Myelodysplastic Syndrome: Diagnosis and Screening
Abstract
:1. Cytopenia
2. Morphology
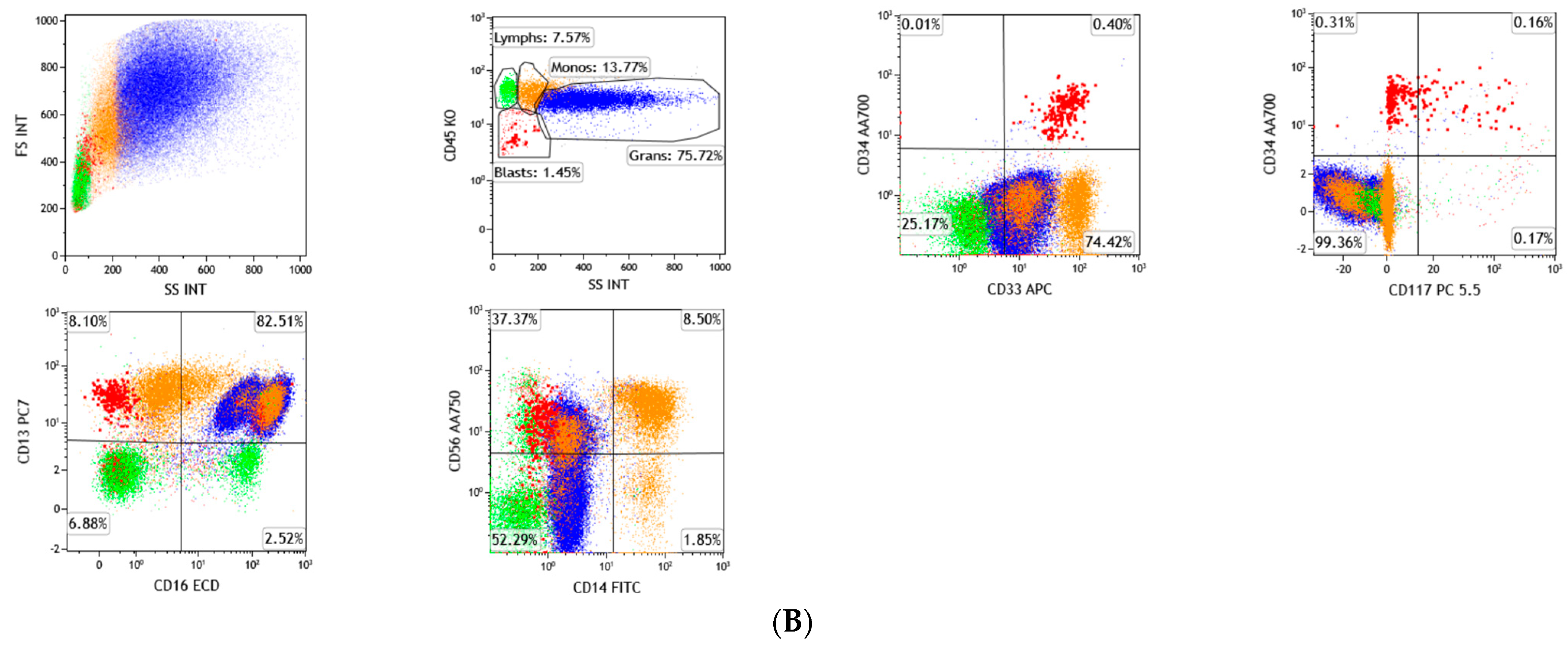
3. Flow Cytometry
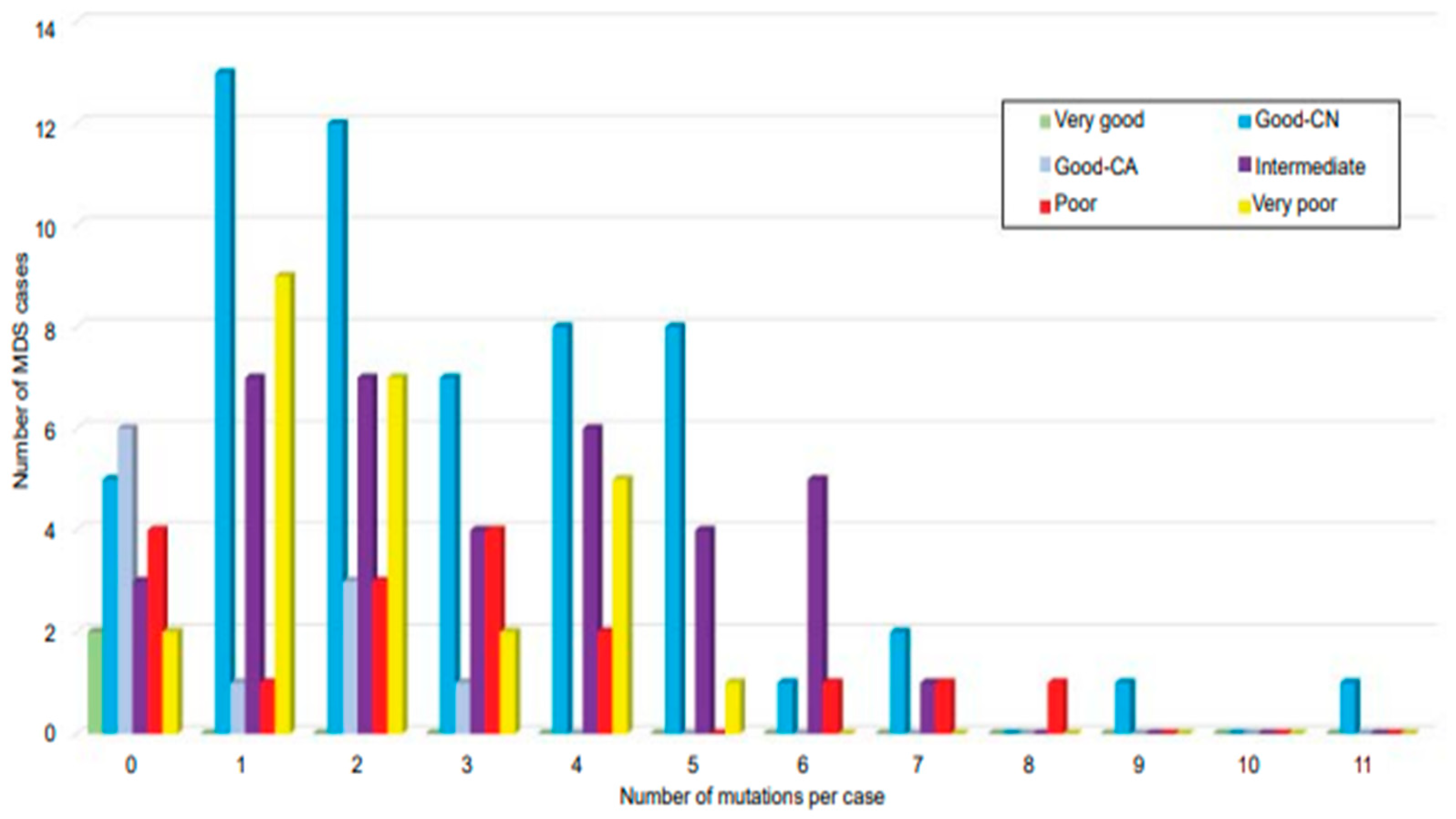
4. Cytogenetics
4.1. del(7q) or Monosomy 7
4.2. del(5q)
4.3. del(20q) and Loss of Y
4.4. Trisomy 8
4.5. del(11q) and del(12p)
4.6. del(9q)
4.7. t(17p) or Isochromosome 17q
4.8. t(11;16)
4.9. inv(3) or t(3;3)
4.10. t(6;9)
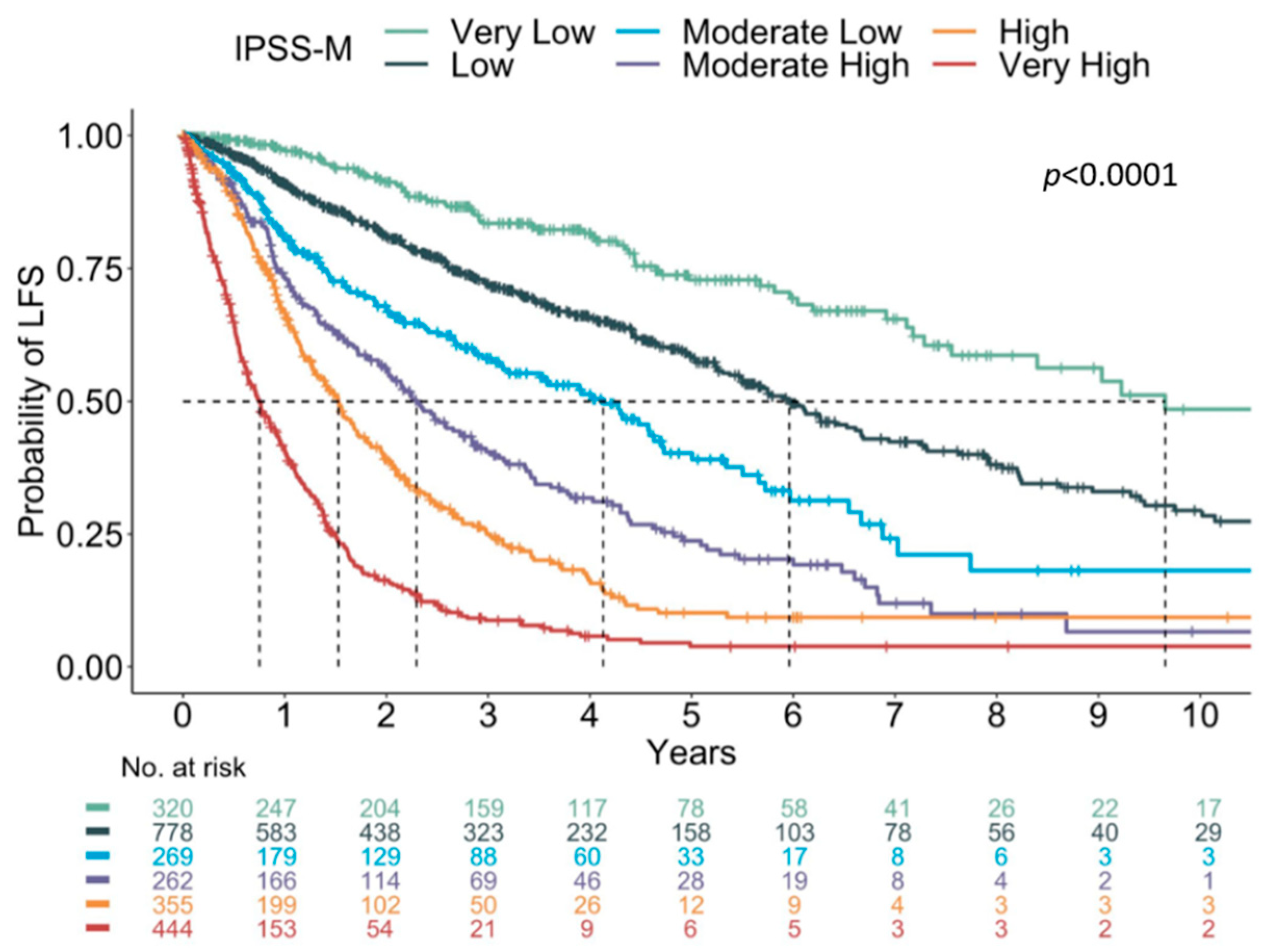
5. Next Generation Sequencing (NGS)
6. Machine Learning and Future Directions
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Olcay, L.; Yetgin, S. Disorders Mimicking Myelodysplastic Syndrome and Difficulties in Its Diagnosis; IntechOpen: London, UK, 2016. [Google Scholar] [CrossRef] [Green Version]
- Steensma, D.P. Dysplasia has A differential diagnosis: Distinguishing genuine myelodysplastic syndromes (MDS) from mimics, imitators, copycats and impostors. Curr. Hematol. Malig. Rep. 2012, 7, 310–320. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef]
- Greenberg, P.; Cox, C.; LeBeau, M.M.; Fenaux, P.; Morel, P.; Sanz, G.; Sanz, M.; Vallespi, T.; Hamblin, T.; Oscier, D.; et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood 1997, 89, 2079–2088. [Google Scholar] [CrossRef]
- Greenberg, P.L.; Tuechler, H.; Schanz, J.; Sanz, G.; Garcia-Manero, G.; Sole, F.; Bennett, J.M.; Bowen, D.; Fenaux, P.; Dreyfus, F.; et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood 2012, 120, 2454–2465. [Google Scholar] [CrossRef]
- Swerdlow, S.; Campo, E.; Harris, N.; Jaffe, E.; Pileri, S.; Stein, H.; Thiele, J.; Arber, D.; Hasserjian, R.; Le Beau, M.; et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th ed.; WHO Press: Geneva, Switzerland, 2017. [Google Scholar]
- Greenberg, P.L.; Tuechler, H.; Schanz, J.; Sanz, G.; Garcia-Manero, G.; Solé, F.; Bennett, J.M.; Bowen, D.; Fenaux, P.; Dreyfus, F.; et al. Cytopenia levels for aiding establishment of the diagnosis of myelodysplastic syndromes. Blood 2016, 128, 2096–2097. [Google Scholar] [CrossRef] [Green Version]
- Estey, E.H.; Hasserjian, R.P.; Döhner, H. Distinguishing AML from MDS: A fixed blast percentage may no longer be optimal. Blood 2022, 139, 323–332. [Google Scholar] [CrossRef]
- Della Porta, M.G.; Travaglino, E.; Boveri, E.; Ponzoni, M.; Malcovati, L.; Papaemmanuil, E.; Rigolin, G.M.; Pascutto, C.; Croci, G.; Gianelli, U.; et al. Minimal morphological criteria for defining bone marrow dysplasia: A basis for clinical implementation of WHO classification of myelodysplastic syndromes. Leukemia 2015, 29, 66–75. [Google Scholar] [CrossRef]
- Bain, B.J. The bone marrow aspirate of healthy subjects. Br. J. Haematol. 1996, 94, 206–209. [Google Scholar] [CrossRef]
- Bento, L.C.; Correia, R.P.; Mangueira, C.L.P.; De Souza Barroso, R.; Rocha, F.A.; Bacal, N.S.; Marti, L.C. The Use of Flow Cytometry in Myelodysplastic Syndromes: A Review. Front. Oncol. 2017, 7, 270. [Google Scholar] [CrossRef] [Green Version]
- van de Loosdrecht, A.A.; Alhan, C.; Béné, M.C.; Della Porta, M.G.; Dräger, A.M.; Feuillard, J.; Font, P.; Germing, U.; Haase, D.; Homburg, C.H.; et al. Standardization of flow cytometry in myelodysplastic syndromes: Report from the first European LeukemiaNet working conference on flow cytometry in myelodysplastic syndromes. Haematologica 2009, 94, 1124–1134. [Google Scholar] [CrossRef] [Green Version]
- Westers, T.M.; Ireland, R.; Kern, W.; Alhan, C.; Balleisen, J.S.; Bettelheim, P.; Burbury, K.; Cullen, M.; Cutler, J.A.; Della Porta, M.G.; et al. Standardization of flow cytometry in myelodysplastic syndromes: A report from an international consortium and the European LeukemiaNet Working Group. Leukemia 2012, 26, 1730–1741. [Google Scholar] [CrossRef] [Green Version]
- van de Loosdrecht, A.A.; Kern, W.; Porwit, A.; Valent, P.; Kordasti, S.; Cremers, E.; Alhan, C.; Duetz, C.; Dunlop, A.; Hobo, W.; et al. Clinical application of flow cytometry in patients with unexplained cytopenia and suspected myelodysplastic syndrome: A report of the European LeukemiaNet International MDS-Flow Cytometry Working Group. Cytom. Part B Clin. Cytom. 2021. [Google Scholar] [CrossRef]
