Challenges for Field-Effect-Transistor-Based Graphene Biosensors
Abstract
1. Introduction
2. Overview of Graphene Biosensing by Field Effects
- 1.
- Graphene has a high specific surface area because all its carbon atoms are present on the surface [78,79]. In addition, as it is a carbon material, it has a wide potential window [80]. Therefore, unlike silicon FET biosensors, which require a SiO2 insulating layer, the target in the aqueous solution is in direct contact with the graphene channel, inducing carriers effectively.
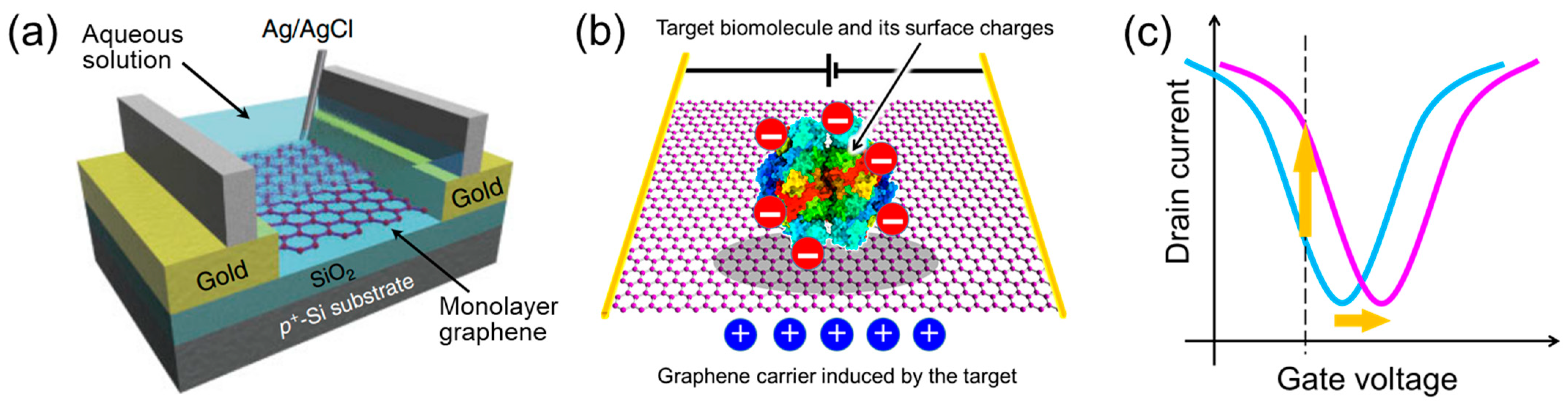
- 2.
- Very few carriers are induced by small amounts of target molecules. However, owing to the high carrier mobility of graphene [82,83], a small modulation of carriers can be converted to a large current change. In addition, the current-based detection method allows for label-free real-time sensing, unlike conventional optical detection methods.
- 3.
- Because the temporal changes in biosensor characteristics, such as current and the charge neutrality point, are monitored, an on/off ratio of the FET is not needed for the biosensing. The lack of a bandgap, which is the greatest weakness of graphene in semiconductor applications, is not an issue in graphene-FET biosensors [6,84,85].
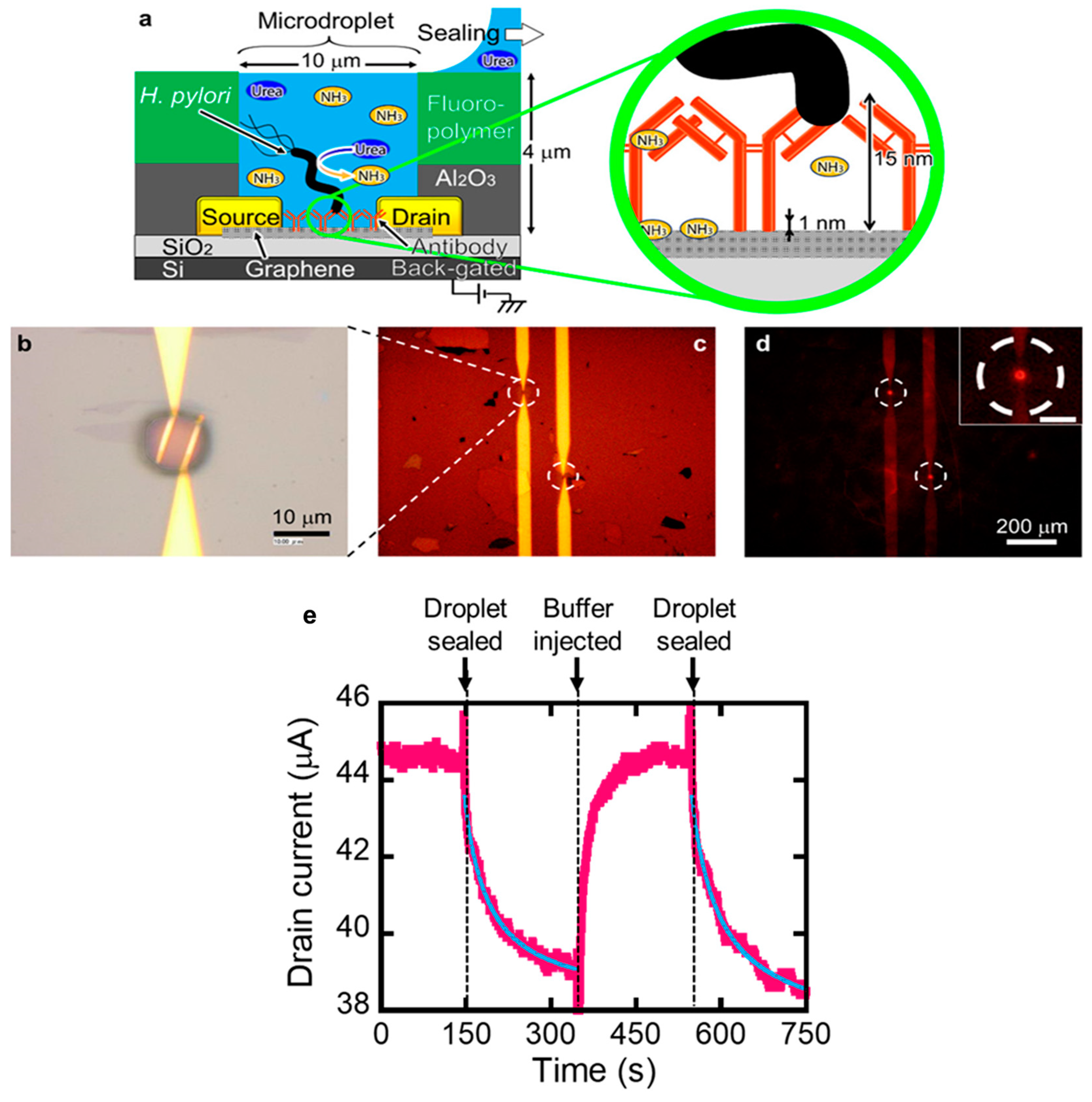
3. Debye Screening
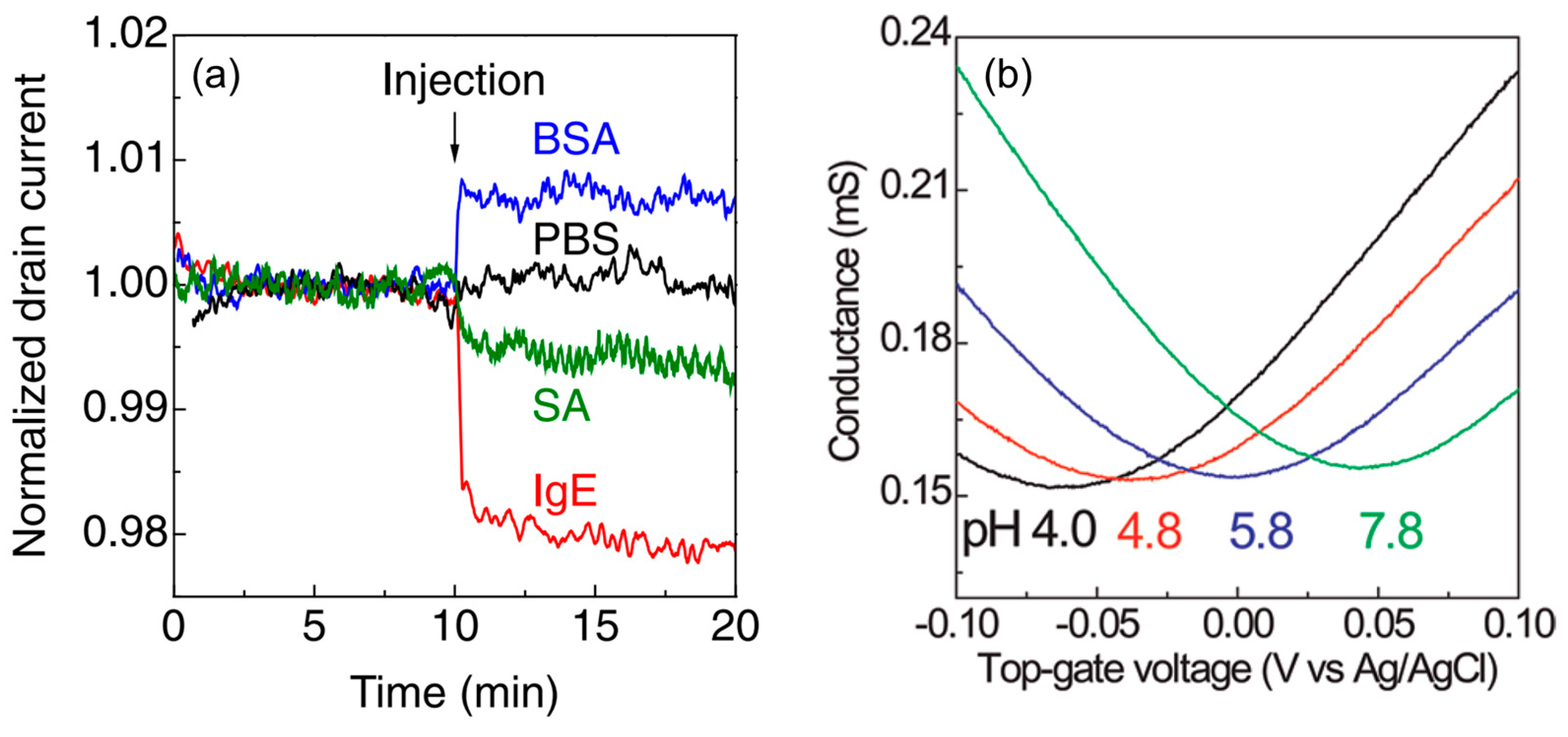
4. Attempt to Rationally Extract Signals from Samples with Complex Components
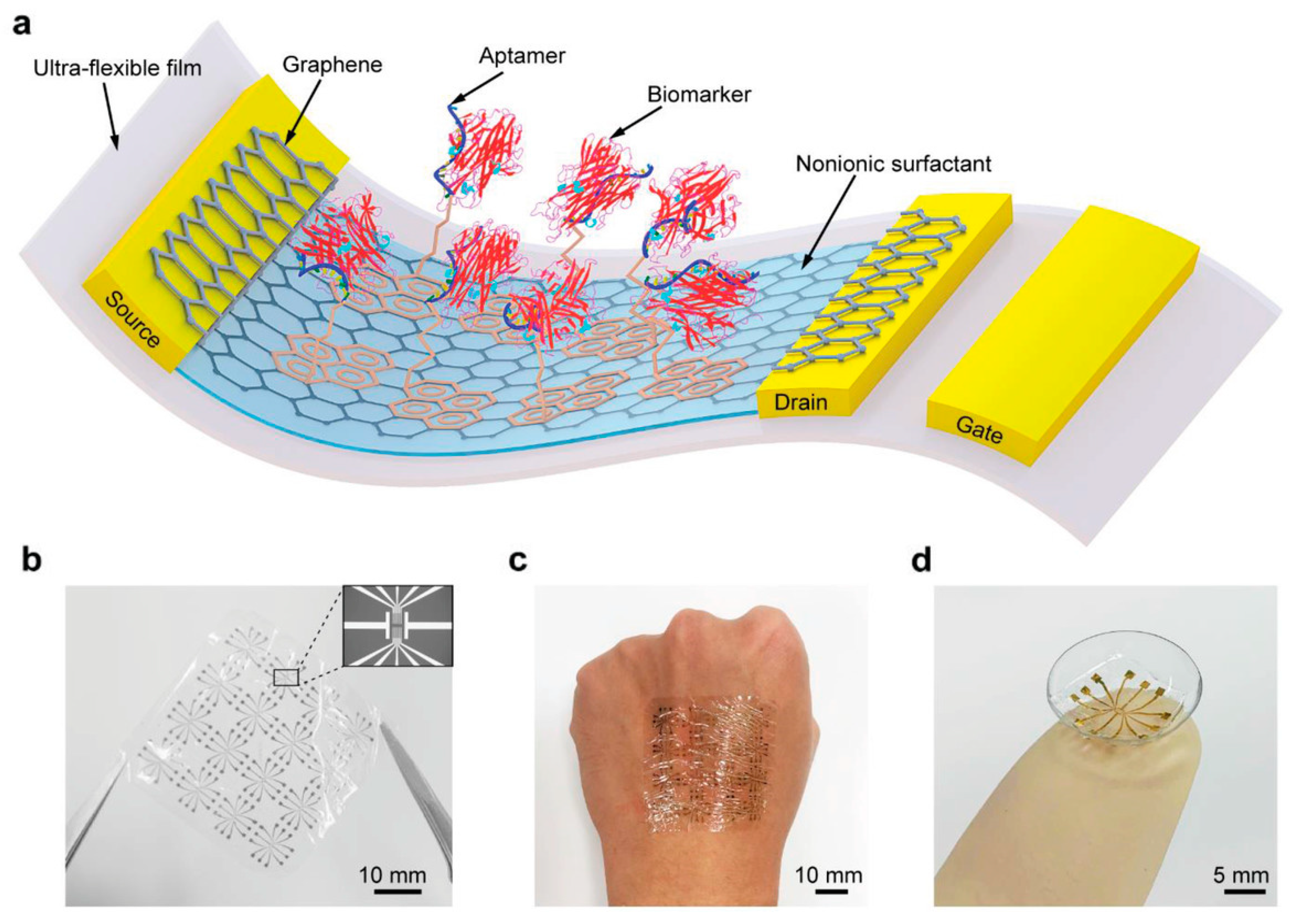
5. Multifaceted Evaluation of Measurement Systems by Combining Multiple Measurement Principles
6. Conclusions and Outlook
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liao, L.; Lin, Y.-C.; Bao, M.; Cheng, R.; Bai, J.; Liu, Y.; Qu, Y.; Wang, K.L.; Huang, Y.; Duan, X. High-Speed Graphene Transistors with a Self-Aligned Nanowire Gate. Nature 2010, 467, 305–308. [Google Scholar] [CrossRef] [PubMed]
- Novoselov, K.S.; Fal′ko, V.I.; Colombo, L.; Gellert, P.R.; Schwab, M.G.; Kim, K. A Roadmap for Graphene. Nature 2012, 490, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.; Bai, J.; Liao, L.; Zhou, H.; Chen, Y.; Liu, L.; Lin, Y.-C.; Jiang, S.; Huang, Y.; Duan, X. High-Frequency Self-Aligned Graphene Transistors with Transferred Gate Stacks. Proc. Natl. Acad. Sci. USA 2012, 109, 11588–11592. [Google Scholar] [CrossRef]
- Franklin, A.D. Nanomaterials in Transistors: From High-Performance to Thin-Film Applications. Science 2015, 349, aab2750. [Google Scholar] [CrossRef]
- Shimazaki, Y.; Yamamoto, M.; Borzenets, I.V.; Watanabe, K.; Taniguchi, T.; Tarucha, S. Generation and Detection of Pure Valley Current by Electrically Induced Berry Curvature in Bilayer Graphene. Nat. Phys. 2015, 11, 1032–1036. [Google Scholar] [CrossRef]
- Chhowalla, M.; Jena, D.; Zhang, H. Two-Dimensional Semiconductors for Transistors. Nat. Rev. Mater. 2016, 1, 16052. [Google Scholar] [CrossRef]
- Sharbati, M.T.; Du, Y.; Torres, J.; Ardolino, N.D.; Yun, M.; Xiong, F. Low-Power, Electrochemically Tunable Graphene Synapses for Neuromorphic Computing. Adv. Mater. 2018, 30, 1802353. [Google Scholar] [CrossRef] [PubMed]
- Muruganathan, M.; Van, N.H.; Schmidt, M.E.; Mizuta, H. Sub 0.5 Volt Graphene-hBN van Der Waals Nanoelectromechanical (NEM) Switches. Adv. Funct. Mater. 2022, 32, 2209151. [Google Scholar] [CrossRef]
