The Santorini Volcanic Complex as a Valuable Source of Enzymes for Bioenergy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Source of the Investigated Metagenomic Data
2.2. List of Genes
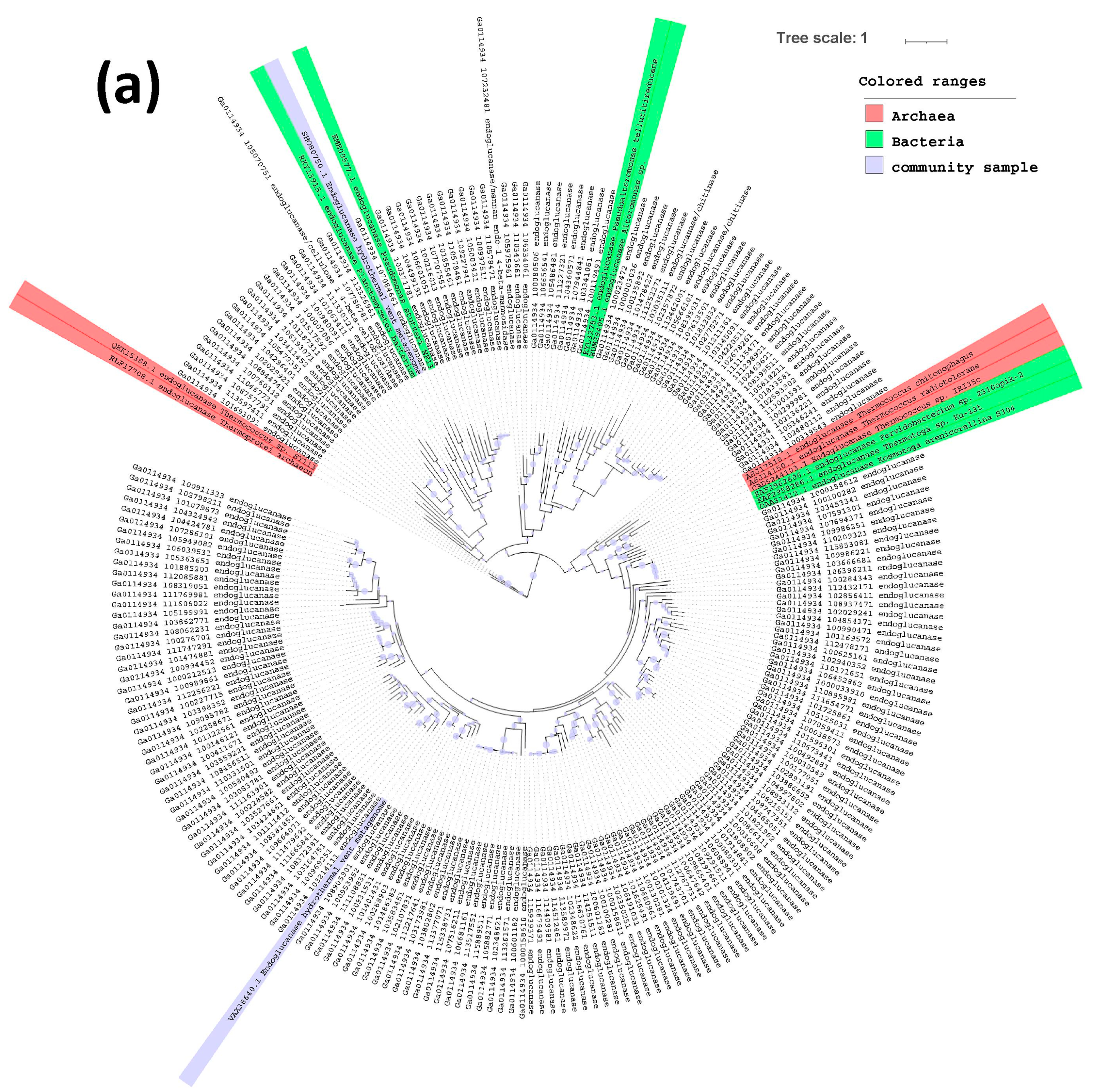
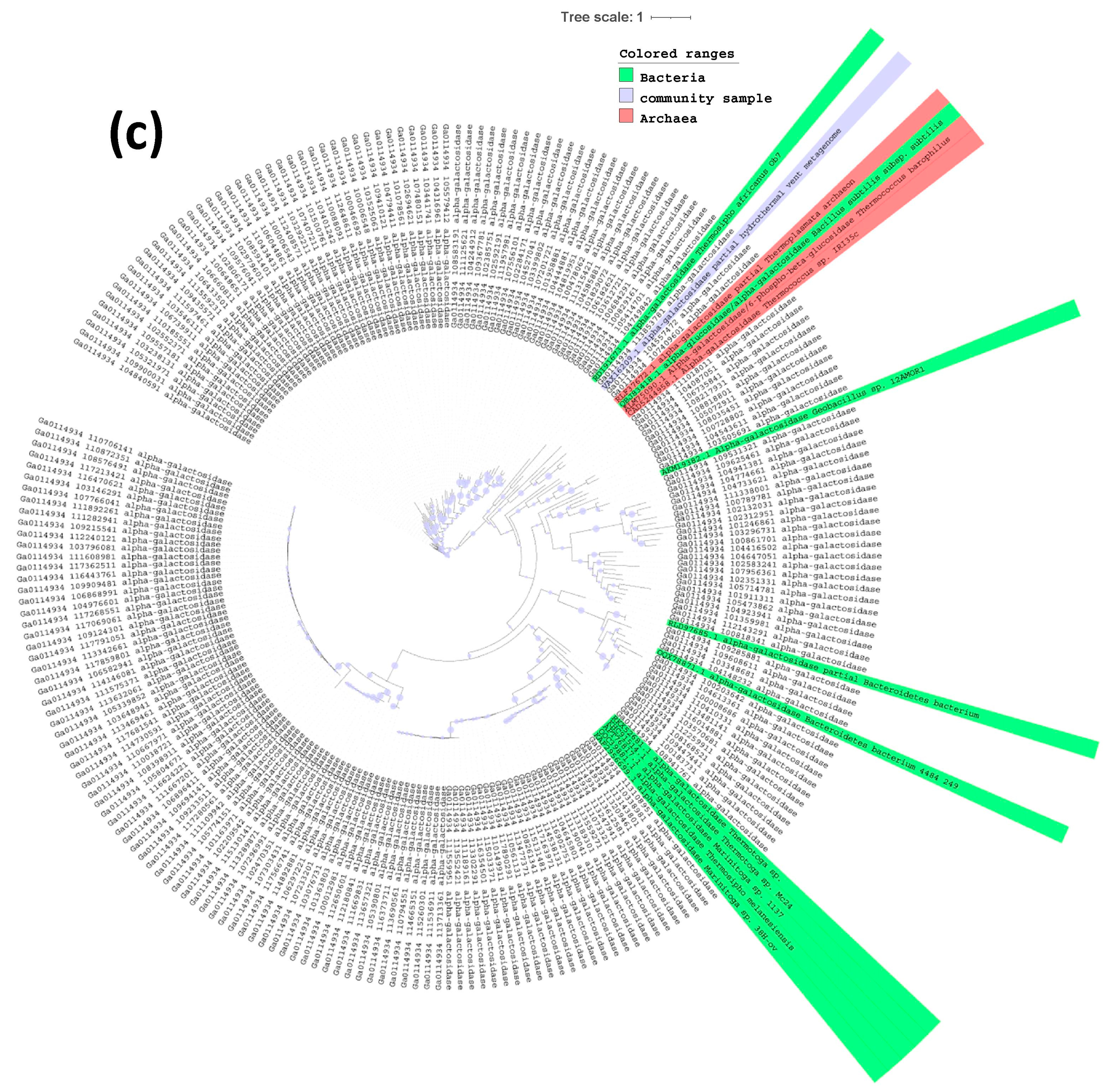
2.3. Phylogenetic Tree of Endoglucanases, Beta-Glucosidases and Alpha-Galactosidases
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dalmazo, G.Z.L.; Ferreira, D.; Vermelho, A.B. Marine Extremophiles: A source of hydrolases for biotechnological applications. Mar. Drugs 2015, 13, 1925–1965. [Google Scholar] [CrossRef] [Green Version]
- Coker, J.A. Extremophiles and biotechnology: Current uses and prospects. F100Research 2016, 5, 396. [Google Scholar] [CrossRef] [PubMed]
- Orcutt, B.; Sylvan, J.B.; Knab, N.J.; Edwards, K.J. Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol. Mol. Biol. Rev. 2011, 75, 361–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivarsson, M.; Broman, C.; Sturkell, E.; Ormö, J.; Siljeström, S.; van Zuilen, M.; Bengtson, S. Fungal colonization of an Ordovician impact-induced hydrothermal system. Nat. Sci. Rep. 2013, 3, 3487. [Google Scholar] [CrossRef]
- Oulas, A.; Polymenakou, P.N.; Seshadri, R.; Tripp, H.J.; Mandalakis, M.; Paez-Espino, D.A.; Pati, A.; Chain, P.; Nomikou, P.; Carey, S.; et al. Metagenomic investigation of the geologically unique Hellenic Volcanic Arc reveals a distinctive ecosystem with unexpected physiology. Env. Microbiol. 2016, 18, 1122–1136. [Google Scholar] [CrossRef] [PubMed]
- Drake, H.; Ivarsson, M.; Bengtson, S.; Heim, C.; Siljeström, S.; Whitehouse, M.J.; Broman, C.; Belivanova, V.; Åström, M.E. Anaerobic consortia of fungi and sulfate reducing bacteria in deep granite fractures. Nat. Commun. 2017, 8, 55. [Google Scholar] [CrossRef] [Green Version]
- Le Pichon, X.; Angelier, J. The Hellenic Arc and Trench system: A key to the neotectonic evolution of the Eastern Mediterranean area. Tectonophysics 1979, 60, 1–42. [Google Scholar] [CrossRef]
- Nomikou, P.; Carey, S.; Papanikolaou, D.; Croff Bell, K.; Sakellariou, D.; Alexandri, M.; Bejelou, K. Submarine Volcanoes of the Kolumbo volcanic zone NE of Santorini Caldera, Greece. Glob. Planet. Chang. 2012, 90–91, 135–151. [Google Scholar] [CrossRef]
- Kilias, S.P.; Nomikou, P.; Papanikolaou, D.; Polymenakou, P.N.; Godelitsas, A.; Argyraki, A.; Carey, S.; Gamaletsos, P.; Mertimekis, T.J.; Stathopoulou, E.; et al. New insights into hydrothermal vent processes in the unique shallow-submarine arc-volcano Kolumbo (Santorini), Greece. Nat. Sci. Rep. 2013, 3, 2421. [Google Scholar] [CrossRef]