- Wells, D.A.; Benesch, M.; Loken, M.R.; Vallejo, C.; Myerson, D.; Leisenring, W.M.; Deeg, H.J. Myeloid and monocytic dyspoiesis as determined by flow cytometric scoring in myelodysplastic syndrome correlates with the IPSS and with outcome after hematopoietic stem cell transplantation. Blood 2003, 102, 394–403. [Google Scholar] [CrossRef] [Green Version]
- Arroyo, J.L.; Fernández, M.E.; Hernández, J.M.; Orfao, A.; Miguel, J.F.S.; Del Cañizo, M.C. Impact of immunophenotype on prognosis of patients with myelodysplastic syndromes. Its value in patients without karyotypic abnormalities. Hematol. J. 2004, 5, 227–233. [Google Scholar] [CrossRef]
- Ogata, K.; Della Porta, M.G.; Malcovati, L.; Picone, C.; Yokose, N.; Matsuda, A.; Yamashita, T.; Tamura, H.; Tsukada, J.; Dan, K. Diagnostic utility of flow cytometry in low-grade myelodysplastic syndromes: A prospective validation study. Haematologica 2009, 94, 1066–1074. [Google Scholar] [CrossRef] [Green Version]
- Della Porta, M.G.; Picone, C.; Pascutto, C.; Malcovati, L.; Tamura, H.; Handa, H.; Czader, M.; Freeman, S.; Vyas, P.; Porwit, A.; et al. Multicenter validation of a reproducible flow cytometric score for the diagnosis of low-grade myelodysplastic syndromes: Results of a European LeukemiaNET study. Haematologica 2012, 97, 1209–1217. [Google Scholar] [CrossRef]
- Aalbers, A.M.; Heuvel-Eibrink, M.M.V.D.; De Haas, V.; Marvelde, J.G.T.; De Jong, A.X.; van der Burg, M.; Dworzak, M.; Hasle, H.; Locatelli, F.; De Moerloose, B.; et al. Applicability of a reproducible flow cytometry scoring system in the diagnosis of refractory cytopenia of childhood. Leukemia 2013, 27, 1923–1925. [Google Scholar] [CrossRef]
- Westers, T.M.; Cremers, E.M.; Oelschlaegel, U.; Johansson, U.; Bettelheim, P.; Matarraz, S.; Orfao, A.; Moshaver, B.; Brodersen, L.E.; Loken, M.R.; et al. Immunophenotypic analysis of erythroid dysplasia in myelodysplastic syndromes. A report from the IMDSFlow working group. Haematologica 2017, 102, 308–319. [Google Scholar] [CrossRef] [Green Version]
- Cremers, E.M.; Westers, T.M.; Alhan, C.; Cali, C.; Visser-Wisselaar, H.A.; Chitu, D.A.; van der Velden, V.H.; Te Marvelde, J.G.; Klein, S.K.; Muus, P.; et al. Implementation of erythroid lineage analysis by flow cytometry in diagnostic models for myelodysplastic syndromes. Haematologica 2017, 102, 320–326. [Google Scholar] [CrossRef] [Green Version]
- Schanz, J.; Tuechler, H.; Solé, F.; Mallo, M.; Luño, E.; Cervera, J.; Granada, I.; Hildebrandt, B.; Slovak, M.L.; Ohyashiki, K.; et al. New comprehensive cytogenetic scoring system for primary myelodysplastic syndromes (MDS) and oligoblastic acute myeloid leukemia after MDS derived from an international database merge. J. Clin. Oncol. 2012, 30, 820–829. [Google Scholar] [CrossRef]
- Haase, D. Cytogenetic features in myelodysplastic syndromes. Ann. Hematol. 2008, 87, 515–526. [Google Scholar] [CrossRef] [Green Version]
- França, I.G.D.F.; De Melo, M.M.L.; Teixeira, M.S.C.; Cordeiro, J.V.A.; Borges, D.D.P.; De Oliveira, R.T.G.; Furtado, S.R.; Magalhães, S.M.M.; Pinheiro, R.F. Role of conventional cytogenetics in sequential karyotype analysis of myelodysplastic syndrome: A patient with der(1;7)(q10;p10). Hematol. Transfus. Cell Ther. 2019, 41, 91–94. [Google Scholar] [CrossRef]
- Neukirchen, J.; Lauseker, M.; Hildebrandt, B.; Nolting, A.-C.; Kaivers, J.; Kobbe, G.; Gattermann, N.; Haas, R.; Germing, U. Cytogenetic clonal evolution in myelodysplastic syndromes is associated with inferior prognosis. Cancer 2017, 123, 4608–4616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haase, D.; Germing, U.; Schanz, J.; Pfeilstöcker, M.; Nösslinger, T.; Hildebrandt, B.; Kundgen, A.; Lübbert, M.; Kunzmann, R.; Giagounidis, A.A.N.; et al. New insights into the prognostic impact of the karyotype in MDS and correlation with subtypes: Evidence from a core dataset of 2124 patients. Blood 2007, 110, 4385–4395. [Google Scholar] [CrossRef] [PubMed]