- Bonaccorso, F.; Sun, Z.; Hasan, T.; Ferrari, A.C. Graphene Photonics and Optoelectronics. Nat. Photonics 2010, 4, 611–622. [Google Scholar] [CrossRef]
- Shimatani, M.; Ogawa, S.; Fujisawa, D.; Okuda, S.; Kanai, Y.; Ono, T.; Matsumoto, K. Photocurrent Enhancement of Graphene Phototransistors Using p–n Junction Formed by Conventional Photolithography Process. Jpn. J. Appl. Phys. 2016, 55, 110307. [Google Scholar] [CrossRef]
- Kufer, D.; Konstantatos, G. Photo-FETs: Phototransistors Enabled by 2D and 0D Nanomaterials. ACS Photonics 2016, 3, 2197–2210. [Google Scholar] [CrossRef]
- Shimatani, M.; Ogawa, S.; Fujisawa, D.; Okuda, S.; Kanai, Y.; Ono, T.; Matsumoto, K. Giant Dirac Point Shift of Graphene Phototransistors by Doped Silicon Substrate Current. AIP Adv. 2016, 6, 035113. [Google Scholar] [CrossRef]
- Fukushima, S.; Shimatani, M.; Okuda, S.; Ogawa, S.; Kanai, Y.; Ono, T.; Matsumoto, K. High Responsivity Middle-Wavelength Infrared Graphene Photodetectors Using Photo-Gating. Appl. Phys. Express 2018, 113, 061102. [Google Scholar] [CrossRef]
- Ogawa, S.; Shimatani, M.; Fukushima, S.; Okuda, S.; Kanai, Y.; Ono, T.; Matsumoto, K. Broadband Photoresponse of Graphene Photodetector from Visible to Long-Wavelength Infrared Wavelengths. Opt. Eng. 2019, 58, 057106. [Google Scholar] [CrossRef]
- Shimatani, M.; Ogawa, S.; Fukushima, S.; Okuda, S.; Kanai, Y.; Ono, T.; Matsumoto, K. Enhanced Photogating via Pyroelectric Effect Induced by Insulator Layer for High-Responsivity Long-Wavelength Infrared Graphene-Based Photodetectors Operating at Room Temperature. Appl. Phys. Express 2019, 12, 025001. [Google Scholar] [CrossRef]
- Fukushima, S.; Shimatani, M.; Okuda, S.; Ogawa, S.; Kanai, Y.; Ono, T.; Inoue, K.; Matsumoto, K. Low Dark Current and High-Responsivity Graphene Mid-Infrared Photodetectors Using Amplification of Injected Photo-Carriers by Photo-Gating. Opt. Lett. 2019, 44, 2598–2601. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, S.; Shimatani, M.; Okuda, S.; Ogawa, S.; Kanai, Y.; Ono, T.; Inoue, K.; Matsumoto, K. Photogating for Small High-Responsivity Graphene Middle-Wavelength Infrared Photodetectors. Opt. Eng. 2020, 59, 037101. [Google Scholar] [CrossRef]
- Tamura, K.; Tang, C.; Ogiura, D.; Suwa, K.; Fukidome, H.; Takida, Y.; Minamide, H.; Suemitsu, T.; Otsuji, T.; Satou, A. Fast and Sensitive Terahertz Detection with a Current-Driven Epitaxial-Graphene Asymmetric Dual-Grating-Gate Field-Effect Transistor Structure. APL Photonics 2022, 7, 126101. [Google Scholar] [CrossRef]
- Hassoun, J.; Bonaccorso, F.; Agostini, M.; Angelucci, M.; Betti, M.G.; Cingolani, R.; Gemmi, M.; Mariani, C.; Panero, S.; Pellegrini, V.; et al. An Advanced Lithium-Ion Battery Based on a Graphene Anode and a Lithium Iron Phosphate Cathode. Nano Lett. 2014, 14, 4901–4906. [Google Scholar] [CrossRef]
- Bonaccorso, F.; Colombo, L.; Yu, G.; Stoller, M.; Tozzini, V.; Ferrari, A.C.; Ruoff, R.S.; Pellegrini, V. Graphene, Related Two-Dimensional Crystals, and Hybrid Systems for Energy Conversion and Storage. Science 2015, 347, 1246501. [Google Scholar] [CrossRef]
- Yu, Z.; Tetard, L.; Zhai, L.; Thomas, J. Supercapacitor Electrode Materials: Nanostructures from 0 to 3 Dimensions. Energy Environ. Sci. 2015, 8, 702–730. [Google Scholar] [CrossRef]