- Dando, P.R.; Aliani, S.; Arab, H.; Bianchi, C.N.; Brehmer, M.; Cocito, S.; Fowlers, S.W.; Gundersen, J.; Hooper, L.E.; Kölbh, R.; et al. Hydrothermal studies in the Aegean Sea. Phys. Chem. Earth B 2000, 25, 1–8. [Google Scholar] [CrossRef]
- Dando, P.R.; Hughes, J.A.; Leahy, Y.; Niven, S.J.; Smith, C. Gas venting rates from submarine hydrothermal areas around the island of Milos, Hellenic Volcanic Arc. Cont. Shelf Res. 1995, 15, 913–929. [Google Scholar] [CrossRef]
- Sigurdsson, H.; Carey, S.; Alexandri, M.; Vougioukalakis, G.; Croff, K.; Roman, C.; Sakellariou, D.; Anagnostou, C.; Rousakis, G.; Ioakim, C.; et al. Marine investigations of Greece’s Santorini volcanic field. Eos 2006, 87, 337–342. [Google Scholar] [CrossRef] [Green Version]
- Hall-Spencer, J.M.; Rodolfo-Metalpa, R.; Martin, S.; Ransome, E.; Fine, M.; Turner, S.M.; Rowley, S.J.; Tedesco, D.; Buia, M.-C. Volcanic carbon dioxide vents show ecosystem effects of ocean acidification. Nature 2008, 454, 96–99. [Google Scholar] [CrossRef] [Green Version]
- Carey, S.; Nomikou, P.; Croff-Bell, K.; Lilley, M.; Lupton, J.; Roman, C.; Stathopoulou, E.; Bejelou, K.; Ballard, R. CO2 degassing from hydrothermal vents at Kolumbo submarine volcano, Greece, and the accumulation of acidic crater water. Geology 2013, 41, 1035–1038. [Google Scholar] [CrossRef]
- Christakis, C.A.; Polymenakou, P.N.; Mandalakis, M.; Nomikou, P.; Kristoffersen, J.B.; Lampridou, D.; Kotoulas, G.; Mandalakis, M. Microbial community differentiation between active and inactive sulfide chimneys of the Kolumbo submarine volcano, Hellenic Volcanic Arc. Extremophiles 2018, 22, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Mandalakis, M.; Gabriilidou, A.; Polymenakou, P.N.; Christakis, C.A.; Nomikou, P.; Medvecký, M.; Kilias, S.P.; Kentouri, M.; Kotoulas, G.; Magoulas, A. Microbial strains isolated from CO2-venting Kolumbo submarine volcano show enhanced co-tolerance to acidity and antibiotics. Mar. Env. Res. 2019, 144, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Christakis, C.A.; Polymenakou, P.N.; Kilias, S.P.; Nomikou, P.; Nielsen, T.; Kyrpides, N.; Kristoffersen, J.B.; Kotoulas, G.; Magoulas, A. Metagenomic evidence of microbial iron oxidation in the iron-mat ecosystem of the Kallisti Limnes low-temperature CO2-rich seafloor hydrothermal pools, Santorini. Unpublished work. 2021. [Google Scholar]