- Zahid, M.F.; Malik, U.A.; Sohail, M.; Hassan, I.N.; Ali, S.; Shaukat, M.H.S. Cytogenetic Abnormalities in Myelodysplastic Syndromes: An Overview. Int. J. Hematol. Oncol. Stem Cell Res. 2017, 11, 231–239. [Google Scholar]
- Le Beau, M.M.; Espinosa, R., 3rd; Davis, E.M.; Eisenbart, J.D.; Larson, R.A.; Green, E.D. Cytogenetic and molecular delineation of a region of chromosome 7 commonly deleted in malignant myeloid diseases. Blood 1996, 88, 1930–1935. [Google Scholar] [CrossRef] [Green Version]
- Liang, H.; Fairman, J.; Claxton, D.F.; Nowell, P.C.; Green, E.D.; Nagarajan, L. Molecular anatomy of chromosome 7q deletions in myeloid neoplasms: Evidence for multiple critical loci. Proc. Natl. Acad. Sci. USA 1998, 95, 3781–3785. [Google Scholar] [CrossRef] [Green Version]
- Kerr, M.C.M.; Adema, V.; Walter, W.; Hutter, S.; Snider, B.C.A.; Nagata, Y.; Meggendorfer, M.; Haferlach, C.; Visconte, V.; Sekeres, M.M.A.; et al. Genetics of Monosomy 7 and Del(7q) in MDS Informs Potential Therapeutic Targets. Blood 2019, 134, 1703. [Google Scholar] [CrossRef]
- McNerney, M.E.; Brown, C.D.; Wang, X.; Bartom, E.T.; Karmakar, S.; Bandlamudi, C.; Yu, S.; Ko, J.; Sandall, B.P.; Stricker, T.; et al. CUX1 is a haploinsufficient tumor suppressor gene on chromosome 7 frequently inactivated in acute myeloid leukemia. Blood 2013, 121, 975–983. [Google Scholar] [CrossRef] [Green Version]
- Sakhdari, A.; Class, C.; Montalban-Bravo, G.; Sasaki, K.; Bueso-Ramos, C.E.; Patel, K.P.; Khoury, J.D.; Kantarjian, H.M.; Garcia-Manero, G.; Medeiros, L.J.; et al. Loss of EZH2 Protein Expression in Myelodysplastic Syndrome Correlates with EZH2 Mutation and Portends a Worse Outcome. Blood 2019, 134, 3016. [Google Scholar] [CrossRef]
- Ernst, T.; Chase, A.J.; Score, J.; Hidalgo-Curtis, C.E.; Bryant, C.; Jones, A.V.; Waghorn, K.; Zoi, K.; Ross, F.M.; Reiter, A.; et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat. Genet. 2010, 42, 722–726. [Google Scholar] [CrossRef] [PubMed]
- Berghe, H.V.D.; Cassiman, J.-J.; David, G.; Fryns, J.-P.; Michaux, J.-L.; Sokal, G. Distinct haematological disorder with deletion of long arm of no. 5 chromosome. Nature 1974, 251, 437–438. [Google Scholar] [CrossRef] [PubMed]
- Adema, V.; Bejar, R. What lies beyond del(5q) in myelodysplastic syndrome? Haematologica 2013, 98, 1819–1821. [Google Scholar] [CrossRef] [PubMed]
- Jerez, A.; Gondek, L.P.; Jankowska, A.M.; Makishima, H.; Przychodzen, B.; Tiu, R.V.; O’Keefe, C.L.; Mohamedali, A.M.; Batista, D.; Sekeres, M.A.; et al. Topography, clinical, and genomic correlates of 5q myeloid malignancies revisited. J. Clin. Oncol. 2012, 30, 1343–1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papaemmanuil, E.; Gerstung, M.; Malcovati, L.; Tauro, S.; Gundem, G.; Van Loo, P.; Yoon, C.J.; Ellis, P.; Wedge, D.C.; Pellagatti, A.; et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013, 122, 3616–3627; quiz 3699. [Google Scholar] [CrossRef]
- Christiansen, D.H.; Andersen, M.K.; Pedersen-Bjergaard, J. Mutations with loss of heterozygosity of p53 are common in therapy-related myelodysplasia and acute myeloid leukemia after exposure to alkylating agents and significantly associated with deletion or loss of 5q, a complex karyotype, and a poor prognosis. J. Clin. Oncol. 2001, 19, 1405–1413. [Google Scholar] [CrossRef]
- Boultwood, J.; Pellagatti, A.; McKenzie, A.N.J.; Wainscoat, J.S. Advances in the 5q- syndrome. Blood 2010, 116, 5803–5811. [Google Scholar] [CrossRef] [Green Version]
- Ebert, B.L.; Pretz, J.; Bosco, J.; Chang, C.Y.; Tamayo, P.; Galili, N.; Raza, A.; Root, D.E.; Attar, E.; Ellis, S.R.; et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature 2008, 451, 335–339. [Google Scholar] [CrossRef]
- Starczynowski, D.T.; Kuchenbauer, F.; Argiropoulos, B.; Sung, S.; Morin, R.; Muranyi, A.; Hirst, M.; Hogge, D.; Marra, M.; Wells, R.A.; et al. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat. Med. 2010, 16, 49–58. [Google Scholar] [CrossRef]
- List, A.; Dewald, G.; Bennett, J.; Giagounidis, A.; Raza, A.; Feldman, E.; Powell, B.; Greenberg, P.; Thomas, D.; Stone, R.; et al. Lenalidomide in the myelodysplastic syndrome with chromosome 5q deletion. N. Engl. J. Med. 2006, 355, 1456–1465. [Google Scholar] [CrossRef] [Green Version]