- Sun, H.; Del Rio Castillo, A.E.; Monaco, S.; Capasso, A.; Ansaldo, A.; Prato, M.; Dinh, D.A.; Pellegrini, V.; Scrosati, B.; Manna, L.; et al. Binder-Free Graphene as an Advanced Anode for Lithium Batteries. J. Mater. Chem. A 2016, 4, 6886–6895. [Google Scholar] [CrossRef]
- Fang, R.; Chen, K.; Yin, L.; Sun, Z.; Li, F.; Cheng, H.-M. The Regulating Role of Carbon Nanotubes and Graphene in Lithium-Ion and Lithium–Sulfur Batteries. Adv. Mater. 2019, 31, 1800863. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.; Kim, Y.-K.; Shin, D.; Ryoo, S.-R.; Hong, B.H.; Min, D.-H. Biomedical Applications of Graphene and Graphene Oxide. Acc. Chem. Res. 2013, 46, 2211–2224. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Zhang, M.; Li, J. Solution-Gated Graphene Transistors for Chemical and Biological Sensors. Adv. Healthc. Mater. 2014, 3, 313–331. [Google Scholar] [CrossRef]
- Cheng, C.; Li, S.; Thomas, A.; Kotov, N.A.; Haag, R. Functional Graphene Nanomaterials Based Architectures: Biointeractions, Fabrications, and Emerging Biological Applications. Chem. Rev. 2017, 117, 1826–1914. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Adkins, M.; Wang, Z. Recent Progress on Semiconductor-Interface Facing Clinical Biosensing. Sensors 2021, 21, 3467. [Google Scholar] [CrossRef] [PubMed]
- Pasinszki, T.; Krebsz, M.; Tung, T.T.; Losic, D. Carbon Nanomaterial Based Biosensors for Non-Invasive Detection of Cancer and Disease Biomarkers for Clinical Diagnosis. Sensors 2017, 17, 1919. [Google Scholar] [CrossRef]
- Ku, M.; Kim, J.; Won, J.-E.; Kang, W.; Park, Y.-G.; Park, J.; Lee, J.-H.; Cheon, J.; Lee, H.H.; Park, J.-U. Smart, Soft Contact Lens for Wireless Immunosensing of Cortisol. Sci. Adv. 2020, 6, eabb2891. [Google Scholar] [CrossRef]
- Mohanty, N.; Berry, V. Graphene-Based Single-Bacterium Resolution Biodevice and DNA Transistor: Interfacing Graphene Derivatives with Nanoscale and Microscale Biocomponents. Nano Lett. 2008, 8, 4469–4476. [Google Scholar] [CrossRef]
- Ohno, Y.; Maehashi, K.; Yamashiro, Y.; Matsumoto, K. Electrolyte-Gated Graphene Field-Effect Transistors for Detecting pH and Protein Adsorption. Nano Lett. 2009, 9, 3318–3322. [Google Scholar] [CrossRef] [PubMed]
- Sofue, Y.; Ohno, Y.; Maehashi, K.; Inoue, K.; Matsumoto, K. Highly Sensitive Electrical Detection of Sodium Ions Based on Graphene Field-Effect Transistors. Jpn. J. Appl. Phys. 2011, 50, 06GE07. [Google Scholar] [CrossRef]
- Fakih, I.; Durnan, O.; Mahvash, F.; Napal, I.; Centeno, A.; Zurutuza, A.; Yargeau, V.; Szkopek, T. Selective Ion Sensing with High Resolution Large Area Graphene Field Effect Transistor Arrays. Nat. Commun. 2020, 11, 3226. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Lee, A.; Ban, D.K.; Wang, K.; Bandaru, P. Femtomolar Level-Specific Detection of Lead Ions in Aqueous Environments, Using Aptamer-Derivatized Graphene Field-Effect Transistors. ACS Appl. Nano Mater. 2023, 6, 2228–2235. [Google Scholar] [CrossRef]
- Schedin, F.; Geim, A.K.; Morozov, S.V.; Hill, E.W.; Blake, P.; Katsnelson, M.I.; Novoselov, K.S. Detection of Individual Gas Molecules Adsorbed on Graphene. Nat. Mater. 2007, 6, 652–655. [Google Scholar] [CrossRef] [PubMed]
