid chain [13,14]. Omega-3 PUFAs are considered essential fatty acids as they cannot be synthesised in the human body or that of animals [14,15] and must be consumed [16]. Recently, the change in diets consisting of increased fast-food intake has overturned the balance and dramatically increased saturated fatty acid consumption as compared to PUFAs. The low intake of essential dietary PUFAs is thought to be one of the main reasons for the increased risk of cardiovascular diseases, Alzheimer’s, and depression [17,18]. Omega-3 PUFAs have a vital role in maintaining the overall health and wellbeing of the human body and in reducing the risk of diseases [19]. Among the essential fatty acids, alpha-linolenic acid (ALA, 18:3, n-3), linoleic acid (18:2, n-6), and Docosahexaenoic acid (DHA, 22:6, n-3) are very important polyunsaturated fatty acids for maintaining functions in mammals [20]. Research performed using alpha-linolenic acid showed that an increased dietary intake of ALA assisted in reducing blood pressure [21]. In addition, an epidemiological study carried out in 2004 suggested that fish oil supplemented with omega-3 (DHA and EPA) demonstrated potential in reducing the risk of heart attacks and cardiovascular-related deaths [22]. Omega-3 supplementation in diets has also been investigated for minimizing the possibility and treatment of breast cancer in women from different countries [23]. Furthermore, the various health benefits of dietary intake of omega-3 fatty acids from different sources (plants, fish, and microbial)—relating to cardiovascular diseases, hormonal balance, inflammatory responses in different developmental stages of life, and neurocognitive and visual development in the early stages—have been discussed by Calder [24]. As such, the regulation of incorporating DHA in infant foods was made compulsory in almost every country of the world [25,26].
Furthermore, the health benefits associated with omega-3 PUFAs, and the low quantities available in diets consumed today, have increased the demand for functional foods’. This has elevated the trend of food fortification, adding small quantities of omega-3 fatty acids in dairy, baked goods, dressings, spreads, meat products, and chocolates [27]. As such, nutraceutical and food industries have become more interested in developing omega-3-enriched foods, which require omega-3 PUFAs in a concentrated form. There are various strategies such as molecular distillation, urea complexation, supercritical fluid extraction, and enzymatic enrichment employed to concentrate omega-3 PUFAs [28,29,30,31].
Among these, enzymatic enrichment is a highly studied alternative as it requires less energy and can be performed at lower temperatures, which prevents omega-3 oxidation and degradation [28]. Marine lipases are increasingly used for this purpose as they possess fatty acid selectivity that allows them to hydrolyse saturated and monounsaturated fatty acids [32]. However, the major drawback of using free enzymes is the loss of enzymatic activity and stability due to denaturation and its limited reuse. Marine microorganisms have been considered recently as preferred sources for lipase extraction due to their specific characteristics; however, further studies are required to identify if lipases from marine microorganisms contain any specific characteristics such as improved stability and ease of large-scale production [8]. For enhancing the stability and activity of enzymes, immobilization techniques can be carried out [33,34,35]. The aim of this review is to provide inference on the recent advancements made in utilising marine lipases as an effective means for enriching omega-3 PUFAs, and future research directions.
2. Lipases and Their Sources
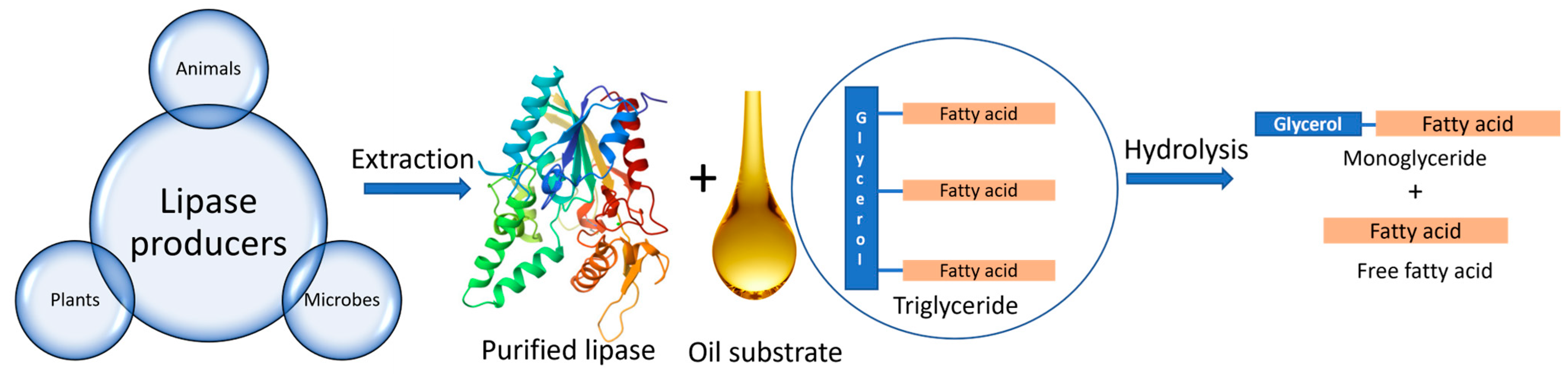
Lipases are basically triacylglycerol ester hydrolases that have the ability to hydrolyse fats and oils [36]. Lipases cleave ester bonds present in triglycerides to form monoglycerides and free fatty acids [37], as seen in Figure 1. Lipases can also catalyse other types of reactions such as esterification, transesterification, interesterification, and amino lysis [38]. Their molecular size ranges between 20 and 60 kDa and comprises 270 to 641 amino acids [38]. Lipases possess a unique property of interfacial activation, which allows the catalysis of lipids at the lipid–water interface. Lipases contain a helical oligopeptide unit referred to as Lid, which assists in activating the active site of the enzyme under specific conditions such as in the presence of micellar substrates [39]. Moreover, the specificity of lipases also depends on the size and hydrophobicity of the catalytic beads. The active site comprises a catalytic triad of three amino acids: serine, histidine, and aspartate [40]. In the active site, there are four substrate-binding pockets for triglycerides that can accommodate fatty acids at the sn-1, sn-2, and sn-3 positions [38]. The selectivity of the lipases can be enhanced by using acylating agents, organic solvents, and additives, such as ethanol, and by changing the operating conditions such as temperature [41]. Lipases can be obtained from various sources such as plants, animals, and microorganisms.
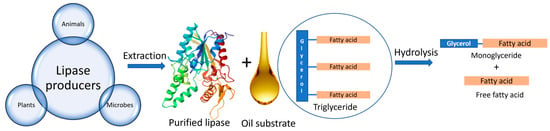
Figure 1.
Various sources of lipase enzyme and its mechanism of action.
2.1. Plant Lipases
Plant lipases are mostly present in seeds like sunflower, castor bean, almond, black cumin, and sesame [42]; moreover, fruit waste from orange, mango, papaya, and palm are also reported as good sources for lipase [43]. Plant lipases are used for various pharmaceutical purposes, such as Carica papaya latex, which is used in the production of canola phylosterol oleate esters that can be further utilised as cholesterol-lowering agents to reduce the risk of coronary disease [44]. However, there is a limitation of lower production of enzymes from plants, which makes purification procedures complicated and prone to activity loss [43].
2.2. Animal Lipases
Animal-derived lipase sources include mammals, insects, and fish. Animals like pigs, cattle, hogs, and sheep are used to extract pancreatic and progastric lipases [45]. Animal-sourced lipases such as pancreatic lipase have been extensively used for catalysing primary alcohol esters hydrolysis [46]. The lipases from animals were also used in the dairy industry for developing flavour in cheese and other products. For this reason, lipases from porcine pancreas have been used to induce flavour in cheddar cheese [47]. However, due to its low stability and complex recovery procedures, its use at a commercial level has been limited [48,49].
2.3. Marine Lipases from Various Microorganisms
Microorganism-based lipases from bacteria and fungi (Table 1) have wider applications in industry and are the most studied source of lipases due to their variety of catalytic specificity, simple genetic modifications, high growth rate, and the ability to grow in laboratory-controlled conditions [50]. The first microbial lipases were identified in bacteria Bacillus prodigiosus, B.pyococyaneus, and B.fluoroscens in 1901 [51]. Later on, various fungi and yeast were also found to synthesise lipases [52]. Microorganism-based lipases hold great commercial value due to their better stability, higher selectivity, and broad substrate specificity [53].

Table 1.
Lipase-producing microorganisms and their lipolytic activity.
2.3.1. Microbial Lipases
Marine microbes have recently achieved increasing attention as a source of bioactive metabolites that have various biomedical potential [74]. Marine microbes have the ability to adapt to various extreme environmental conditions and possess high genetic plasticity that can positively influence compound and secondary metabolite production [75]. Various marine bacteria and yeasts are used to extract cytotoxic compounds, enzyme inhibitors, and anti-inflammatory agents which have broad clinical importance [76].