- Ravindran, A.; He, R.; Ketterling, R.P.; Jawad, M.D.; Chen, D.; Oliveira, J.L.; Nguyen, P.L.; Viswanatha, D.S.; Reichard, K.K.; Hoyer, J.D.; et al. The significance of genetic mutations and their prognostic impact on patients with incidental finding of isolated del(20q) in bone marrow without morphologic evidence of a myeloid neoplasm. Blood Cancer J. 2020, 10, 7. [Google Scholar] [CrossRef] [PubMed]
- Martín, I.; Villamón, E.; Abellán, R.; Calasanz, M.J.; Irigoyen, A.; Sanz, G.; Such, E.; Mora, E.; Gutiérrez, M.; Collado, R.; et al. Myelodysplastic syndromes with 20q deletion: Incidence, prognostic value and impact on response to azacitidine of ASXL1 chromosomal deletion and genetic mutations. Br. J. Haematol. 2021, 194, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Asada, S.; Fujino, T.; Goyama, S.; Kitamura, T. The role of ASXL1 in hematopoiesis and myeloid malignancies. Cell. Mol. Life Sci. 2019, 76, 2511–2523. [Google Scholar] [CrossRef] [PubMed]
- Bejar, R.; Stevenson, K.; Abdel-Wahab, O.; Galili, N.; Nilsson, B.; Garcia-Manero, G.; Kantarjian, H.; Raza, A.; Levine, R.L.; Neuberg, D.; et al. Clinical effect of point mutations in myelodysplastic syndromes. N. Engl. J. Med. 2011, 364, 2496–2506. [Google Scholar] [CrossRef] [Green Version]
- Brezinova, J.; Sarova, I.; Svobodova, K.; Lhotska, H.; Ransdorfova, S.; Izakova, S.; Pavlistova, L.; Lizcova, L.; Skipalova, K.; Hodanova, L.; et al. ASXL1 gene alterations in patients with isolated 20q deletion. Neoplasma 2019, 66, 627–630. [Google Scholar] [CrossRef] [PubMed]
- Navada, S.C.; Chatalbash, A.; Silverman, L.R. Clinical Significance of Cytogenetic Manifestations in Myelodysplastic Syndromes. Lab. Med. 2013, 44, 103–107. [Google Scholar] [CrossRef] [Green Version]
- Stone, J.F.; Sandberg, A.A. Sex chromosome aneuploidy and aging. Mutat. Res. 1995, 338, 107–113. [Google Scholar] [CrossRef]
- Ganster, C.; Kämpfe, D.; Jung, K.; Braulke, F.; Shirneshan, K.; Machherndl-Spandl, S.; Suessner, S.; Bramlage, C.P.; Legler, T.J.; Koziolek, M.J.; et al. New data shed light on Y-loss-related pathogenesis in myelodysplastic syndromes. Genes Chromosom. Cancer 2015, 54, 717–724. [Google Scholar] [CrossRef]
- Saumell, S.; Florensa, L.; Luño, E.; Sanzo, C.; Cañizo, C.; Hernández, J.M.; Cervera, J.; Gallart, M.A.; Carbonell, F.; Collado, R.; et al. Prognostic value of trisomy 8 as a single anomaly and the influence of additional cytogenetic aberrations in primary myelodysplastic syndromes. Br. J. Haematol. 2012, 159, 311–321. [Google Scholar] [CrossRef]
- Saumell, S.; Solé, F.; Arenillas, L.; Montoro, J.; Valcarcel, D.; Pedro, C.; Sanzo, C.; Luño, E.; Giménez, T.; Arnan, M.; et al. Trisomy 8, a Cytogenetic Abnormality in Myelodysplastic Syndromes, Is Constitutional or Not? PLoS ONE 2015, 10, e0129375. [Google Scholar] [CrossRef]
- Maserati, E.; Aprili, F.; Vinante, F.; Locatelli, F.; Amendola, G.; Zatterale, A.; Milone, G.; Minelli, A.; Bernardi, F.; Lo Curto, F.; et al. Trisomy 8 in myelodysplasia and acute leukemia is constitutional in 15–20% of cases. Genes Chromosom. Cancer 2002, 33, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Vardiman, J.W.; Thiele, J.; Arber, D.A.; Brunning, R.D.; Borowitz, M.J.; Porwit, A.; Harris, N.L.; Le Beau, M.M.; Hellström-Lindberg, E.; Tefferi, A.; et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: Rationale and important changes. Blood 2009, 114, 937–951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sloand, E.M.; Mainwaring, L.; Fuhrer, M.; Ramkissoon, S.; Risitano, A.M.; Keyvanafar, K.; Lu, J.; Basu, A.; Barrett, A.J.; Young, N.S. Preferential suppression of trisomy 8 compared with normal hematopoietic cell growth by autologous lymphocytes in patients with trisomy 8 myelodysplastic syndrome. Blood 2005, 106, 841–851. [Google Scholar] [CrossRef]
- Cortes, J.; O’Brien, S.; Kantarjian, H.; Cork, A.; Stass, S.; Freireich, E.J.; Keating, M.; Pierce, S.; Estey, E. Abnormalities in the long arm of chromosome 11 (11q) in patients with de novo and secondary acute myelogenous leukemias and myelodysplastic syndromes. Leukemia 1994, 8, 2174–2178. [Google Scholar]