- Nallon, E.C.; Schnee, V.P.; Bright, C.J.; Polcha, M.P.; Li, Q. Discrimination Enhancement with Transient Feature Analysis of a Graphene Chemical Sensor. Anal. Chem. 2016, 88, 1401–1406. [Google Scholar] [CrossRef]
- Kulkarni, G.S.; Reddy, K.; Zhong, Z.; Fan, X. Graphene Nanoelectronic Heterodyne Sensor for Rapid and Sensitive Vapour Detection. Nat. Commun. 2014, 5, 4376. [Google Scholar] [CrossRef]
- Tehrani, Z.; Burwell, G.; Azmi, M.A.M.; Castaing, A.; Rickman, R.; Almarashi, J.; Dunstan, P.; Beigi, A.M.; Doak, S.H.; Guy, O.J. Generic Epitaxial Graphene Biosensors for Ultrasensitive Detection of Cancer Risk Biomarker. 2D Mater. 2014, 1, 025004. [Google Scholar] [CrossRef]
- Torrente-Rodríguez, R.M.; Tu, J.; Yang, Y.; Min, J.; Wang, M.; Song, Y.; Yu, Y.; Xu, C.; Ye, C.; IsHak, W.W.; et al. Investigation of Cortisol Dynamics in Human Sweat Using a Graphene-Based Wireless mHealth System. Matter 2020, 2, 921–937. [Google Scholar] [CrossRef]
- Xu, G.; Abbott, J.; Qin, L.; Yeung, K.Y.M.; Song, Y.; Yoon, H.; Kong, J.; Ham, D. Electrophoretic and Field-Effect Graphene for All-Electrical DNA Array Technology. Nat. Commun. 2014, 5, 4866. [Google Scholar] [CrossRef]
- Li, H.; Yang, J.; Wu, G.; Weng, Z.; Song, Y.; Zhang, Y.; Vanegas, J.A.; Avery, L.; Gao, Z.; Sun, H.; et al. Amplification-Free Detection of SARS-CoV-2 and Respiratory Syncytial Virus Using CRISPR Cas13a and Graphene Field-Effect Transistors. Angew. Chem. Int. Ed. Engl. 2022, 61, e202203826. [Google Scholar] [CrossRef]
- Haslam, C.; Damiati, S.; Whitley, T.; Davey, P.; Ifeachor, E.; Awan, S.A. Label-Free Sensors Based on Graphene Field-Effect Transistors for the Detection of Human Chorionic Gonadotropin Cancer Risk Biomarker. Diagnostics 2018, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.; Chauhan, N.; Islam, S.; Mahari, S.; Ghawri, B.; Gandham, R.K.; Majumdar, S.S.; Ghosh, A.; Gandhi, S. Graphene Functionalized Field-Effect Transistors for Ultrasensitive Detection of Japanese Encephalitis and Avian Influenza Virus. Sci. Rep. 2020, 10, 14546. [Google Scholar] [CrossRef] [PubMed]
- Torrente-Rodríguez, R.M.; Lukas, H.; Tu, J.; Min, J.; Yang, Y.; Xu, C.; Rossiter, H.B.; Gao, W. SARS-CoV-2 RapidPlex: A Graphene-Based Multiplexed Telemedicine Platform for Rapid and Low-Cost COVID-19 Diagnosis and Monitoring. Matter 2020, 3, 1981–1998. [Google Scholar] [CrossRef]
- Sengupta, J.; Hussain, C.M. Graphene-Based Field-Effect Transistor Biosensors for the Rapid Detection and Analysis of Viruses: A Perspective in View of COVID-19. Carbon Trends 2021, 2, 100011. [Google Scholar] [CrossRef]
- Mannoor, M.S.; Tao, H.; Clayton, J.D.; Sengupta, A.; Kaplan, D.L.; Naik, R.R.; Verma, N.; Omenetto, F.G.; McAlpine, M.C. Graphene-Based Wireless Bacteria Detection on Tooth Enamel. Nat. Commun. 2012, 3, 763. [Google Scholar] [CrossRef]
- Cohen-Karni, T.; Qing, Q.; Li, Q.; Fang, Y.; Lieber, C.M. Graphene and Nanowire Transistors for Cellular Interfaces and Electrical Recording. Nano Lett. 2010, 10, 1098–1102. [Google Scholar] [CrossRef]
- Hess, L.H.; Jansen, M.; Maybeck, V.; Hauf, M.V.; Seifert, M.; Stutzmann, M.; Sharp, I.D.; Offenhäusser, A.; Garrido, J.A. Graphene Transistor Arrays for Recording Action Potentials from Electrogenic Cells. Adv. Mater. 2011, 23, 5045–5049. [Google Scholar] [CrossRef]