The marine yeasts, Candida antartica and Candida rugosa, are the most widely used sources of lipases and are categorised as GRAS (generally regarded as safe). The GRAS status makes these lipases more suitable for various applications in food and clinical industries, such as flavouring agents and for the production of antioxidants [77]. Nine yeast strains—Candida intermedia YA01a, Pichia guilliermondii N12c, Candida parapsilosis 3eA2, Lodderomyces elongisporus YF12c, Candida quercitrusa JHSb, Candia rugosa wl8, Yarrowia lipolytica N9a, Rhodotorula mucilaginosa L10-2, and Aureobasidium pullulans HN2.3—were identified to produce stable lipases in the pH range of 6.0 to 8.5 and temperature range of 35–40 °C by Wang et al. [54]. C. rugosa lipases are sold by companies such as Sigma, Roche, and Amano in immobilized and lyophilic powder forms [78]. Various studies are carried out to improve the thermostability and activity of yeast-based lipases through immobilization, medium engineering, and protein engineering approaches [79,80]. The unique property of these lipases is their broad specificity towards long-chain triacylglycerols as they can hydrolyse shorter fatty acids at faster rates; therefore, they can be used for the enrichment of long-chain fatty acids [81].
Marine microorganisms are reported to have the ability to produce lipases with a varying range of lipolytic activity (Table 1). The marine bacteria, Oceanobacillus caeni, isolated from the east coast of India, has the unique property of being stable at a wide range of pH from 3 to 11 and temperatures of 10 to 70 °C [67]. Moreover, lipases from Bacillus sonorensis were studied for their efficacy as a detergent additive for the efficient removal of corn oil stains, where it was found that it was stable at a temperature ranging from 23 to 60 °C [68]. Furthermore, lipase from Bacillus pumilus B106, associated with the South China Sea Sponge Halichondria rugosa, has the appealing feature of tolerance towards high salinity, which is considered an important factor in producing biodiesel derived from marine organisms [82]. These studies suggest increased interest in lipase-producing marine bacteria in various industrial applications.
2.3.2. Microalgae Lipases
There are various other studies that provide evidence of marine microalgae as lipase producers. A genomic study performed on the microalgae Chlamydomonas reinhardtii and Chlorella vulgaris revealed that they have lipase-encoding genes. In the case of C. reinhardtii, the galactoglycerolipid lipase-encoding gene CrLIP1 was identified using E. coli as a protein expression system and was obtained in purified form [83]. While Chlorella vulgaris demonstrated a total of 14 lipase-encoding genes, characterised using sequence homologies and bioinformatics tools, further attempts to extract and purify the lipases were not performed [84]. Moreover, Savvidou et al. [72] confirmed the presence of thermostable lipase enzymes in Nannochloropsis oceanica for the first time and were successful in extracting the lipases from the cell surface and cell debris fraction [72]. Similarly, extracellular lipases produced from Botryococcus sudeticus, a phototrophic microalgae, were purified and reported to exhibit various properties such as resistance towards a broad range of temperatures, pH tolerance, and showed variation in specificities for different substrates [73]. However, the presence of easily available commercial lipases on the market has limited investigations into microalgae-based lipase sources [84].
3. Lipase Commercial Market and Applications
3.1. The Lipase Commercial Market
After protease and carbohydrates, lipases make up the third-largest group of enzymes, based on their market value [85,86,87]. In 2019, the global market size for microbial lipases was valued at USD 349.8 million and is expected to maintain a compound annual growth rate (CARG) of 5.2%, reaching USD 428.6 million by 2025 [88]. This demonstrates the increased demand for lipases globally, and according to a review performed in 2021, the animal- and microbial-based lipase segments in 2020 held the largest market shares of 26.6% and 61.64%, respectively [89]. As of 2021, North America was the largest market producer of lipases, making up 38% of the total shares worth USD 263 million, followed by Europe, which accounted for 31% of global market share [90].
3.2. Marine Lipase Applications
The marine ecosystem is one of the major sources of biodiversity, especially due to the harsh conditions underwater, and various microorganisms have unique and sophisticated genetics and characteristics with the ability to produce bioactive compounds [91]. Recently, marine fungi have gained increased attention for lipase production, as they produce extracellular lipases, which have reduced fat and oil contents by 92% in oil-polluted effluent [86]. In 2014, alkaline-stable lipase enzymes were used in milk flavour production [92]. They are also used in the acceleration of cheese ripening, to modify flavours in food through the synthesis of ester short-chain fatty acids and alcohols [7,93]. They are used in food industries to process food such as fruits, meat, beer, and milk products, and to improve the flavour of dairy products [94]. Similarly, lipases are used in the textile industry, to help in the removal of lubricants to provide a fabric with high absorbency for improved dyeing [95]. They are also used for the production of paper and pulp by hydrolysing the wood triglycerides or waxes [93]. Lipases have their importance in biofuel production by transesterification of fats and vegetable oils with short alcohol chains [85]. Lipases are environmentally friendly as they can be used in detergents that allow lower wash temperatures with less toxic residues, fewer chemicals in the detergents, no threat to aquatic life, and no adverse effects on wastewater [96]. In addition, lipase solvents or detergents remove fats and greases from leather, which makes it soft and easy to use for further processing [97]. Furthermore, lipases also play a role in the pharmaceutical industry as they have the ability to prevent epimerization, rearrangement, racemization, and isomerization [96,98]. They also have several applications in the medical industry and have been utilised for their therapeutic and diagnostic uses in digestive tract disorder, pancreatic damage, and as a measuring tool for serum lipid profiles [97].
4. Omega-3 Enrichment Techniques
There are various methods used for concentrating omega-3 PUFAs from fish and algal oils. Most industries use fish oils like sardines to obtain omega-3 PUFA concentrates. Before concentrating omega-3 PUFAs at an industrial level, fat-soluble contaminants are removed by either adsorption processes or chromatographic methods [99]. This is then followed by the removal of the glycerol backbone in the presence of an alkaline catalyst to convert it into ethyl esters or free fatty acids. After removing the glycerol backbone, the enrichment process can be carried out using urea precipitation, supercritical fluid extraction, molecular distillation, and enzymatic enrichment, as seen in Table 2.

Table 2.
Omega-3 PUFA-enrichment techniques.
4.1. Urea Precipitation
Urea precipitation is a method that is based on the property of urea crystals to form complexes with straight-chain and monounsaturated fatty acids (MUFAs). To achieve this, urea is first dissolved into an organic solvent such as methanol or ethanol, which is allowed to cool down in the presence of decontaminated oil. While cooling, the formation of urea crystals occurs, which in turn traps the saturated and monounsaturated fatty acids allowing for the separation of PUFAs. Urea can be filtered to obtain a concentrated form of PUFAs [103]. This process can be helpful in obtaining omega-3 PUFA concentrates with 45–60% EPA (Eicosapetanoic acid) plus DHA (Docosahexanoic caid) content; however, the major bottleneck of this process is dealing with flammable solvents in larger volumes and the disposal of urea saturated with fatty acids, which make it a highly expensive process [99].
4.2. Supercritical Fluid Extraction
This method is primarily known for its ability to selectively separate Methyl stearate (C18), Methyl eicosapentanoate (C20), and Methyl docosahexanoate (C22) fatty acids using the supercritical form of carbon dioxide (SC-CO2), which can easily solubilize fatty acids with low molecular weight [104]. The additional and most important advantage of this technique is that SC-CO2 is non-toxic, non-flammable, and a clean solvent alternative [105]. DHA with high purity can be enriched at a ratio of 60% using this method [106]; however, carbon dioxide needs to be compressed to a pressure of more than 73 bar and temperatures of 32 °C to reach the supercritical state [100]. Moreover, SC-CO2 is pumped through a vertical column from the bottom to the top under high pressure, along with the fish oil ethyl ester continuously in the counter-current direction of the CO2. Thus, the major drawback of this technique is the high capital cost associated with the pressure-generating equipment [99].
4.3. Molecular Distillation
Molecular distillation is the most commonly used technique in industry nowadays to obtain concentrated omega-3 PUFAs. This method is based on the principle of differences in molecular weight and boiling point of the different fatty acids under low pressure [32]. At lower pressures and higher temperatures ranging from 140 to 170 °C, EPA and DHA content can be enriched by up to 60%, as compared to the original content of 30% in fish oil; however, the high temperatures used in this technique lead to the loss of fatty acid saturation and causes hydrolysis, isomerization, and thermal degradation of omega-3 PUFAs at certain levels [101].
4.4. Enzymatic Enrichment Method
The utilisation of enzymes at an industrial level is a recent and more advantageous option compared to other alternatives discussed above due to its simplicity in processing and low capital requirements; moreover, it eliminates the need for using organic solvents and high temperatures that can deteriorate the quality of omega-3 PUFAs. These positive aspects of enzymatic enrichment have promoted the interest of researchers to make this technology more efficient by developing different strategies. The use of enzymes such as protease, exopeptidase, endopeptidase, and lipase has shown better enrichment of omega-3 PUFAs [102]. Among these, lipases can catalyse hydrolysis and esterification reactions with high selectivity towards omega-3 PUFAs. The selectivity of the lipases is due to the greater number of double bonds present in EPA and DHA, which cause steric hindernce that acts as a barrier for the enzyme denying the hydrolyses of these fatty acids [107]. Thus, using lipase-based hydrolysis, free-form fatty acids, monoacylglycerols, diacyglycerols, and triacylglycerols are formed, which can be further separated by solvent extraction method [108]. This property of lipase enzymes makes them a highly suitable alternative for omega-3 PUFA enrichment through a two-step enzymatic method; however, the major drawback is the enzymes’ nature of getting easily denatured and their limited reusability. To mitigate these problems, intensive research has been carried out by researchers to develop different immobilization strategies to improve the stability, activity, and reusability of lipase enzymes [109] (Section 5).