- Wang, S.A.; Abruzzo, L.V.; Hasserjian, R.P.; Zhang, L.; Hu, Y.; Zhang, Y.; Zhao, M.; Galili, N.; Raza, A.; Medeiros, L.J.; et al. Myelodysplastic syndromes with deletions of chromosome 11q lack cryptic MLL rearrangement and exhibit characteristic clinicopathologic features. Leuk. Res. 2011, 35, 351–357. [Google Scholar] [CrossRef]
- Tria, F., IV; Raess, P.W.; Ang, D.; Andal, J.J.; Press, R.; Tran, N.; Dunlap, J.; Wiszniewska, J.; Anekpuritanang, T.; Fan, G. Somatic Mutational Analysis using Next Generation Sequencing in Predicting Disease Behavior of Cytogenetically Normal Myelodysplastic Syndromes. Int. J. Blood Res. Disord. 2021, 8, 063. [Google Scholar] [CrossRef]
- Barresi, V.; Palumbo, G.A.; Musso, N.; Consoli, C.; Capizzi, C.; Meli, C.R.; Romano, A.; Di Raimondo, F.; Condorelli, D.F. Clonal selection of 11q CN-LOH and CBL gene mutation in a serially studied patient during MDS progression to AML. Leuk. Res. 2010, 34, 1539–1542. [Google Scholar] [CrossRef]
- Braulke, F.; Müller-Thomas, C.; Götze, K.; Platzbecker, U.; Germing, U.; Hofmann, W.-K.; Giagounidis, A.A.N.; Lübbert, M.; Greenberg, P.L.; Bennett, J.M.; et al. Frequency of del(12p) is commonly underestimated in myelodysplastic syndromes: Results from a German diagnostic study in comparison with an international control group. Genes Chromosomes Cancer 2015, 54, 809–817. [Google Scholar] [CrossRef]
- Kobayashi, H.; Montgomery, K.T.; Bohlander, S.K.; Adra, C.N.; Lim, B.L.; Kucherlapati, R.S.; Donis-Keller, H.; Holt, M.S.; Le Beau, M.M.; Rowley, J.D. Fluorescence in situ hybridization mapping of translocations and deletions involving the short arm of human chromosome 12 in malignant hematologic diseases. Blood 1994, 84, 3473–3482. [Google Scholar] [CrossRef] [Green Version]
- Wall, M.; Rayeroux, K.C.; MacKinnon, R.; Zordan, A.; Campbell, L.J. ETV6 deletion is a common additional abnormality in patients with myelodysplastic syndromes or acute myeloid leukemia and monosomy 7. Haematologica 2012, 97, 1933–1936. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, L.; Haferlach, T.; Meggendorfer, M.; Kern, W.; Haferlach, C.; Stengel, A. Comprehensive molecular characterization of myeloid malignancies with 9q deletion. Leuk. Lymphoma 2019, 60, 2591–2593. [Google Scholar] [CrossRef]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Buchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef] [Green Version]
- Diamantopoulos, P.; Koumbi, D.; Kotsianidis, I.; Pappa, V.; Symeonidis, A.; Galanopoulos, A.; Zikos, P.; Papadaki, H.A.; Panayiotidis, P.; Dimou, M.; et al. The prognostic significance of chromosome 17 abnormalities in patients with myelodysplastic syndrome treated with 5-azacytidine: Results from the Hellenic 5-azacytidine registry. Cancer Med. 2019, 8, 2056–2063. [Google Scholar] [CrossRef] [Green Version]
- Welch, J.S.; Petti, A.A.; Miller, C.A.; Fronick, C.C.; O’Laughlin, M.; Fulton, R.S.; Wilson, R.K.; Baty, J.D.; Duncavage, E.J.; Tandon, B.; et al. TP53 and Decitabine in Acute Myeloid Leukemia and Myelodysplastic Syndromes. N. Engl. J. Med. 2016, 375, 2023–2036. [Google Scholar] [CrossRef]
- Cumbo, C.; Tota, G.; Anelli, L.; Zagaria, A.; Specchia, G.; Albano, F. TP53 in Myelodysplastic Syndromes: Recent Biological and Clinical Findings. Int. J. Mol. Sci. 2020, 21, 3432. [Google Scholar] [CrossRef]
- Tamai, H.; Inokuchi, K. 11q23/MLL acute leukemia: Update of clinical aspects. J. Clin. Exp. Hematop. 2010, 50, 91–98. [Google Scholar] [CrossRef] [Green Version]
- Kwok, R.P.S.; Lundblad, J.R.; Chrivia, J.C.; Richards, J.P.; Bächinger, H.P.; Brennan, R.G.; Roberts, S.G.; Green, M.R.; Goodman, R.H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 1994, 370, 223–226. [Google Scholar] [CrossRef]
- Xie, W.; Tang, G.; Wang, E.; Kim, Y.; Cloe, A.; Shen, Q.; Zhou, Y.; Garcia-Manero, G.; Loghavi, S.; Hu, A.Y.; et al. t(11;16)(q23;p13)/KMT2A-CREBBP in hematologic malignancies: Presumptive evidence of myelodysplasia or therapy-related neoplasm? Ann. Hematol. 2020, 99, 487–500. [Google Scholar] [CrossRef]