- Novodchuk, I.; Bajcsy, M.; Yavuz, M. Graphene-Based Field Effect Transistor Biosensors for Breast Cancer Detection: A Review on Biosensing Strategies. Carbon 2021, 172, 431–453. [Google Scholar] [CrossRef]
- Béraud, A.; Sauvage, M.; Bazán, C.M.; Tie, M.; Bencherif, A.; Bouilly, D. Graphene Field-Effect Transistors as Bioanalytical Sensors: Design, Operation and Performance. Analyst 2021, 146, 403–428. [Google Scholar] [CrossRef]
- Thangamani, J.G.; Deshmukh, K.; Kumar Sadasivuni, K.; Chidambaram, K.; Basheer Ahamed, M.; Ponnamma, D.; Al-Ali AlMaadeed, M.; Khadheer Pasha, S.K. Recent Advances in Electrochemical Biosensor and Gas Sensors Based on Graphene and Carbon Nanotubes (CNT)—A Review. Adv. Mater. Lett. 2017, 8, 196–205. [Google Scholar] [CrossRef]
- Lawal, A.T. Progress in Utilisation of Graphene for Electrochemical Biosensors. Biosens. Bioelectron. 2018, 106, 149–178. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Park, S.-J.; Choi, J.-W. Electrical Property of Graphene and Its Application to Electrochemical Biosensing. Nanomaterials 2019, 9, 297. [Google Scholar] [CrossRef] [PubMed]
- Stobiecka, M.; Dworakowska, B.; Jakiela, S.; Lukasiak, A.; Chalupa, A.; Zembrzycki, K. Sensing of Survivin mRNA in Malignant Astrocytes Using Graphene Oxide Nanocarrier-Supported Oligonucleotide Molecular Beacons. Sens. Actuators B Chem. 2016, 235, 136–145. [Google Scholar] [CrossRef]
- Ratajczak, K.; Krazinski, B.; Kowalczyk, A.; Dworakowska, B.; Jakiela, S.; Stobiecka, M. Optical Biosensing System for the Detection of Survivin mRNA in Colorectal Cancer Cells Using a Graphene Oxide Carrier-Bound Oligonucleotide Molecular Beacon. Nanomaterials 2018, 8, 510. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chen, T.; Gooding, J.J.; Liu, J. Review of Carbon and Graphene Quantum Dots for Sensing. ACS Sens. 2019, 4, 1732–1748. [Google Scholar] [CrossRef]
- Nesakumar, N.; Srinivasan, S.; Alwarappan, S. Graphene Quantum Dots: Synthesis, Properties, and Applications to the Development of Optical and Electrochemical Sensors for Chemical Sensing. Microchim. Acta 2022, 189, 258. [Google Scholar] [CrossRef]
- Nurrohman, D.T.; Chiu, N.-F. A Review of Graphene-Based Surface Plasmon Resonance and Surface-Enhanced Raman Scattering Biosensors: Current Status and Future Prospects. Nanomaterials 2021, 11, 216. [Google Scholar] [CrossRef]
- Patil, P.O.; Pandey, G.R.; Patil, A.G.; Borse, V.B.; Deshmukh, P.K.; Patil, D.R.; Tade, R.S.; Nangare, S.N.; Khan, Z.G.; Patil, A.M.; et al. Graphene-Based Nanocomposites for Sensitivity Enhancement of Surface Plasmon Resonance Sensor for Biological and Chemical Sensing: A Review. Biosens. Bioelectron. 2019, 139, 111324. [Google Scholar] [CrossRef]
- Paulus, G.L.C.; Nelson, J.T.; Lee, K.Y.; Wang, Q.H.; Reuel, N.F.; Grassbaugh, B.R.; Kruss, S.; Landry, M.P.; Kang, J.W.; Vander Ende, E.; et al. A Graphene-Based Physiometer Array for the Analysis of Single Biological Cells. Sci. Rep. 2014, 4, 6865. [Google Scholar] [CrossRef]
- Ishraq, S.; Liu, Y. Synthesis, Characterization and Bioapplications of Pristine Graphene: A Review. Univers. J. Carbon Res. 2023, 1, 1–21. [Google Scholar] [CrossRef]
- Jung, J.H.; Cheon, D.S.; Liu, F.; Lee, K.B.; Seo, T.S. A Graphene Oxide Based Immuno-Biosensor for Pathogen Detection. Angew. Chem. Int. Ed. Engl. 2010, 122, 5844–5847. [Google Scholar] [CrossRef]