5. Marine Lipases in Enriching Omega-3
Lipases have the ability to selectively enrich EPA and DHA content as they have a partial selective ability towards chain length, position of the fatty acids, and cis-double bonds [110]. Conventionally, ethanol-based transesterification was used at an industrial level to produce concentrated PUFA esters [111]; however, this type of chemical transesterification can lead to deteriorating effects on the oxidative stability of oil [112]. Recently, various studies have carried out the enrichment of EPA and DHA from different oil sources, with the majority being fish oil using lipases, see Table 3.

Table 3.
Lipases derived from marine microorganisms and their PUFA-enrichment ability using different oil substrates.
In a study conducted by Yang et al. [113], the commercially available lipases AY “Amano” 400SD (free lipase from Candida cylindracea, 400 U/mg), AY “Amano” 30SD (free lipase from Candida cylindracea, 30 U/mg), AY “Amano” S (free lipase from Candida cylindracea, 30 U/mg), DF “Amano” 15 (free lipase from Rhizopus oryzea, 150 U/mg), G “Amano” 50 (free lipase from Aspergillus oryzae, 50 U/mg), and nonspecific Lipozyme 435 (immobilized lipase from Candida antarctica, 10 U/mg, particle form) were tested for their omega-3 enrichment specificity using tuna oil. It was concluded that AY “Amano” 400SD possesses the most effective selectivity to hydrolyse saturated fatty acids and monounsaturated fatty acids by increasing the omega-3 PUFA content from 34.3% to 57.7%, while the other lipases accounted for PUFA content increases from 34.3% to less than 50% [113].
Lipases from marine natural sources are also extensively studied for their enrichment ability. For instance, in the study of Baloch et al. [114], lipases were produced from three different yeast strains C. rugosa TISTR 5627, Y. lipolytica TISTR 5212, and P. guilliermondii TISTR 5142 and a comparison of their ability to concentrate EPA and DHA was made; it was reported that C. rugosa lipase had better enrichment ability compared to the lipases from the other two strains [114].
Interestingly, various researchers have also attempted to produce recombinant lipase with better properties compared to lipases obtained from natural sources. In a recent study, a recombinant LipB from Pseudomonas fluorescens was cloned into Bacillus subtillis. It was reported that the recombinant lipase had excellent properties such as solvent tolerance against acetonitrile, isopropanol, acetone, and DMSO; moreover, it was able to increase the PUFA concentration from 43.2% to 72.2% using fish oil and had thermostability at 70 °C [115].
The study of López et al. [11] investigated the omega-3 enrichment of microalgal oil from Nannochloropsis species using the commercial lipases Lipozyme TL 100 L, Lecitase® Ultra, Lipozyme® CALB, Quara® LowP, Lipase D, Lipase DF, Lipase MER, Lipase AY, and Lipase QLM®. It was found that the QLM lipase, extracted from Alcaligenes species, was highly efficient in enriching EPA polar lipids as it exhibited 1,3 positional specificity and this lipase was further used for a selecting solvent system. Moreover, lipase QLM was immobilized on Accurel MP 1000 and was able to enrich EPA from 48.5% to 70% with a recovery rate of 92% [11]. This study suggests that enzyme immobilization can be used to reduce the cost of the process by recovering and reusing the enzyme.
Lipases can also be used for enriching PUFAs in fortified oils. The research group at Deakin University used pure omega-3 ethyl esters that were then reacted with commercially available immobilized lipases from Rhizomucor miehei (Lipozyme RMIM), Thermomyces lanuginosus (Lipozyme TLIM), and Candida antarctica B (CALB, Novozym® 435), and the monoacylglycerols and diacyglycerols obtained as products after the lipase Novozyme 435-mediated glycerolysis reaction were further fortified with extra virgin olive oil. The resulting fortified oil treated with lipase accounted for an increase in EPA content from 0.75% to 13% and a DHA content of 16% [116]. Novozyme 435 was also used for producing glycerides containing concentrated levels of omega-3 PUFAs, and it was reported that the synthesised glycerides had 1.21 and 2.71 times more EPA and DHA, respectively, as compared to the crude fish oil [117]. Moreover, Novozyme 435’s ability to catalyse acidolysis to produce DHA/EPA ethyl esters was also evaluated and analysed, and it was found that the enzyme was able to produce concentrated DHA/EPA ethyl esters with a 94% conversion yield ratio [118].
These studies demonstrate the ability of marine lipases to enrich omega-3 essential fatty acids using a simple process. The only bottlenecks are the risk of losing the lipase’s stability and activity after a single use, and the recovery of the free form of lipases is difficult to achieve. These constraints result in increased overall costs for producing enriched omega-3 concentrates. To mitigate this, lipases have been employed in their immobilized form in order to enhance reusability and recovery; moreover, the immobilization of lipases also tends to improve their overall efficiency [119].
6. Enzyme Immobilization for Advancing the Enrichment of Omega-3 Fatty Acids
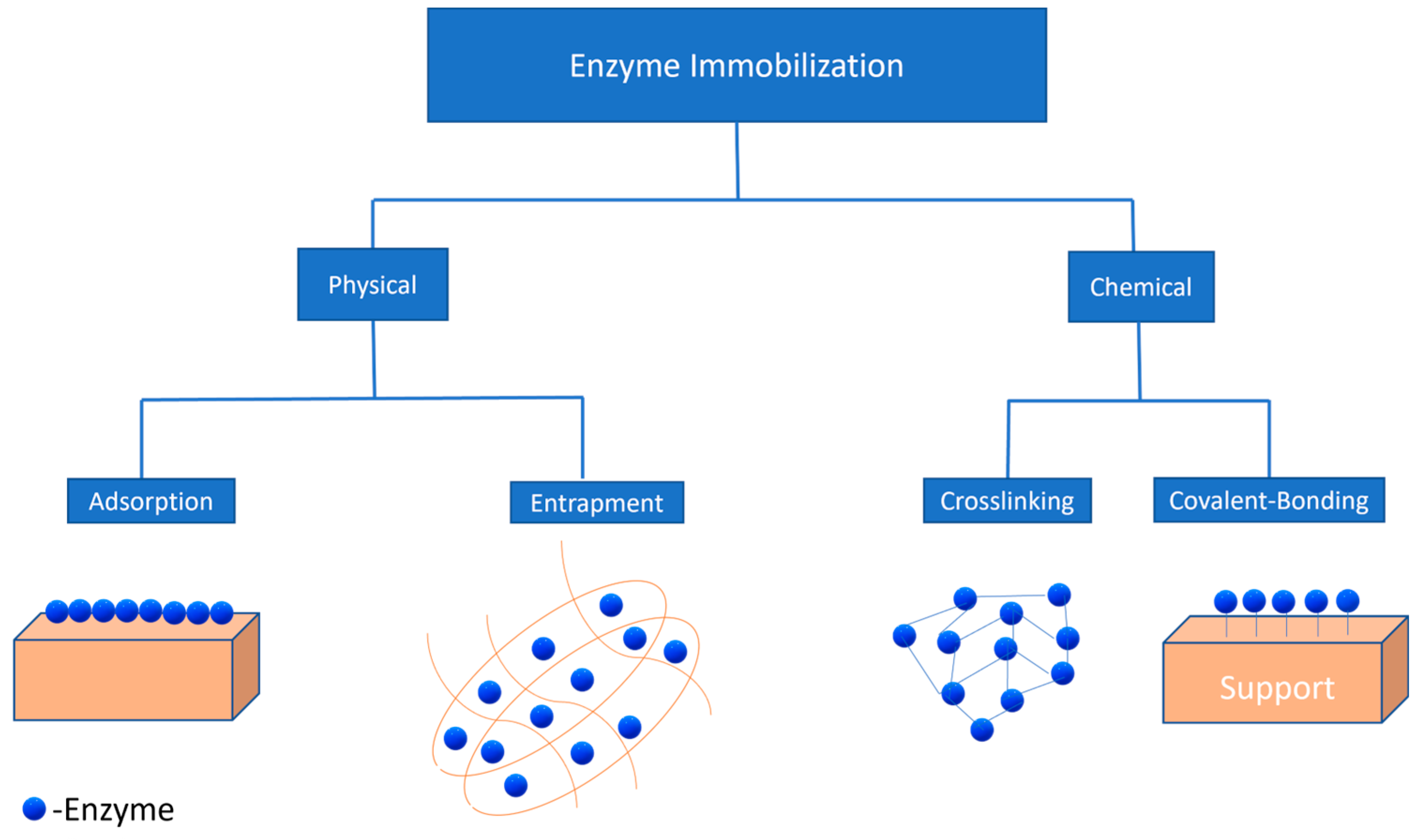
Over the last five decades, enzyme immobilization has been used to enhance the ability of enzymes to catalyse reactions in a controlled manner. Immobilization of enzymes has allowed enhanced reusability of enzymes in order to reduce the overall cost of the enrichment process. Different immobilization strategies used to date are mentioned below (Figure 2).