- Testoni, N.; Borsaru, G.; Martinelli, G.; Carboni, C.; Ruggeri, D.; Ottaviani, E.; Pelliconi, S.; Ricci, P.; Pastano, R.; Visani, G.; et al. 3q21 and 3q26 cytogenetic abnormalities in acute myeloblastic leukemia: Biological and clinical features. Haematologica 1999, 84, 690–694. [Google Scholar]
- Bitter, M.A.; Neilly, M.E.; Le Beau, M.M.; Pearson, M.G.; Rowley, J.D. Rearrangements of chromosome 3 involving bands 3q21 and 3q26 are associated with normal or elevated platelet counts in acute nonlymphocytic leukemia. Blood 1985, 66, 1362–1370. [Google Scholar] [CrossRef]
- Martinelli, G.; Ottaviani, E.; Buonamici, S.; Isidori, A.; Borsaru, G.; Visani, G.; Piccaluga, P.P.; Malagola, M.; Testoni, N.; Rondoni, M.; et al. Association of 3q21q26 syndrome with different RPN1/EVI1 fusion transcripts. Haematologica 2003, 88, 1221–1228. [Google Scholar]
- Lahortiga, I.; Vázquez, I.; Agirre, X.; Larrayoz, M.J.; Vizmanos, J.L.; Gozzetti, A.; Calasanz, M.J.; Odero, M.D. Molecular heterogeneity in AML/MDS patients with 3q21q26 rearrangements. Genes Chromosomes Cancer 2004, 40, 179–189. [Google Scholar] [CrossRef]
- Nucifora, G.; Laricchia-Robbio, L.; Senyuk, V. EVI1 and hematopoietic disorders: History and perspectives. Gene 2006, 368, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Slovak, M.L.; Gundacker, H.; Bloomfield, C.D.; Dewald, G.W.; Appelbaum, F.R.; Larson, R.A.; Tallman, M.S.; Bennett, J.M.; Stirewalt, D.L.; Meshinchi, S.; et al. A retrospective study of 69 patients with t(6;9)(p23;q34) AML emphasizes the need for a prospective, multicenter initiative for rare ‘poor prognosis’ myeloid malignancies. Leukemia 2006, 20, 1295–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, H.; Yabe, M.; Zhang, X.; Kim, Y.; Wu, X.; Wei, P.; Chi, S.; Zheng, L.; Garcia-Manero, G.; Shao, L.; et al. Myelodysplastic syndrome with t(6;9)(p22;q34.1)/DEK-NUP214 better classified as acute myeloid leukemia? A multicenter study of 107 cases. Mod. Pathol. 2021, 34, 1143–1152. [Google Scholar] [CrossRef]
- Genovese, G.; Kähler, A.K.; Handsaker, R.E.; Lindberg, J.; Rose, S.A.; Bakhoum, S.F.; Chambert, K.; Mick, E.; Neale, B.M.; Fromer, M.; et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 2014, 371, 2477–2487. [Google Scholar] [CrossRef] [Green Version]
- Jaiswal, S.; Fontanillas, P.; Flannick, J.; Manning, A.; Grauman, P.V.; Mar, B.G.; Lindsley, R.C.; Mermel, C.H.; Burtt, N.; Chavez, A.; et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014, 371, 2488–2498. [Google Scholar] [CrossRef] [Green Version]
- Xie, M.; Lu, C.; Wang, J.; McLellan, M.D.; Johnson, K.J.; Wendl, M.C.; McMichael, J.F.; Schmidt, H.K.; Yellapantula, V.; Miller, C.A.; et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 2014, 20, 1472–1478. [Google Scholar] [CrossRef]
- Valent, P. ICUS, IDUS, CHIP and CCUS: Diagnostic Criteria, Separation from MDS and Clinical Implications. Pathobiology 2019, 86, 30–38. [Google Scholar] [CrossRef]
- Valent, P.; Horny, H.-P.; Bennett, J.M.; Fonatsch, C.; Germing, U.; Greenberg, P.; Haferlach, T.; Haase, D.; Kolb, H.-J.; Krieger, O.; et al. Definitions and standards in the diagnosis and treatment of the myelodysplastic syndromes: Consensus statements and report from a working conference. Leuk. Res. 2007, 31, 727–736. [Google Scholar] [CrossRef]
- Malcovati, L.; Galli, A.; Travaglino, E.; Ambaglio, I.; Rizzo, E.; Molteni, E.; Elena, C.; Ferretti, V.V.; Catricala, S.; Bono, E.; et al. Clinical significance of somatic mutation in unexplained blood cytopenia. Blood 2017, 129, 3371–3378. [Google Scholar] [CrossRef] [PubMed]
- Lindsley, R.C.; Mar, B.G.; Mazzola, E.; Grauman, P.V.; Shareef, S.; Allen, S.L.; Pigneux, A.; Wetzler, M.; Stuart, R.K.; Erba, H.P.; et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 2015, 125, 1367–1376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steensma, D.P.; Bejar, R.; Jaiswal, S.; Lindsley, R.C.; Sekeres, M.A.; Hasserjian, R.P.; Ebert, B.L. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 2015, 126, 9–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bejar, R.; Lord, A.; Stevenson, K.; Bar-Natan, M.; Pérez-Ladaga, A.; Zaneveld, J.; Wang, H.; Caughey, B.; Stojanov, P.; Getz, G.; et al. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood 2014, 124, 2705–2712. [Google Scholar] [CrossRef]