- Christakis, C.A.; Polymenakou, P.N.; Kilias, S.P.; Nomikou, P.; Nielsen, T.; Kyrpides, N.; Kristoffersen, J.B.; Kotoulas, G.; Magoulas, A. Metagenomic exploration of three different microbial mat layers of a polymetallic gas chimney, Kolumbo volcano, Hellenic Volcanic Arc. Unpublished work. 2021. [Google Scholar]
- Barrangou, R.; Horvarth, P. A decade of discovery: CRISPR functions and applications. Nat. Microbiol. 2017, 2, 17092. [Google Scholar] [CrossRef]
- Lauria, M.; Molinari, F.; Motto, M. Genetic Strategies to Enhance Plant Biomass Yield and Quality—Related Traits for Bio-Renewable Fuel and Chemical Productions; Intechopen: London, UK, 2014. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.C.; Chen, Y.C.; Hseu, R.S. Purification and characterization of a cellulolytic multienzyme complex produced by Neocallimastix patriciarum J11. Biochem. Biophys. Res. Commun. 2014, 451, 190–195. [Google Scholar] [CrossRef]
- Sukumaran, K.; Abraham, A.; Mathew, A.K. Enzymes for Bioenergy. In Bioresources and Bioprocess in Biotechnology; Sugathan, S., Pradeep, N.S., Abdulhameed, S., Eds.; Springer: Singapore, 2017; Volume 2, pp. 3–44. [Google Scholar]
- Barruetabeña, N.; Alonso-Lerma, B.; Galera-Prat, A.; Joudeh, N.; Barandiaran, L.; Aldazabal, L.; Arbulu, M.; Alcalde, M.; De Sancho, D.; Gavira, J.A.; et al. Resurrection of efficient Precambrian endoglucanases for lignocellulosic biomass hydrolysis. Comm. Chem. 2019, 2, 76. [Google Scholar] [CrossRef] [Green Version]
- Sticklen, M.B. Plant genetic engineering for biofuel production: Towards affordable cellulosic ethanol. Nat. Rev. Genet. 2008, 9, 433–443. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, L.; Liu, D. Biomass recalcitrance Part I: The chemical compositions and physical structures affecting the enzymatic hydrolysis of lignocellulose. Biofuels Bioprod. Biorefin. 2012, 6, 465–482. [Google Scholar] [CrossRef]
- Gírio, F.M.; Fonseca, C.; Carvalheiro, F.; Duarte, L.C.; Marques, S.; Bogel-Łukasik, R. Hemicelluloses for fuel ethanol: A review. Bioresour. Technol. 2010, 101, 4775–4800. [Google Scholar] [CrossRef]
- Kumar, P.; Barrett, D.M.; Delwiche, M.J.; Stroeve, P. Methods for pretreatment of lignocellulosic biomass for efficient hydrolysis and biofuel production. Ind. Eng. Chem. Res. 2009, 48, 3713–3729. [Google Scholar] [CrossRef]
- Schulein, M. Cellulases of Trichoderma reesei. In Methods in Enzymology; Wodd, W.A., Abelson, J.N., Eds.; Elsevier: New York, NY, USA, 1998; Volume 160, pp. 234–242. [Google Scholar]
- Jonsson, V. Statistical Analysis and Modelling of Gene Count Data in Metagenomics. Ph.D. Thesis, Clamers University of Technology and Göteborg University, Gothenburg, Sweden, 2017. [Google Scholar]