- Hasegawa, M.; Hirayama, Y.; Ohno, Y.; Maehashi, K.; Matsumoto, K. Characterization of Reduced Graphene Oxide Field-Effect Transistor and Its Application to Biosensor. Jpn. J. Appl. Phys. 2014, 53, 05FD05. [Google Scholar] [CrossRef]
- Rowley-Neale, S.J.; Randviir, E.P.; Abo Dena, A.S.; Banks, C.E. An Overview of Recent Applications of Reduced Graphene Oxide as a Basis of Electroanalytical Sensing Platforms. Appl. Mater. Today 2018, 10, 218–226. [Google Scholar] [CrossRef]
- Fan, Z.; Li, S.; Yuan, F.; Fan, L. Fluorescent Graphene Quantum Dots for Biosensing and Bioimaging. RSC Adv. 2015, 5, 19773–19789. [Google Scholar] [CrossRef]
- Xie, R.; Wang, Z.; Zhou, W.; Liu, Y.; Fan, L.; Li, Y.; Li, X. Graphene Quantum Dots as Smart Probes for Biosensing. Anal. Methods 2016, 8, 4001–4016. [Google Scholar] [CrossRef]
- Shende, P.; Augustine, S.; Prabhakar, B. A Review on Graphene Nanoribbons for Advanced Biomedical Applications. Carbon Lett. 2020, 30, 465–475. [Google Scholar] [CrossRef]
- Luo, S.; Chen, X.; He, Y.; Gu, Y.; Zhu, C.; Yang, G.-H.; Qu, L.-L. Recent Advances in Graphene Nanoribbons for Biosensing and Biomedicine. J. Mater. Chem. B 2021, 9, 6129–6143. [Google Scholar] [CrossRef]
- Lawal, A.T. Graphene-Based Nano Composites and Their Applications. A Review. Biosens. Bioelectron. 2019, 141, 111384. [Google Scholar] [CrossRef]
- Kumar Krishnan, S.; Singh, E.; Singh, P.; Meyyappan, M.; Singh Nalwa, H. A Review on Graphene-Based Nanocomposites for Electrochemical and Fluorescent Biosensors. RSC Adv. 2019, 9, 8778–8881. [Google Scholar] [CrossRef]
- Iftikhar, T.; Iftikhar, N.; Chi, G.; Qiu, W.; Xie, Y.; Liang, Z.; Huang, C.; Su, L. Unlocking the Future of Brain Research: MOFs, TMOs, and MOFs/TMOs for Electrochemical NTMs Detection and Analysis. Talanta 2024, 267, 125146. [Google Scholar] [CrossRef] [PubMed]
- Kanai, Y.; Ishibashi, Y.; Ono, T.; Inoue, K.; Ohno, Y.; Maehashi, K.; Matsumoto, K. Dynamical Thermodiffusion Model of Graphene Synthesis on Polymer Films by Laser Irradiation and Application to Strain Sensors. Jpn. J. Appl. Phys. 2017, 56, 075102. [Google Scholar] [CrossRef]
- Qiao, Y.; Li, X.; Hirtz, T.; Deng, G.; Wei, Y.; Li, M.; Ji, S.; Wu, Q.; Jian, J.; Wu, F.; et al. Graphene-Based Wearable Sensors. Nanoscale 2019, 11, 18923–18945. [Google Scholar] [CrossRef] [PubMed]
- Mudhulu, S.; Channegowda, M.; Balaji, S.; Khosla, A.; Sekhar, P. Trends in Graphene-Based E-Skin and Artificial Intelligence for Biomedical Applications—A Review. IEEE Sens. J. 2023, 23, 18963–18976. [Google Scholar] [CrossRef]
- Akter, M.; Anik, H.R.; Tushar, S.I.; Tania, I.S.; Chowdhury, M.K.H.; Hasan, S.M.M.; Bristy, B.F. Advances in Functionalized Applications of Graphene-Based Wearable Sensors in Healthcare. Adv. Sens. Res. 2023, 2300120. [Google Scholar] [CrossRef]
- Kostarelos, K. Translating Graphene and 2D Materials into Medicine. Nat. Rev. Mater. 2016, 1, 16084. [Google Scholar] [CrossRef]
- Park, S. The Puzzle of Graphene Commercialization. Nat. Rev. Mater. 2016, 1, 16085. [Google Scholar] [CrossRef]
- Züttel, A.; Sudan, P.; Mauron, P.; Wenger, P. Model for the Hydrogen Adsorption on Carbon Nanostructures. Appl. Phys. A 2004, 78, 941–946. [Google Scholar] [CrossRef]