- Haferlach, T.; Nagata, Y.; Grossmann, V.; Okuno, Y.; Bacher, U.; Nagae, G.; Schnittger, S.; Sanada, M.; Kon, A.; Alpermann, T.; et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2014, 28, 241–247. [Google Scholar] [CrossRef] [Green Version]
- Itzykson, R.; Thépot, S.; Quesnel, B.; Dreyfus, F.; Beyne-Rauzy, O.; Turlure, P.; Vey, N.; Recher, C.; Dartigeas, C.; Legros, L.; et al. Prognostic factors for response and overall survival in 282 patients with higher-risk myelodysplastic syndromes treated with azacitidine. Blood 2011, 117, 403–411. [Google Scholar] [CrossRef] [Green Version]
- Patnaik, M.M.; Hanson, C.A.; Hodnefield, J.M.; Lasho, T.L.; Finke, C.M.; Knudson, R.A.; Ketterling, R.P.; Pardanani, A.; Tefferi, A. Differential prognostic effect of IDH1 versus IDH2 mutations in myelodysplastic syndromes: A Mayo Clinic study of 277 patients. Leukemia 2012, 26, 101–105. [Google Scholar] [CrossRef]
- Dinardo, C.D.; Jabbour, E.; Ravandi, F.; Takahashi, K.; Daver, N.; Routbort, M.J.; Patel, K.P.; Brandt, M.L.; Pierce, S.; Kantarjian, H.M.; et al. IDH1 and IDH2 mutations in myelodysplastic syndromes and role in disease progression. Leukemia 2016, 30, 980–984. [Google Scholar] [CrossRef] [Green Version]
- Pardanani, A.; Patnaik, M.M.; Lasho, T.L.; Mai, M.; Knudson, R.A.; Finke, C.; Ketterling, R.P.; McClure, R.F.; Tefferi, A. Recurrent IDH mutations in high-risk myelodysplastic syndrome or acute myeloid leukemia with isolated del(5q). Leukemia 2010, 24, 1370–1372. [Google Scholar] [CrossRef] [Green Version]
- Molenaar, R.J.; Thota, S.; Nagata, Y.; Patel, B.B.; Clemente, M.J.; Przychodzen, B.; Hirsh, C.; Viny, A.; Hosano, N.; Bleeker, F.E.; et al. Clinical and biological implications of ancestral and non-ancestral IDH1 and IDH2 mutations in myeloid neoplasms. Leukemia 2015, 29, 2134–2142. [Google Scholar] [CrossRef]
- Thol, F.; Weissinger, E.M.; Krauter, J.; Wagner, K.; Damm, F.; Wichmann, M.; Gohring, G.; Schumann, C.; Bug, G.; Ottmann, O.; et al. IDH1 mutations in patients with myelodysplastic syndromes are associated with an unfavorable prognosis. Haematologica 2010, 95, 1668–1674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DiNardo, C.D.; Stein, E.M.; de Botton, S.; Roboz, G.J.; Altman, J.K.; Mims, A.S.; Swords, R.; Collins, R.H.; Mannis, G.N.; Pollyea, D.A.; et al. Durable Remissions with Ivosidenib in IDH1-Mutated Relapsed or Refractory AML. N. Engl. J. Med. 2018, 378, 2386–2398. [Google Scholar] [CrossRef] [PubMed]
- Nazha, A.; Komrokji, R.; Meggendorfer, M.; Jia, X.; Radakovich, N.; Shreve, J.; Hilton, C.B.; Nagata, Y.; Hamilton, B.K.; Mukherjee, S.; et al. Personalized Prediction Model to Risk Stratify Patients with Myelodysplastic Syndromes. J. Clin. Oncol. 2021, 39, 3737–3746. [Google Scholar] [CrossRef]
- Fenaux, P.; Platzbecker, U.; Mufti, G.J.; Garcia-Manero, G.; Buckstein, R.; Santini, V.; Díez-Campelo, M.; Finelli, C.; Cazzola, M.; Ilhan, O.; et al. Luspatercept in Patients with Lower-Risk Myelodysplastic Syndromes. N. Engl. J. Med. 2020, 382, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Sanada, M.; Shiraishi, Y.; Nowak, D.; Nagata, Y.; Yamamoto, R.; Sato, Y.; Sato-Otsubo, A.; Kon, A.; Nagasaki, M.; et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011, 478, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Young, A.L.; Wong, T.N.; Hughes, A.E.; Heath, S.E.; Ley, T.J.; Link, D.C.; Druley, T.E. Quantifying ultra-rare pre-leukemic clones via targeted error-corrected sequencing. Leukemia 2015, 29, 1608–1611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernard, E.; Tuechler, H.; Greenberg, P.; Hasserjian, R.; Ossa, J.A.; Nannya, Y.; Devlin, S.; Creignou, M.; Pinel, P.; Monnier, L.; et al. Molecular International Prognosis Scoring System for Myelodysplastic Syndromes. NEJM Evid. 2021, 1, EVIDoa2200008. [Google Scholar]