- Nielsen, H.; Tsirigos, K.D.; Brunak, S.; von Heijne, G. A brief history of protein sorting prediction. Protein J. 2019, 38, 200–216. [Google Scholar] [CrossRef] [Green Version]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef] [Green Version]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidson, S. Sustainable bioenergy: Genomics and biofuels development. Nat. Educ. 2008, 1, 175. [Google Scholar]
- Sukumaran, R.K.; Singhania, R.R.; Pandey, A. Microbial cellulases—Production, applications and challenges. J. Sci. Ind. Res. 2005, 64, 832–844. [Google Scholar]
- Yennamalli, R.M.; Rader, A.J.; Kenny, A.J.; Wolt, J.D.; Sen, T.Z. Endoglucanases: Insights into thermostability for biofuel applications. Biotechnol. Biofuels 2013, 6, 136. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Wang, L. Chapter 4—Enzymatic hydrolysis of pretreated biomass. In Technologies for Biochemical Conversion of Biomass; Academic Press: Cambridge, CA, USA, 2017; pp. 65–99. [Google Scholar]
- Xue, C.; Cheng, C. Chapter Two—Butanol production by Clostridium. Adv. Bioenergy 2019, 4, 35–77. [Google Scholar]
- Kim, D.-S.; Chi, W.-J.; Hong, S.-K. Molecular characterization of an endo-β-1,4-glucanase, CelAJ93, from the recently isolated marine bacterium, Cellulophaga sp. J9-3. Appl. Sci. 2019, 9, 4061. [Google Scholar] [CrossRef] [Green Version]
- Thomas, L.; Ram, H.; Singh, V.P. Inducible cellulase production from an organic solvent tolerant Bacillus sp. SV1 and evolutionary divergence of endoglucanase in different species of the genus Bacillus. Braz. J. Microbiol. 2018, 49, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Pang, H.; Zhang, P.; Duan, C.-J.; Mo, X.-C.; Tang, J.-L.; Feng, J.-X. Identification of cellulase genes from the metagenomes of compost soils and functional characterization of one novel endoglucanase. Curr. Microbiol. 2009, 58, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Fan, Y.; Zheng, H.; Du, F.; Zhang, K.-Q.; Huang, X.; Wang, L.; Zhang, M.; Niu, Q. Isolation and characterization of a novel endoglucanase from a Bursaphelenchus xylophilus metagenomic library. PLoS ONE 2013, 8, e82437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.W.; Ishikawa, K. Structure of hyperthermophilic endocellulase from Pyrococcus horikoshii. Proteins Struct. Funct. Bioinforma. 2009, 78, 496–500. [Google Scholar] [CrossRef]
- Suleiman, M.; Schröder, C.; Klippel, B.; Schäfers, C.; Krüger, A.; Antranikian, G. Extremely thermoactive archaeal endoglucanase from a shallow marine hydrothermal vent from Vulcano Island. Appl. Microbiol. Biotechnol. 2019, 103, 1267–1274. [Google Scholar] [CrossRef]