- Montes-Navajas, P.; Asenjo, N.G.; Santamaría, R.; Menéndez, R.; Corma, A.; García, H. Surface Area Measurement of Graphene Oxide in Aqueous Solutions. Langmuir 2013, 29, 13443–13448. [Google Scholar] [CrossRef]
- Zhou, M.; Zhai, Y.; Dong, S. Electrochemical Sensing and Biosensing Platform Based on Chemically Reduced Graphene Oxide. Anal. Chem. 2009, 81, 5603–5613. [Google Scholar] [CrossRef]
- Ohno, Y.; Maehashi, K.; Inoue, K.; Matsumoto, K. Label-Free Aptamer-Based Immunoglobulin Sensors Using Graphene Field-Effect Transistors. Jpn. J. Appl. Phys. 2011, 50, 070120. [Google Scholar] [CrossRef]
- Hwang, E.H.; Adam, S.; Sarma, S.D. Carrier Transport in Two-Dimensional Graphene Layers. Phys. Rev. Lett. 2007, 98, 186806. [Google Scholar] [CrossRef] [PubMed]
- Bolotin, K.I.; Sikes, K.J.; Jiang, Z.; Klima, M.; Fudenberg, G.; Hone, J.; Kim, P.; Stormer, H.L. Ultrahigh Electron Mobility in Suspended Graphene. Solid State Commun. 2008, 146, 351–355. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, T.-T.; Girit, C.; Hao, Z.; Martin, M.C.; Zettl, A.; Crommie, M.F.; Shen, Y.R.; Wang, F. Direct Observation of a Widely Tunable Bandgap in Bilayer Graphene. Nature 2009, 459, 820–823. [Google Scholar] [CrossRef] [PubMed]
- Chaves, A.; Azadani, J.G.; Alsalman, H.; Da Costa, D.R.; Frisenda, R.; Chaves, A.J.; Song, S.H.; Kim, Y.D.; He, D.; Zhou, J.; et al. Bandgap Engineering of Two-Dimensional Semiconductor Materials. NPJ 2D Mater. Appl. 2020, 4, 29. [Google Scholar] [CrossRef]
- Salvo, P.; Melai, B.; Calisi, N.; Paoletti, C.; Bellagambi, F.; Kirchhain, A.; Trivella, M.G.; Fuoco, R.; Di Francesco, F. Graphene-Based Devices for Measuring pH. Sens. Actuators B Chem. 2018, 256, 976–991. [Google Scholar] [CrossRef]
- Ang, P.K.; Chen, W.; Wee, A.T.S.; Loh, K.P. Solution-Gated Epitaxial Graphene as pH Sensor. J. Am. Chem. Soc. 2008, 130, 14392–14393. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Nef, C.; Knopfmacher, O.; Tarasov, A.; Weiss, M.; Calame, M.; Schönenberger, C. Graphene Transistors Are Insensitive to pH Changes in Solution. Nano Lett. 2011, 11, 3597–3600. [Google Scholar] [CrossRef]
- Kwon, S.S.; Yi, J.; Lee, W.W.; Shin, J.H.; Kim, S.H.; Cho, S.H.; Nam, S.; Park, W.I. Reversible and Irreversible Responses of Defect-Engineered Graphene-Based Electrolyte-Gated pH Sensors. ACS Appl. Mater. Interfaces 2016, 8, 834–839. [Google Scholar] [CrossRef]
- Heller, I.; Chatoor, S.; Männik, J.; Zevenbergen, M.A.G.; Dekker, C.; Lemay, S.G. Influence of Electrolyte Composition on Liquid-Gated Carbon Nanotube and Graphene Transistors. J. Am. Chem. Soc. 2010, 132, 17149–17156. [Google Scholar] [CrossRef]
- Tan, X.; Chuang, H.-J.; Lin, M.-W.; Zhou, Z.; Cheng, M.M.-C. Edge Effects on the pH Response of Graphene Nanoribbon Field Effect Transistors. J. Phys. Chem. C 2013, 117, 27155–27160. [Google Scholar] [CrossRef]
- Kim, D.-J.; Sohn, I.Y.; Jung, J.-H.; Yoon, O.J.; Lee, N.-E.; Park, J.-S. Reduced Graphene Oxide Field-Effect Transistor for Label-Free Femtomolar Protein Detection. Biosens. Bioelectron. 2013, 41, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Angizi, S.; Yu, E.Y.C.; Dalmieda, J.; Saha, D.; Selvaganapathy, P.R.; Kruse, P. Defect Engineering of Graphene to Modulate pH Response of Graphene Devices. Langmuir 2021, 37, 12163–12178. [Google Scholar] [CrossRef] [PubMed]