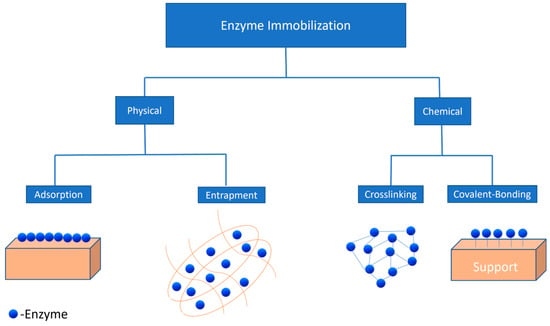
Figure 2.
Physical and chemical methods for enzyme immobilization.
6.1. Physical Methods
Physical methods for enzyme immobilization consist of two strategies, physical adsorption and entrapment. In the former, enzyme adsorption occurs on a support material (Figure 2) through weak forces such as hydrophobic interactions, ionic bonding, and Van der Waals forces [120]. Hydrophobic supports, such as octyl-sepharose, have been used to immobilize lipases via physical adsorption, which were further used for the hydrolysis of sardine oil [121]. Moreover, it has been reported that ionic supports such as carboxymethyl and sulfopropyl derivatives tested with immobilizing lipases had better selectivity towards EPA and DHA during fish oil hydrolysis [122]. On the other hand, the latter entrapment technique involves the confinement of the enzyme into a matrix without any chemical reaction, which can reduce the distortion in the structure of the enzyme, affecting the lipase activity [123]. Various sol–gel formulations such as tetramethoxysila, methyltrimethoxysila, and ethytrimethoxysila can also be used for encapsulating lipases and the hydrolysis of oil substrates such as olive oil [124].
New encapsulation approaches such as electrospinning and metal–organic frameworks (MOFs) have also been employed for immobilizing lipases. Recently, lipase from Burkholderia cepacia was immobilized via double-needle electrospinning and the gelation approach, collectively, by utilising hydrogel fibre–hydrophobic fibre hybrid membranes (hg-HMs), and showed elevation in the specific activity compared to free enzyme [125]. Moreover, lipase immobilization using Zeolitic imidazolate frameworks (ZIF), a type of MOF, was also used to immobilize lipases from genetically modified Thermomyces lanuginose, which was demonstrated to have better catalytic activity [126]. However, electrospinning and MOFs have not been investigated in lipase immobilization for the enrichment of omega-3. The most important drawback of physical methods is the ease of disintegration of the enzymes from the support material, leading to enzyme leaching and the requirement for more enzymes.
6.2. Chemical Methods
Chemical methods are further classified into two categories: crosslinking and covalent bonding (Figure 2). Crosslinking is the technique that uses crosslinking agents such as glutaraldehyde in order to improve the interaction between the enzyme and support material. Different support materials such as polyolefin [127] and chitosan-chitin [128] have been used to immobilize enzymes by crosslinking. In one study, crosslinking of C. rugosa lipases was carried out using glutaraldehyde, and it was found that the enantioselectivity of the crosslinked enzyme had improved when used for the hydrolysis of olive oil [129]. However, there are various limitations in this technique, such as poor mechanical stability and inefficient reproducibility, which make it less desirable for lipase immobilization.
On the other hand, covalent bonding has been proven to provide better mechanical stability with support that extremely reduces the chances of enzyme leaching into the media and also allows the reactivation of enzymes [130]; moreover, through covalent binding immobilization, the highest enzymatic activity was reported when compared to other methods. Various support materials were used for immobilizing enzymes through covalent bonding, as shown in Table 4.

Table 4.
Different materials used for covalent bonding of lipases for immobilization with a comparison of stability, activity, reusability, and omega-3 enrichment as compared to free enzyme form.
Covalent Bonding-Based Immobilization of Lipase for the Enrichment of Omega-3 PUFAs
Covalent bonding is highly efficient in terms of enzyme stability due to its strong interaction with the support, which reduces enzyme loss. This valuable characteristic of covalent bonding has led to an increased number of studies carried out, which are associated with the immobilization of lipase through covalent bonding (Table 4). The amination of lipases from Geotrichum candidum was carried out to facilitate covalent bonding with carboxymethyl and sulfopropyl agarose beads for efficient immobilization, and it was found that the immobilized enzyme was stable up to two cycles and its overall stability had increased by 3.2-fold in hydrolysis compared to the free aminated enzyme [131]. Furthermore, epoxy-functionalised silica particles were also used for immobilizing lipases from Rhizomucor miehei and it was reported that the immobilized enzyme can be reused up to five cycles, where it was able to enrich EPA and DHA content with a ratio of 6.8:1, which was higher compared to the free enzyme [132]. In 2019, ferrous nanoparticles were used for easier recyclability of the enzyme through magnetic separation. The research concluded that the immobilized enzyme had shown around 65% activity up to seven reuse cycles and had better thermostability, even at higher temperatures of 95 °C [133].
Moreover, the application of an ecofriendly support matrix for immobilizing enzymes is also gaining interest as it can assist in mitigating various environmental problems. For instance, silica from agricultural waste such as rice husk was utilised to create amino-functionalised nano–silica systems that were used to immobilize lipases derived from Candida rugosa through covalent bonding. A 2-fold increase in relative enzyme activity was obtained at 45 °C and a 2.5-fold increase in EPA content was reported using this immobilized enzyme for the hydrolysis of oil derived from N. oceanica [134]. In a recent study, crosslinker azelaic acid was employed, which allowed the covalent bond between lipase molecules to form crosslinked enzymes, and a selectivity ratio of 22 was obtained for EPA and DHA that retained activity up to 60% for five cycles [135]. Thus, covalent bonding-based immobilization can be ascribed as a promising method for enzyme immobilization; however, further studies are required for the immobilization of lipases generated from marine-derived sources that can support their claim of being suitable lipases for pharmaceutical and nutraceutical applications.
7. Concluding Remarks and Future Directions
The amount of research carried out in the area of omega-3 enrichment clearly suggests the importance of PUFAs in the medical and commercial sectors. Lipases are enzymes able to enrich omega-3 from different substrates such as fish oils and algal oils. Lipases from marine microorganisms hold an advantage due to their unique properties such as thermostability and tolerance against organic solvents; however, there is always a risk of loss in stability and activity of lipases over time. The method of covalent bonding-based immobilization of lipases to various support materials is the current solution that is applied to overcome this problem; however, the support materials used should have a minimal negative impact on the environment. Thus, greener and more inexpensive options for this method need to be investigated, such as the immobilization of lipases on plasma-polymerised surfaces as it does not demand the use of any organic solvents during the process. Furthermore, omega-3-enriched concentrates generated from marine microorganism lipases—that will further be used in food fortification—need to be investigated for their effects on the human gut microbiome as it can be helpful to understand the biochemical route through which omega-3 can infer positive effects on human health. Nevertheless, there are very limited studies carried out using oil from sources such as microalgae to meet the demand for essential omega-3 concentrates. Microalgae can be considered a suitable and sustainable oil source as it falls under the category of GRAS, can be grown in a controlled manner, and various species of microalgae such as thraustochytrids have been reported with higher amounts of EPA and DHA content. In this way, enriched omega-3 concentrates can also sustain the market demand for populations that follow a vegetarian diet. In summary, further development and studies are required to improve enzymatic enrichment using cost-effective and sustainable approaches and to divert the focus towards using oils from sources that can be consumed by a larger number of the population.
Author Contributions
M.P. and M.A. conceived the presented idea. M.K. (Mahejbin Karia) and M.K. (Mona Kaspal) participated in the collection of the data from the literature and in the drafting of the manuscript. M.K. (Mahejbin Karia) contributed to enzyme immobilization and enrichment Sections, and M.K. (Mona Kaspal) contributed to the sources of lipase and role of lipids. M.P. and M.A. reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Data are contained within the article.