- Tallapragada, P.; Dikshit, R. Chapter 11—Microbial Production of Secondary Metabolites As Food Ingredients. In Microbial Production of Food Ingredients and Additives; Handbook of Food Bioengineering; Academic Press: Cambridge, CA, USA, 2017; pp. 317–345. [Google Scholar]
- Yeoman, C.J.; Han, Y.; Dodd, D.; Schroeder, C.M.; Mackie, R.I.; Cann, I.K.O. Thermostable enzymes as biocatalysts in the biofuel industry. Adv. Appl. Microbiol. 2010, 70, 1–55. [Google Scholar]
- Molina, G.; Contesini, F.J.; de Melo, R.R.; Sato, H.H.; Pastore, G.M. Chapter 11—β-Glucosidase from Aspergillus. In New and Future Developments in Microbial Biotechnology and Bioengineering; Gupta, V.K., Ed.; Elsevier: New York, NY, USA, 2016; pp. 155–169. ISBN 9780444635051. [Google Scholar]
- Dombrowski, N.; Teske, A.P.; Baker, B.J. Expansive microbial metabolic versatility and biodiversity in dynamic Guaymas Basin hydrothermal sediments. Nat. Commun. 2018, 9, 4999. [Google Scholar] [CrossRef]
- Seitz, K.W.; Dombrowski, N.; Eme, L.; Spang, A.; Lombard, J.; Sieber, J.R.; Teske, A.P.; Ettema, T.J.G.; Baker, B.J. Asgard archaea capable of anaerobic hydrocarbon cycling. Nat. Commun. 2019, 10, 1822. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Cao, M.E.; Cerdán, M.E.; González-Siso, M.I.; Becerra, M. Bioconversion of beet molasses to slpha-galactosidase and ethanol. Front Microbiol. 2019, 10, 405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wissuwa, J.; Stokke, R.; Fedoy, A.E.; Lian, K.; Smalas, A.O.; Steen, I.H. Isolation and complete genome sequence of the thermophilic Geobacillus sp. 12AMOR1 from an Arctic deep-sea hydrothermal vent site. Stand Genom. Sci. 2016, 11, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marteinsson, V.T.; Birrien, J.L.; Reysenbach, A.L.; Vernet, M.; Marie, D.; Gambacorta, A.; Messner, P.; Sleytr, U.B.; Prieur, D. Thermococcus barophilus sp. nov., a new barophilic and hyperthermophilic archaeon isolated under high hydrostatic pressure from a deep-sea hydrothermal vent. Int. J. Syst. Bacteriol. 1999, 49, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Kao, M.R.; Kuo, H.W.; Lee, C.C.; Huang, K.-Y.; Huang, T.-Y.; Li, C.-W.; Chen, C.W.; Wang, A.H.-J.; Yu, S.M.; Ho, T.-H.D. Chaetomella Raphigera β-glucosidase D2-BGL has intriguing structural features and a high substrate affinity that renders it an efficient cellulase supplement for lignocellulosic biomass hydrolysis. Biotechnol. Biofuels 2019, 12, 258. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues-Dutra, T.; Monteze-Guimarães, V.; Mascarenhas-Varela, E.; da Silva-Fialho, L.; Ferreira-Milagres, A.M.; Falkoski, D.L.; Zanuncio, J.C.; de Rezende, S.T. A Chrysoporthe cubensis enzyme cocktail produced from a low-cost carbon source with high biomass hydrolysis efficiency. Sci. Rep. 2017, 7, 3893. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, N.; Rathour, R.; Jha, S.; Pandey, K.; Srivastava, M.; Thakur, V.K.; Sengar, R.S.; Gupta, V.K.; Mazumder, P.B.; Khan, A.F.; et al. Microbial beta glucosidase enzymes: Recent advances in biomass conversation for biofuels application. Biomolecules 2019, 9, 220. [Google Scholar] [CrossRef] [Green Version]
- Gusakov, A.V.; Salanovich, T.M.; Antonov, A.I.; Ustinov, B.B.; Okunev, O.N.; Burlingam, R.P.; Emalfarb, M.A.; Baez, M.A.; Sinitsyn, A.P.; Arkady, P. Construction of Highly Efficient Cellulase Compositions for Enzymatic Hydrolysis of Cellulose. U.S. Patent 8916363, 23 December 2014. [Google Scholar]
- Maki, M.; Leung, K.T.; Qin, W. The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass. Int. J. Biol. Sci. 2009, 5, 500–516. [Google Scholar] [CrossRef]



| EC Number | WH1 | WH2 | RED1 | RED2 | RED3 | CH1-V16c.out | CH1-V16c.mid | CH1-V16c.in | CH2 | WAT1 | WAT2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. Cellulases | ||||||||||||
| endoglucanases (EG) | EC 3.2.1.4 | 637 | 222 | 1646 | 60 | 72 | 29 | 32 | 17 | 372 | 119 | 178 |
| beta-glucosidase (BGL) bgIX | EC 3.2.1.21 | 586 | 226 | 1164 | 33 | 98 | 21 | 19 | 22 | 282 | 262 | 296 |
| beta-glucosidase (BGL) E3.2.1.21 | EC 3.2.1.21 | 163 | 4 | 271 | 0 | 1 | 0 | 0 | 0 | 5 | 1 | 5 |
| beta-glucosidase (BGL) bgIB | EC 3.2.1.21 | 124 | 70 | 450 | 17 | 27 | 4 | 8 | 7 | 164 | 82 | 125 |
| cellulose 1,4-beta-cellobiosidase CBHA_B & CBHC | EC 3.2.1.91 | 18 | 21 | 36 | 3 | 3 | 0 | 2 | 0 | 9 | 9 | 12 |
| exoglucanases/cellobiohydrolases (CBH) | EC 3.2.1.176 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| exoglucanases/cellobiohydrolases (CBH) | EC 3.2.1.74 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2. Hemicellulases | ||||||||||||
| alpha-galactosidases E3.2.1.22B, gaIA, rafA | EC 3.2.1.22 | 153 | 216 | 530 | 9 | 23 | 14 | 13 | 10 | 180 | 86 | 179 |
| endo-1,4-beta-xylanase xynA | EC 3.2.1.8 | 129 | 38 | 285 | 10 | 11 | 4 | 7 | 5 | 114 | 38 | 55 |
| alpha-galactosidases melA | EC 3.2.1.22 | 111 | 50 | 445 | 25 | 48 | 3 | 4 | 1 | 88 | 102 | 206 |
| alpha-N-arabinofuranosidases abfA | EC 3.2.1.55 | 92 | 28 | 487 | 20 | 18 | 0 | 0 | 1 | 57 | 50 | 63 |