- Choi, R.Y.; Coyner, A.S.; Kalpathy-Cramer, J.; Chiang, M.F.; Campbell, J.P. Introduction to Machine Learning, Neural Networks, and Deep Learning. Transl. Vis. Sci. Technol. 2020, 9, 14. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the dimensionality of data with neural networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef] [Green Version]
- Mori, J.; Kaji, S.; Kawai, H.; Kida, S.; Tsubokura, M.; Fukatsu, M.; Harada, K.; Noji, H.; Ikezoe, T.; Maeda, T.; et al. Assessment of dysplasia in bone marrow smear with convolutional neural network. Sci. Rep. 2020, 10, 14734. [Google Scholar] [CrossRef] [PubMed]
- Herbig, M.; Jacobi, A.; Wobus, M.; Weidner, H.; Mies, A.; Krater, M.; Otto, O.; Thiede, C.; Weickert, M.T.; Gotze, K.S.; et al. Machine learning assisted real-time deformability cytometry of CD34+ cells allows to identify patients with myelodysplastic syndromes. Sci. Rep. 2022, 12, 870. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, C.A.; Bill, M.; Rodrigues, M.A.; Hauerslev, M.; Kerndrup, G.B.; Hokland, P.; Ludvigsen, M. Exploring dyserythropoiesis in patients with myelodysplastic syndrome by imaging flow cytometry and machine-learning assisted morphometrics. Cytom. Part B Clin. Cytom. 2021, 100, 554–567. [Google Scholar] [CrossRef] [PubMed]





| Prognostic Subgroup | Defining Cytogenetic Abnormalities |
|---|---|
| Very good | Loss of Y chromosome |
| del(11q) | |
| Good | Normal |
| del(5q) | |
| del(12p) | |
| del(20q) | |
| Double, including del(5q) | |
| Intermediate | del(7q) |
| Gain of chromosome 8 | |
| Gain of chromosome 19 | |
| Isochromosome 17q | |
| Single or double abnormalities not specified in other subgroups | |
| Two or more independent non-complex clones | |
| Poor | Loss of chromosome 7 |
| Inv(3), t(3q) or del(3q) | |
| Double including loss of chromosome 7 or del(7q) | |
| Complex (three abnormalities) | |
| Very poor | Complex (>three abnormalities |
| Prognostic Variable | Score Values | ||||||
|---|---|---|---|---|---|---|---|
| 0 | 0.5 | 1 | 1.5 | 2 | 3 | 4 | |
| Karyotype (CCSS) | Very good | - | Good | - | Intermediate | Poor | Very poor |
| BM blast percentage | ≤2% | - | >2% to <5% | - | 5–10% | >10% | - |
| Hemoglobin concentration (g/dL) | ≥10 | - | 8 to <10 | <8 | - | - | - |
| Platelets (×109/L) | ≥100 | 50 to <100 | <50 | - | - | - | - |
| Absolute neutrophil count (×109/L) | ≥0.8 | <0.8 | - | - | - | - | - |
| Five risk groups are defined on the basis of total score of the parameters listed above: Very low: ≤1.5 Low: >1.5 to 3 Intermediate: >3 to 4.5 High: >4.5 to 6 Very high: >6 | |||||||
| - Indicates not applicable | |||||||
| Chromosomal Abnormality | Frequency | ||
|---|---|---|---|
| MDS Overall | Therapy-Related MDS | ||
| Unbalanced | Gain of chromosome 8 * | 10% | |
| Loss of chromosome 7 or del(7q) | 10% | 50% | |
| del(5q) | 10% | 40% | |
| del(20q) * | 5% to 8% | ||
| Loss of Y chromosome * | 5% | ||
| Isochromosome 17q or t(17p) | 3% to 5% | 25% to 30% | |
| Loss of chromosome 13 or del(13q) | 3% | ||
| del(11q) | 3% | ||
| del(12p) or t(12p) | 3% | ||
| idic(X)(q13) | 1% to 2% | ||
| Balanced | t(11;16)(q23.3;p13.3) | 3% | |
| t(3:21)(q23.2;q22.1) | 2% | ||
| t(1:3)(p36.3;q21.2) | 1% | ||
| t(2;11)(p21;q23.3) | 1% | ||
| inv(3)(q21.3;q26.2)/t(3;3)(q21.3;q26.2) | 1% | ||
| t(6;9)(p23;q34.1) | 1% | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tria, F.P., IV; Ang, D.C.; Fan, G. Myelodysplastic Syndrome: Diagnosis and Screening. Diagnostics 2022, 12, 1581. https://doi.org/10.3390/diagnostics12071581
Tria FP IV, Ang DC, Fan G. Myelodysplastic Syndrome: Diagnosis and Screening. Diagnostics. 2022; 12(7):1581. https://doi.org/10.3390/diagnostics12071581
Chicago/Turabian StyleTria, Francisco P., IV, Daphne C. Ang, and Guang Fan. 2022. "Myelodysplastic Syndrome: Diagnosis and Screening" Diagnostics 12, no. 7: 1581. https://doi.org/10.3390/diagnostics12071581
APA StyleTria, F. P., IV, Ang, D. C., & Fan, G. (2022). Myelodysplastic Syndrome: Diagnosis and Screening. Diagnostics, 12(7), 1581. https://doi.org/10.3390/diagnostics12071581