Acknowledgments
The authors gratefully acknowledge the funding support provided by Flinders University to M.K. to pursue her research work in the bioprocessing group. The efforts of Emeritus Professor Christopher Franco in the editing of the manuscript are greatly acknowledged.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Satpute, S.K.; Banat, I.M.; Dhakephalkar, P.K.; Banpurkar, A.G.; Chopade, B.A. Biosurfactants, bioemulsifiers and exopolysaccharides from marine microorganisms. Biotechnol. Adv. 2010, 28, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Debashish, G.; Malay, S.; Barindra, S.; Joydeep, M. Marine enzymes. Mar. Biotechnol. I 2005, 96, 189–218. [Google Scholar]
- Maneerat, S.; Phetrong, K. Isolation of biosurfactant-producing marine bacteria and characteristics of selected biosurfactant. Songklanakarin J. Sci. Technol. 2007, 29, 781–791. [Google Scholar]
- Li, S.; Tan, E.C.; Dutta, A.; Snowden-Swan, L.J.; Thorson, M.R.; Ramasamy, K.K.; Bartling, A.W.; Brasington, R.; Kass, M.D.; Zaimes, G.G. Techno-economic analysis of sustainable biofuels for marine transportation. Environ. Sci. Technol. 2022, 56, 17206–17214. [Google Scholar] [CrossRef] [PubMed]
- Poli, A.; Finore, I.; Romano, I.; Gioiello, A.; Lama, L.; Nicolaus, B. Microbial diversity in extreme marine habitats and their biomolecules. Microorganisms 2017, 5, 25. [Google Scholar] [CrossRef] [PubMed]
- Niyonzima, F.N.; More, V.S.; Nsanganwimana, F.; Rao, A.S.; Nair, A.; Anantharaju, K.; More, S.S. Microbial enzymes used in textile industry. In Biotechnology of Microbial Enzymes; Elsevier: Amsterdam, The Netherlands, 2023; pp. 649–684. [Google Scholar]
- Choudhury, P.; Bhunia, B. Industrial application of lipase: A review. Biopharm. J. 2015, 1, 41–47. [Google Scholar]
- Navvabi, A.; Razzaghi, M.; Fernandes, P.; Karami, L.; Homaei, A. Novel lipases discovery specifically from marine organisms for industrial production and practical applications. Process Biochem. 2018, 70, 61–70. [Google Scholar] [CrossRef]
- Dokania, P.; Fopase, R.; Swagathnath, G.; Vivekanand; Gupta, K.; Pabari, P.; Sahoo, K.K.; Sarkar, A. Future Marine Microbial Products for the Pharmaceuticals Industry. In Microbial Products for Future Industrialization; Springer: Berlin/Heidelberg, Germany, 2023; pp. 199–221. [Google Scholar]
- Deng, D.; Zhang, Y.; Sun, A.; Liang, J.; Hu, Y. Functional characterization of a novel marine microbial GDSL lipase and its utilization in the resolution of (±)-1-phenylethanol. Appl. Biochem. Biotechnol. 2016, 179, 75–93. [Google Scholar] [CrossRef] [PubMed]
- López, E.N.; Callejón, M.J.J.; Sánchez, M.D.M.; Moreno, P.A.G.; Medina, A.R. Obtaining eicosapentaenoic acid-enriched polar lipids from microalga Nannochloropsis sp. by lipase-catalysed hydrolysis. Algal Res. 2023, 71, 103073. [Google Scholar] [CrossRef]
- MarketsandMarkets. Omega-3 Market by Type (DHA, EPA, AND ALA), Application (Dietary Supplements, Functional Foods & Beverages, Pharmaceuticals, Infant Formula, and Pet Food & Feed), Source (Marine and Plant), and Region–Global Forecast to 2029; Market Research Report. Available online: https://www.marketsandmarkets.com/Market-Reports/omega-3-omega-6-227.html (accessed on 23 February 2024).
- Lee, J.M.; Lee, H.; Kang, S.; Park, W.J. Fatty acid desaturases, polyunsaturated fatty acid regulation, and biotechnological advances. Nutrients 2016, 8, 23. [Google Scholar] [CrossRef]
- Gupta, A.; Barrow, C.J.; Puri, M. Omega-3 biotechnology: Thraustochytrids as a novel source of omega-3 oils. Biotechnol. Adv. 2012, 30, 1733–1745. [Google Scholar] [CrossRef]
- Qin, J.; Kurt, E.; LBassi, T.; Sa, L.; Xie, D. Biotechnological production of omega-3 fatty acids: Current status and future perspectives. Front. Microbiol. 2023, 14, 1280296. [Google Scholar] [CrossRef]
- Bazinet, R.P.; Layé, S. Polyunsaturated fatty acids and their metabolites in brain function and disease. Nat. Rev. Neurosci. 2014, 15, 771–785. [Google Scholar] [CrossRef] [PubMed]
- Swanson, D.; Block, R.; Mousa, S.A. Omega-3 fatty acids EPA and DHA: Health benefits throughout life. Adv. Nutr. 2012, 3, 1–7. [Google Scholar] [CrossRef]
- Alhattab, M.; Moorthy, L.S.; Patel, D.; Franco, C.M.; Puri, M. Oleaginous Microbial Lipids’ Potential in the Prevention and Treatment of Neurological Disorders. Mar. Drugs 2024, 22, 80. [Google Scholar] [CrossRef]
- Shahidi, F.; Ambigaipalan, P. Omega-3 polyunsaturated fatty acids and their health benefits. Annu. Rev. Food Sci. Technol. 2018, 9, 345–381. [Google Scholar] [CrossRef] [PubMed]
- Kris-Etherton, P.M.; Hecker, K.D.; Binkoski, A.E. Polyunsaturated fatty acids and cardiovascular health. Nutr. Rev. 2004, 62, 414–426. [Google Scholar] [CrossRef]
- Paschos, G.; Magkos, F.; Panagiotakos, D.; Votteas, V.; Zampelas, A. Dietary supplementation with flaxseed oil lowers blood pressure in dyslipidaemic patients. Eur. J. Clin. Nutr. 2007, 61, 1201–1206. [Google Scholar] [CrossRef] [PubMed]
- Holub, D.J.; Holub, B.J. Omega-3 fatty acids from fish oils and cardiovascular disease. Mol. Cell. Biochem. 2004, 263, 217–225. [Google Scholar] [CrossRef]
- Liu, J.; Ma, D.W. The role of n-3 polyunsaturated fatty acids in the prevention and treatment of breast cancer. Nutrients 2014, 6, 5184–5223. [Google Scholar] [CrossRef]
- Calder, P.C. Health benefits of omega-3 fatty acids. In Omega-3 Delivery Systems; Elsevier: Amsterdam, The Netherlands, 2021; pp. 25–53. [Google Scholar]
- Lien, E.; Richard, C.; Hoffman, D. DHA and ARA addition to infant formula: Current status and future research directions. Prostaglandins Leukot. Essent. Fat. Acids 2018, 128, 26–40. [Google Scholar] [CrossRef] [PubMed]
- Goed. Global Organization For EPA and DHA Omega-3. 2024. Available online: https://goedomega3.com/about (accessed on 9 March 2024).
- Ghelichi, S.; Hajfathalian, M.; García-Moreno, P.J.; Yesiltas, B.; Moltke-Sørensen, A.-D.; Jacobsen, C. Food enrichment with omega-3 polyunsaturated fatty acids. In Omega-3 Delivery Systems; Elsevier: Amsterdam, The Netherlands, 2021; pp. 395–425. [Google Scholar]
- Liu, Y.; Dave, D. Recent progress on immobilization technology in enzymatic conversion of marine by-products to concentrated omega-3 fatty acids. Green Chem. 2022, 24, 1049–1066. [Google Scholar] [CrossRef]
- Let, M.B.; Jacobsen, C.; Meyer, A.S. Lipid oxidation in milk, yoghurt, and salad dressing enriched with neat fish oil or pre-emulsified fish oil. J. Agric. Food Chem. 2007, 55, 7802–7809. [Google Scholar] [CrossRef] [PubMed]
- Serfert, Y.; Drusch, S.; Schwarz, K. Chemical stabilisation of oils rich in long-chain polyunsaturated fatty acids during homogenisation, microencapsulation and storage. Food Chem. 2009, 113, 1106–1112. [Google Scholar] [CrossRef]
- Jain, P.; Minhas, A.K.; Shukla, S.; Puri, M.; Barrow, C.J.; Mandal, S. Bioprospecting indigenous marine microalgae for polyunsaturated fatty acids under different media conditions. Front. Bioeng. Biotechnol. 2022, 10, 842797. [Google Scholar] [CrossRef]
- Shahidi, F.; Wanasundara, U.N. Omega-3 fatty acid concentrates: Nutritional aspects and production technologies. Trends Food Sci. Technol. 1998, 9, 230–240. [Google Scholar] [CrossRef]
- Puri, M.; Barrow, C.J.; Verma, M.L. Enzyme immobilization on nanomaterials for biofuel production. Trends Biotechnol. 2013, 31, 215–216. [Google Scholar] [CrossRef] [PubMed]
- Verma, M.L.; Chaudhary, R.; Tsuzuki, T.; Barrow, C.J.; Puri, M. Immobilization of β-glucosidase on a magnetic nanoparticle improves thermostability: Application in Cellobiose Hydrolysis. Bioresour. Technol. 2013, 135, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Maghraby, Y.R.; El-Shabasy, R.M.; Ibrahim, A.H.; Azzazy, H.M.E.-S. Enzyme immobilization technologies and industrial applications. ACS Omega 2023, 8, 5184–5196. [Google Scholar] [CrossRef]