| beta-glucuronidase uidA, GUSB | EC 3.2.1.31 | 78 | 17 | 388 | 6 | 21 | 0 | 3 | 0 | 80 | 12 | 16 |
| xylan 1,4-beta-xylosidase xynB | EC 3.2.1.37 | 60 | 8 | 163 | 0 | 1 | 0 | 0 | 0 | 21 | 14 | 6 |
| mannans endo 1,4-beta-mannosidase gmuG | EC 3.2.1.78 | 38 | 5 | 159 | 2 | 6 | 0 | 1 | 0 | 28 | 6 | 5 |
| feruloyl esterase | EC 3.1.1.73 | 21 | 4 | 83 | 0 | 0 | 5 | 12 | 6 | 28 | 53 | 71 |
| acetylxylan esterases AXE1 | EC 3.1.1.72 | 7 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| licheninase | EC 3.2.1.73 | 7 | 1 | 23 | 1 | 1 | 0 | 0 | 1 | 2 | 1 | 7 |
| arabinoxylan arabinofuranohydrolase xynD | EC 3.2.1.55 | 2 | 2 | 13 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| alpha-galactosidases GLA | EC 3.2.1.22 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| beta-D-xylosidase 4 XYL4 | EC 3.2.1.37 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 3 |
| xyloglucan-specific endo-beta-1,4-glucanase XEG | EC 3.2.1.151 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| mannans endo 1,4-beta-mannosidase MAN | EC 3.2.1.78 | 0 | 4 | 0 | 0 | 3 | 0 | 0 | 0 | 6 | 1 | 4 |
| 3. Lignin-degrading enzymes | ||||||||||||
| laccases | EC 1.10.3.2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 |
| lignin peroxidase (LiP) | EC 1.11.1.14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| manganese peroxidase (MnP) | EC 1.11.1.13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Polymenakou, P.N.; Nomikou, P.; Zafeiropoulos, H.; Mandalakis, M.; Anastasiou, T.I.; Kilias, S.; Kyrpides, N.C.; Kotoulas, G.; Magoulas, A. The Santorini Volcanic Complex as a Valuable Source of Enzymes for Bioenergy. Energies 2021, 14, 1414. https://doi.org/10.3390/en14051414
Polymenakou PN, Nomikou P, Zafeiropoulos H, Mandalakis M, Anastasiou TI, Kilias S, Kyrpides NC, Kotoulas G, Magoulas A. The Santorini Volcanic Complex as a Valuable Source of Enzymes for Bioenergy. Energies. 2021; 14(5):1414. https://doi.org/10.3390/en14051414
Chicago/Turabian StylePolymenakou, Paraskevi N., Paraskevi Nomikou, Haris Zafeiropoulos, Manolis Mandalakis, Thekla I. Anastasiou, Stephanos Kilias, Nikos C. Kyrpides, Georgios Kotoulas, and Antoniοs Magoulas. 2021. "The Santorini Volcanic Complex as a Valuable Source of Enzymes for Bioenergy" Energies 14, no. 5: 1414. https://doi.org/10.3390/en14051414
APA StylePolymenakou, P. N., Nomikou, P., Zafeiropoulos, H., Mandalakis, M., Anastasiou, T. I., Kilias, S., Kyrpides, N. C., Kotoulas, G., & Magoulas, A. (2021). The Santorini Volcanic Complex as a Valuable Source of Enzymes for Bioenergy. Energies, 14(5), 1414. https://doi.org/10.3390/en14051414