- Schmid, R.D.; Verger, R. Lipases: Interfacial enzymes with attractive applications. Angew. Chem. Int. Ed. 1998, 37, 1608–1633. [Google Scholar] [CrossRef]
- Javadi, A.; Dowlati, S.; Shourni, S.; Rusli, S.; Eckert, K.; Miller, R.; Kraume, M. Enzymatic hydrolysis of triglycerides at the water–oil interface studied via interfacial rheology analysis of lipase adsorption layers. Langmuir 2021, 37, 12919–12928. [Google Scholar] [CrossRef] [PubMed]
- Hari Krishna, S.; Karanth, N. Lipases and lipase-catalyzed esterification reactions in nonaqueous media. Catal. Rev. 2002, 44, 499–591. [Google Scholar] [CrossRef]
- Filho, D.G.; Silva, A.G.; Guidini, C.Z. Lipases: Sources, immobilization methods, and industrial applications. Appl. Microbiol. Biotechnol. 2019, 103, 7399–7423. [Google Scholar] [CrossRef] [PubMed]
- Bordes, F.; Barbe, S.; Escalier, P.; Mourey, L.; André, I.; Marty, A.; Tranier, S. Exploring the conformational states and rearrangements of Yarrowia lipolytica lipase. Biophys. J. 2010, 99, 2225–2234. [Google Scholar] [CrossRef] [PubMed]
- Reetz, M.T. Lipases as practical biocatalysts. Curr. Opin. Chem. Biol. 2002, 6, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Mounguengui, R.W.M.; Brunschwig, C.; Baréa, B.; Villeneuve, P.; Blin, J. Are plant lipases a promising alternative to catalyze transesterification for biodiesel production? Prog. Energy Combust. Sci. 2013, 39, 441–456. [Google Scholar] [CrossRef]
- Sundaramahalingam, M.; Amrutha, C.; Sivashanmugam, P.; Rajeshbanu, J. An encapsulated report on enzyme-assisted transesterification with an allusion to lipase. 3 Biotech 2021, 11, 481. [Google Scholar] [CrossRef] [PubMed]
- Seth, S.; Chakravorty, D.; Dubey, V.K.; Patra, S. An insight into plant lipase research–challenges encountered. Protein Expr. Purif. 2014, 95, 13–21. [Google Scholar] [CrossRef]
- Kurtovic, I.; Marshall, S.N.; Zhao, X.; Simpson, B.K. Lipases from mammals and fishes. Rev. Fish. Sci. 2009, 17, 18–40. [Google Scholar] [CrossRef]
- Pahoja, V.M.; Sethar, M.A. A review of enzymatic properties of lipase in plants, animals and microorganisms. J. Appl. Sci. 2002, 2, 474–484. [Google Scholar]
- Kheadr, E.; Vuillemard, J.C.; El-Deeb, S. Acceleration of Cheddar cheese lipolysis by using liposome-entrapped lipases. J. Food Sci. 2002, 67, 485–492. [Google Scholar] [CrossRef]
- Kiran, K.; Babu, C.S.; Divakar, S. Thermostability of porcine pancreas lipase in non-aqueous media. Process Biochem. 2001, 36, 885–892. [Google Scholar] [CrossRef]
- Mendes, A.A.; Oliveira, P.C.; de Castro, H.F. Properties and biotechnological applications of porcine pancreatic lipase. J. Mol. Catal. B Enzym. 2012, 78, 119–134. [Google Scholar] [CrossRef]
- Reetz, M.T. Biocatalysis in organic chemistry and biotechnology: Past, present, and future. J. Am. Chem. Soc. 2013, 135, 12480–12496. [Google Scholar] [CrossRef] [PubMed]
- Chandra, P.; Enespa; Singh, R.; Arora, P.K. Microbial lipases and their industrial applications: A comprehensive review. Microb. Cell Factories 2020, 19, 169. [Google Scholar]
- Treichel, H.; De Oliveira, D.; Mazutti, M.A.; Di Luccio, M.; Oliveira, J.V. A review on microbial lipases production. Food Bioprocess Technol. 2010, 3, 182–196. [Google Scholar] [CrossRef]
- Griebeler, N.; Polloni, A.E.; Remonatto, D.; Arbter, F.; Vardanega, R.; Cechet, J.L.; Di Luccio, M.; de Oliveira, D.; Treichel, H.; Cansian, R.L. Isolation and screening of lipase-producing fungi with hydrolytic activity. Food Bioprocess Technol. 2011, 4, 578–586. [Google Scholar] [CrossRef]
- Wang, L.; Chi, Z.; Wang, X.; Liu, Z.; Li, J. Diversity of lipase-producing yeasts from marine environments and oil hydrolysis by their crude enzymes. Ann. Microbiol. 2007, 57, 495–501. [Google Scholar] [CrossRef]
- Tokak, S.; Kiliç, I.H.; Yalçin, H.T.; Duran, T. Detection of extracellular lipases and genotypic identification from yeast causing spoilage of some dairy products produced in Gaziantep. Kahramanmaraş Sütçü İmam Üniversitesi Tarım Ve Doğa Derg. 2019, 22, 206–211. [Google Scholar] [CrossRef]
- Cardoso, V.M.; Borelli, B.M.; Lara, C.A.; Soares, M.A.; Pataro, C.; Bodevan, E.C.; Rosa, C.A. The influence of seasons and ripening time on yeast communities of a traditional Brazilian cheese. Food Res. Int. 2015, 69, 331–340. [Google Scholar] [CrossRef]
- Abu, M.L.; Nooh, H.M.; Oslan, S.N.; Salleh, A.B. Optimization of physical conditions for the production of thermostable T1 lipase in Pichia guilliermondii strain SO using response surface methodology. BMC Biotechnol. 2017, 17, 78. [Google Scholar] [CrossRef] [PubMed]
- Tóth, R.; Alonso, M.F.; Bain, J.M.; Vágvölgyi, C.; Erwig, L.-P.; Gácser, A. Different Candida parapsilosis clinical isolates and lipase deficient strain trigger an altered cellular immune response. Front. Microbiol. 2015, 6, 156578. [Google Scholar] [CrossRef] [PubMed]
- Kuncharoen, N.; Techo, S.; Savarajara, A.; Tanasupawat, S. Identification and lipolytic activity of yeasts isolated from foods and wastes. Mycology 2020, 11, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Adel, A.; El-Baz, A.; Shetaia, Y.; Sorour, N.M. Biosynthesis of polyunsaturated fatty acids by two newly cold-adapted Egyptian marine yeast. 3 Biotech 2021, 11, 461. [Google Scholar] [CrossRef]
- De Maria, P.D.; Sánchez-Montero, J.M.; Sinisterra, J.V.; Alcántara, A.R. Understanding Candida rugosa lipases: An overview. Biotechnol. Adv. 2006, 24, 180–196. [Google Scholar] [CrossRef]
- Dalmau, E.; Montesinos, J.; Lotti, M.; Casas, C. Effect of different carbon sources on lipase production by Candida rugosa. Enzym. Microb. Technol. 2000, 26, 657–663. [Google Scholar] [CrossRef]
- Coşkun, G.; Çıplak, Z.; Yıldız, N.; Mehmetoğlu, Ü. Immobilization of Candida antarctica lipase on nanomaterials and investigation of the enzyme activity and enantioselectivity. Appl. Biochem. Biotechnol. 2021, 193, 430–445. [Google Scholar] [CrossRef] [PubMed]
- Brígida, A.I.; Amaral, P.F.; Coelho, M.A.; Goncalves, L.R. Lipase from Yarrowia lipolytica: Production, characterization and application as an industrial biocatalyst. J. Mol. Catal. B Enzym. 2014, 101, 148–158. [Google Scholar] [CrossRef]
- Corzo, G.; Revah, S. Production and characteristics of the lipase from Yarrowia lipolytica 681. Bioresour. Technol. 1999, 70, 173–180. [Google Scholar] [CrossRef]
- Kayanadath, S.; Nathan, V.K.; Ammini, P. Anti-biofilm activity of biosurfactant derived from Halomonas sp. a Lipolytic marine bacterium from the Bay of Bengal. Microbiology 2019, 88, 585–599. [Google Scholar] [CrossRef]
- Seghal Kiran, G.; Lipton, A.N.; Kennedy, J.; Dobson, A.D.; Selvin, J. A halotolerant thermostable lipase from the marine bacterium Oceanobacillus sp. PUMB02 with an ability to disrupt bacterial biofilms. Bioengineered 2014, 5, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Nerurkar, M.; Joshi, M.; Pariti, S.; Adivarekar, R. Application of lipase from marine bacteria Bacillus sonorensis as an additive in detergent formulation. J. Surfactants Deterg. 2013, 16, 435–443. [Google Scholar] [CrossRef]
- Bhosale, H.; Uzma, S.; Kadam, T. Optimization of Process Parameters for Improved Lipase Production by Hyperthermophilic Bacillus sonorensis 4R. J. Adv. Biol. Biotechnol. 2019, 11, 1–10. [Google Scholar] [CrossRef]
- Hassan, S.W.; El Latif, H.H.A.; Ali, S.M. Production of cold-active lipase by free and immobilized marine Bacillus cereus HSS: Application in wastewater treatment. Front. Microbiol. 2018, 9, 412412. [Google Scholar] [CrossRef] [PubMed]
- Mazhar, H.; Abbas, N.; Zamir, T.; Hussain, Z.; Ali, S.S. Optimization study of lipolytic enzyme from Bacillus cereus, PCSIR NL-37. Punjab Univ. J. Zool. 2018, 33, 217–224. [Google Scholar] [CrossRef]
- Savvidou, M.G.; Sotiroudis, T.G.; Kolisis, F.N. Cell surface and cellular debris-associated heat-stable lipolytic enzyme activities of the marine alga Nannochloropsis oceanica. Biocatal. Biotransform. 2016, 34, 24–32. [Google Scholar] [CrossRef]
- Yong, S.K.; Lim, B.H.; Saleh, S.; Tey, L.-H. Optimisation, purification and characterisation of extracellular lipase from Botryococcus sudeticus (UTEX 2629). J. Mol. Catal. B Enzym. 2016, 126, 99–105. [Google Scholar] [CrossRef]
- Waters, A.L.; Hill, R.T.; Place, A.R.; Hamann, M.T. The expanding role of marine microbes in pharmaceutical development. Curr. Opin. Biotechnol. 2010, 21, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Luna, G.M. Biotechnological potential of marine microbes. In Springer Handbook of Marine Biotechnology; Springer: Berlin/Heidelberg, Germany, 2015; pp. 651–661. [Google Scholar]
- Nagarajan, M.; Kumar, R.R.; Sundaram, K.M.; Sundararaman, M. Marine biotechnology: Potentials of marine microbes and algae with reference to pharmacological and commercial values. In Plant Biology and Biotechnology: Volume II: Plant Genomics and Biotechnology; Springer: Berlin/Heidelberg, Germany, 2015; pp. 685–723. [Google Scholar]
- Chi, Z.; Liu, Z.; Gao, L.; Gong, F.; Ma, C.; Wang, X.; Li, H. Marine yeasts and their applications in mariculture. J. Ocean Univ. China 2006, 5, 251–256. [Google Scholar]
- Ferrer, P.; Montesinos, J.L.; Valero, F.; Solà, C. Production of native and recombinant lipases by Candida rugosa: A review. Appl. Biochem. Biotechnol. 2001, 95, 221–255. [Google Scholar] [CrossRef]
- Ulbert, O.; Bélafi-Bakó, K.; Tonova, K.; Gubicza, L. Thermal stability enhancement of Candida rugosa lipase using ionic liquids. Biocatal. Biotransform. 2005, 23, 177–183. [Google Scholar] [CrossRef]
- Akanbi, T.O.; Barrow, C.J. Candida antarctica lipase A effectively concentrates DHA from fish and thraustochytrid oils. Food Chem. 2017, 229, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Subroto, E.; Indiarto, R.; Pangawikan, A.; Huda, S.; Yarlina, V. Characteristics, immobilization, and application of Candida rugosa lipase. Food Res. 2020, 4, 1391–1401. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, F.; Li, Z. Gene analysis, optimized production and property of marine lipase from Bacillus pumilus B106 associated with South China Sea sponge Halichondria rugosa. World J. Microbiol. Biotechnol. 2009, 25, 1267–1274. [Google Scholar] [CrossRef]
- Li, X.; Benning, C.; Kuo, M.-H. Rapid triacylglycerol turnover in Chlamydomonas reinhardtii requires a lipase with broad substrate specificity. Eukaryot. Cell 2012, 11, 1451–1462. [Google Scholar] [CrossRef] [PubMed]
- Ben Hlima, H.; Dammak, M.; Karray, A.; Drira, M.; Michaud, P.; Fendri, I.; Abdelkafi, S. Molecular and structural characterizations of lipases from Chlorella by functional genomics. Mar. Drugs 2021, 19, 70. [Google Scholar] [CrossRef] [PubMed]
- Hasan, F.; Shah, A.A.; Hameed, A. Industrial applications of microbial lipases. Enzym. Microb. Technol. 2006, 39, 235–251. [Google Scholar] [CrossRef]
- Basheer, S.M.; Chellappan, S.; Beena, P.; Sukumaran, R.K.; Elyas, K.; Chandrasekaran, M. Lipase from marine Aspergillus awamori BTMFW032: Production, partial purification and application in oil effluent treatment. New Biotechnol. 2011, 28, 627–638. [Google Scholar] [CrossRef]
- Zhao, J.; Ma, M.; Zeng, Z.; Wan, D.; Yan, X.; Xia, J.; Yu, P.; Gong, D. Production, purification, properties and current perspectives for modification and application of microbial lipases. Prep. Biochem. Biotechnol. 2024, 1–16, online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Khan, S.A.; Hamayun, M.; Lee, I.-J. The recent advances in the utility of microbial lipases: A review. Microorganisms 2023, 11, 510. [Google Scholar] [CrossRef]
- Zambare, V.; Patankar, R.; Bhusare, B.; Christopher, L. Recent advances in feedstock and lipase research and development towards commercialization of enzymatic biodiesel. Processes 2021, 9, 1743. [Google Scholar] [CrossRef]
- Lipase Market: A Detailed Analysis of the Lipase Market by Liquid, Powder, and Gel. 2023. Available online: https://www.futuremarketinsights.com/reports/lipase-market (accessed on 2 June 2024).
- Ghattavi, S.; Homaei, A. Marine enzymes: Classification and application in various industries. Int. J. Biol. Macromol. 2023, 230, 123136. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Wang, X.; Shang, M.; Huang, J.; Guan, G.; Li, Y.; Shi, B. Isolation of a novel alkaline-stable lipase from a metagenomic library and its specific application for milkfat flavor production. Microb. Cell Factories 2014, 13, 1. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.; Kuila, A. Lipase and their different industrial applications: A review. Braz. J. Biol. Sci. 2018, 5, 237–247. [Google Scholar] [CrossRef]
- Geethanjali, P.; Viswanathan, S.; Gokilavani, K.; Sathya, T.; Kolar, A.B. Metagenomic Lipases and their Applications in the Food Industry. Environ. Sci. Arch. 2024, 3, 132–141. [Google Scholar]
- Gurung, N.; Ray, S.; Bose, S.; Rai, V. A broader view: Microbial enzymes and their relevance in industries, medicine, and beyond. BioMed Res. Int. 2013, 2013, 329121. [Google Scholar] [CrossRef] [PubMed]
- Fahim, Y.A.; El-Khawaga, A.M.; Sallam, R.M.; Elsayed, M.A.; Assar, M.F.A. A Review on Lipases: Sources, Assays, Immobilization Techniques on Nanomaterials and Applications. BioNanoScience 2024, 1–18. [Google Scholar] [CrossRef]
- Kumar, A.; Verma, V.; Dubey, V.K.; Srivastava, A.; Garg, S.K.; Singh, V.P.; Arora, P.K. Industrial applications of fungal lipases: A review. Front. Microbiol. 2023, 14, 1142536. [Google Scholar] [CrossRef]
- Dilip, C.V.; Mulaje, S.; Mohalkar, R. A review on actinomycetes and their biotechnological application. Int. J. Pharm. Sci. Res. 2013, 4, 1730. [Google Scholar]
- Lembke, P. Production techniques for omega-3 concentrates. In Omega-6/3 Fatty Acids: Functions, Sustainability Strategies and Perspectives; Springer: Berlin/Heidelberg, Germany, 2012; pp. 353–364. [Google Scholar]
- Rubio-Rodríguez, N.; Beltrán, S.; Jaime, I.; de Diego, S.M.; Sanz, M.T.; Carballido, J.R. Production of omega-3 polyunsaturated fatty acid concentrates: A review. Innov. Food Sci. Emerg. Technol. 2010, 11, 1–12. [Google Scholar] [CrossRef]
- Senanayake, S.N. Methods of concentration and purification of omega-3 fatty acids. In Separation, Extraction and Concentration Processes in the Food, Beverage and Nutraceutical Industries; Elsevier: Amsterdam, The Netherlands, 2013; pp. 483–505. [Google Scholar]
- Šližyte, R.; Daukšas, E.; Falch, E.; Storrø, I.; Rustad, T. Yield and composition of different fractions obtained after enzymatic hydrolysis of cod (Gadus morhua) by-products. Process Biochem. 2005, 40, 1415–1424. [Google Scholar] [CrossRef]
- Schlenk, H. Urea inclusion compounds of fatty acids. Prog. Chem. Fats Other Lipids 1954, 2, 243–267. [Google Scholar] [CrossRef]
- Tiegs, C.; Riha, V.; Brunner, G.; Steiner, K. Separation of multicomponent mixtures of fatty acid ethyl esters from fishoil by countercurrent SFE. In Process Technology Proceedings; Elsevier: Amsterdam, The Netherlands, 1996. [Google Scholar]
- Létisse, M.; Rozières, M.; Hiol, A.; Sergent, M.; Comeau, L. Enrichment of EPA and DHA from sardine by supercritical fluid extraction without organic modifier: I. Optimization of extraction conditions. J. Supercrit. Fluids 2006, 38, 27–36. [Google Scholar] [CrossRef]
- Tang, S.; Qin, C.; Wang, H.; Li, S.; Tian, S. Study on supercritical extraction of lipids and enrichment of DHA from oil-rich microalgae. J. Supercrit. Fluids 2011, 57, 44–49. [Google Scholar] [CrossRef]
- Bottino, N.R.; Vandenburg, G.A.; Reiser, R. Resistance of certain long-chain polyunsaturated fatty acids of marine oils to pancreatic lipase hydrolysis. Lipids 1967, 2, 489–493. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, K.; Yang, Z.; Chang, M.; Wang, X.; Wang, X. Lipase-catalyzed two-step hydrolysis for concentration of acylglycerols rich in ω-3 polyunsaturated fatty acids. Food Chem. 2023, 400, 134115. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Galan, C.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R.; Rodrigues, R.C. Potential of different enzyme immobilization strategies to improve enzyme performance. Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar] [CrossRef]
- Ranjan Moharana, T.; Byreddy, A.R.; Puri, M.; Barrow, C.; Rao, N.M. Selective enrichment of omega-3 fatty acids in oils by phospholipase A1. PLoS ONE 2016, 11, e0151370. [Google Scholar] [CrossRef] [PubMed]
- Yi, M.; You, Y.; Zhang, Y.; Wu, G.; Karrar, E.; Zhang, L.; Zhang, H.; Jin, Q.; Wang, X. Highly valuable fish oil: Formation process, enrichment, subsequent utilization, and storage of eicosapentaenoic acid ethyl esters. Molecules 2023, 28, 672. [Google Scholar] [CrossRef]
- Melgosa, R.; Sanz, M.T.; Beltrán, S. Supercritical CO2 processing of omega-3 polyunsaturated fatty acids–Towards a biorefinery for fish waste valorization. J. Supercrit. Fluids 2021, 169, 105121. [Google Scholar] [CrossRef]
- Yang, Z.; Jin, W.; Cheng, X.; Dong, Z.; Chang, M.; Wang, X. Enzymatic enrichment of n-3 polyunsaturated fatty acid glycerides by selective hydrolysis. Food Chem. 2021, 346, 128743. [Google Scholar] [CrossRef] [PubMed]
- Baloch, K.A.; Singh, A.; Pudtikajorn, K.; Benjakul, S. Lipases from different yeast strains: Production and application for n-3 fatty acid enrichment of tuna eyeball oil. Biocatal. Agric. Biotechnol. 2023, 48, 102651. [Google Scholar] [CrossRef]
- Hu, Z.; Jiao, L.; Xie, X.; Xu, L.; Yan, J.; Yang, M.; Yan, Y. Characterization of a new thermostable and organic solution-tolerant lipase from Pseudomonas fluorescens and its application in the enrichment of polyunsaturated fatty acids. Int. J. Mol. Sci. 2023, 24, 8924. [Google Scholar] [CrossRef]
- Gunathilake, T.; Akanbi, T.O.; Barrow, C.J. Lipase-produced omega-3 acylglycerols for the fortification and stabilization of extra virgin olive oil using hydroxytyrosyl palmitate. Future Foods 2021, 4, 100045. [Google Scholar] [CrossRef]
- Wang, W.; Li, T.; Ning, Z.; Wang, Y.; Yang, B.; Ma, Y.; Yang, X. A process for the synthesis of PUFA-enriched triglycerides from high-acid crude fish oil. J. Food Eng. 2012, 109, 366–371. [Google Scholar] [CrossRef]
- Kuo, C.-H.; Huang, C.-Y.; Lee, C.-L.; Kuo, W.-C.; Hsieh, S.-L.; Shieh, C.-J. Synthesis of DHA/EPA ethyl esters via lipase-catalyzed acidolysis using Novozym® 435: A kinetic study. Catalysts 2020, 10, 565. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Virgen-Ortíz, J.J.; Santos, J.C.D.; Berenguer-Murcia, Á.; Alcantara, A.R.; Barbosa, O.; Ortiz, C.; Fernandez-Lafuente, R. Immobilization of lipases on hydrophobic supports: Immobilization mechanism, advantages, problems, and solutions. Biotechnol. Adv. 2019, 37, 746–770. [Google Scholar] [CrossRef] [PubMed]
- Facin, B.R.; Melchiors, M.S.; Vale, A.; Oliveira, J.V.; Oliveira, D.B.D. Driving immobilized lipases as biocatalysts: 10 years state of the art and future prospects. Ind. Eng. Chem. Res. 2019, 58, 5358–5378. [Google Scholar] [CrossRef]
- Fernández-Lorente, G.; Betancor, L.; Carrascosa, A.V.; Guisán, J.M. Release of omega-3 fatty acids by the hydrolysis of fish oil catalyzed by lipases immobilized on hydrophobic supports. J. Am. Oil Chem. Soc. 2011, 88, 1173–1178. [Google Scholar] [CrossRef]
- Morais Júnior, W.G.; Fernández-Lorente, G.; Guisán, J.M.; Ribeiro, E.J.; De Resende, M.M.; Pessela, B.C. Production of omega-3 polyunsaturated fatty acids through hydrolysis of fish oil by Candida rugosa lipase immobilized and stabilized on different supports. Biocatal. Biotransform. 2017, 35, 63–73. [Google Scholar] [CrossRef]
- Hanefeld, U.; Gardossi, L.; Magner, E. Understanding enzyme immobilisation. Chem. Soc. Rev. 2009, 38, 453–468. [Google Scholar] [CrossRef] [PubMed]
- Reetz, M.T.; Zonta, A.; Simpelkamp, J. Efficient immobilization of lipases by entrapment in hydrophobic sol-gel materials. Biotechnol. Bioeng. 1996, 49, 527–534. [Google Scholar] [CrossRef]
- Zhang, Q.; Lin, S.; Wang, Z.; Li, J.; Xiao, J. Lipase-interfacial catalytic systems based on hybrid membranes constructed via electrospinning and gelation. LWT 2023, 183, 114956. [Google Scholar] [CrossRef]
- Shomal, R.; Du, W.; Al-Zuhair, S. Immobilization of Lipase on Metal-Organic frameworks for biodiesel production. J. Environ. Chem. Eng. 2022, 10, 107265. [Google Scholar] [CrossRef]
- Abd Manan, F.M.; Attan, N.; Zakaria, Z.; Mahat, N.A.; Wahab, R.A. Insight into the Rhizomucor miehei lipase supported on chitosan-chitin nanowhiskers assisted esterification of eugenol to eugenyl benzoate. J. Biotechnol. 2018, 280, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Anand, A.; Weatherley, L.R. The performance of microbial lipase immobilized onto polyolefin supports for hydrolysis of high oleate sunflower oil. Process Biochem. 2018, 68, 100–107. [Google Scholar] [CrossRef]
- Chuan Wu, J.; Selvam, V.; Teo, H.H.; Chow, Y.; Talukder, M.; Choi, W.J. Immobilization of Candida rugosa lipase by cross-linking with glutaraldehyde followed by entrapment in alginate beads. Biocatal. Biotransform. 2006, 24, 352–357. [Google Scholar] [CrossRef]
- Rueda, N.; Santos, J.C.D.; Torres, R.; Barbosa, O.; Ortiz, C.; Fernandez-Lafuente, R. Reactivation of lipases by the unfolding and refolding of covalently immobilized biocatalysts. RSC Adv. 2015, 5, 55588–55594. [Google Scholar] [CrossRef]
- de Morais Júnior, W.G.; Terrasan, C.R.F.; Fernández-Lorente, G.; Guisán, J.M.; Ribeiro, E.J.; de Resende, M.M.; Pessela, B.C. Solid-phase amination of Geotrichum candidum lipase: Ionic immobilization, stabilization and fish oil hydrolysis for the production of Omega-3 polyunsaturated fatty acids. Eur. Food Res. Technol. 2017, 243, 1375–1384. [Google Scholar] [CrossRef]
- Mohammadi, M.; Habibi, Z.; Dezvarei, S.; Yousefi, M.; Ashjari, M. Selective enrichment of polyunsaturated fatty acids by hydrolysis of fish oil using immobilized and stabilized Rhizomucor miehei lipase preparations. Food Bioprod. Process. 2015, 94, 414–421. [Google Scholar] [CrossRef]
- Verma, M.L.; Rao, N.M.; Tsuzuki, T.; Barrow, C.J.; Puri, M. Suitability of recombinant lipase immobilised on functionalised magnetic nanoparticles for fish oil hydrolysis. Catalysts 2019, 9, 420. [Google Scholar] [CrossRef]
- Jain, P.; Mandal, S.; Minhas, A.K.; Puri, M.; Barrow, C.J. Concentrating omega-3 fatty acids in Nannochloropsis oceanica oil by using enzyme immobilized nano-silica systems. J. Clean. Prod. 2023, 406, 137030. [Google Scholar] [CrossRef]
- Ahrari, F.; Yousefi, M.; Mohammadi, M. The effect of carbon chain length of cross-linking agent on the functionality of carrier-free immobilized Thermomyces lanuginosa lipase particles. Int. J. Biol. Macromol. 2024, 270, 132076. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).