- Angizi, S.; Huang, X.; Hong, L.; Akbar, M.A.; Selvaganapathy, P.R.; Kruse, P. Defect Density-Dependent pH Response of Graphene Derivatives: Towards the Development of pH-Sensitive Graphene Oxide Devices. Nanomaterials 2022, 12, 1801. [Google Scholar] [CrossRef] [PubMed]
- Yates, D.E.; Levine, S.; Healy, T.W. Site-Binding Model of the Electrical Double Layer at the Oxide/Water Iiiterface. J. Chem. Soc. Faraday Trans. 1 1974, 70, 1807. [Google Scholar] [CrossRef]
- Maehashi, K.; Sofue, Y.; Okamoto, S.; Ohno, Y.; Inoue, K.; Matsumoto, K. Selective Ion Sensors Based on Ionophore-Modified Graphene Field-Effect Transistors. Sens. Actuators B Chem. 2013, 187, 45–49. [Google Scholar] [CrossRef]
- Xie, H.; Li, Y.-T.; Lei, Y.-M.; Liu, Y.-L.; Xiao, M.-M.; Gao, C.; Pang, D.-W.; Huang, W.-H.; Zhang, Z.-Y.; Zhang, G.-J. Real-Time Monitoring of Nitric Oxide at Single-Cell Level with Porphyrin-Functionalized Graphene Field-Effect Transistor Biosensor. Anal. Chem. 2016, 88, 11115–11122. [Google Scholar] [CrossRef] [PubMed]
- Sawada, K.; Tanaka, T.; Yokoyama, T.; Yamachi, R.; Oka, Y.; Chiba, Y.; Masai, H.; Terao, J.; Uchida, K. Co-Porphyrin Functionalized CVD Graphene Ammonia Sensor with High Selectivity to Disturbing Gases: Hydrogen and Humidity. Jpn. J. Appl. Phys. 2020, 59, SGGG09. [Google Scholar] [CrossRef]
- Fenoy, G.E.; Marmisollé, W.A.; Azzaroni, O.; Knoll, W. Acetylcholine Biosensor Based on the Electrochemical Functionalization of Graphene Field-Effect Transistors. Biosens. Bioelectron. 2020, 148, 111796. [Google Scholar] [CrossRef]
- Li, C.; Adamcik, J.; Mezzenga, R. Biodegradable Nanocomposites of Amyloid Fibrils and Graphene with Shape-Memory and Enzyme-Sensing Properties. Nat. Nanotechnol. 2012, 7, 421–427. [Google Scholar] [CrossRef]
- Georgakilas, V.; Tiwari, J.N.; Kemp, K.C.; Perman, J.A.; Bourlinos, A.B.; Kim, K.S.; Zboril, R. Noncovalent Functionalization of Graphene and Graphene Oxide for Energy Materials, Biosensing, Catalytic, and Biomedical Applications. Chem. Rev. 2016, 116, 5464–5519. [Google Scholar] [CrossRef] [PubMed]
- Shahriari, S.; Sastry, M.; Panjikar, S.; Singh Raman, R.K. Graphene and Graphene Oxide as a Support for Biomolecules in the Development of Biosensors. Nanotechnol. Sci. Appl. 2021, 14, 197–220. [Google Scholar] [CrossRef] [PubMed]
- Chand, R.; Neethirajan, S. Microfluidic Platform Integrated with Graphene-Gold Nano-Composite Aptasensor for One-Step Detection of Norovirus. Biosens. Bioelectron. 2017, 98, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Huang, L.; Zhang, H.; Sun, Z.; Zhang, Z.; Zhang, G.-J. Gold Nanoparticles-Decorated Graphene Field-Effect Transistor Biosensor for Femtomolar MicroRNA Detection. Biosens. Bioelectron. 2015, 74, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Danielson, E.; Sontakke, V.A.; Porkovich, A.J.; Wang, Z.; Kumar, P.; Ziadi, Z.; Yokobayashi, Y.; Sowwan, M. Graphene Based Field-Effect Transistor Biosensors Functionalized Using Gas-Phase Synthesized Gold Nanoparticles. Sens. Actuators B Chem. 2020, 320, 128432. [Google Scholar] [CrossRef]
- Kim, J.; Park, S.-J.; Min, D.-H. Emerging Approaches for Graphene Oxide Biosensor. Anal. Chem. 2017, 89, 232–248. [Google Scholar] [CrossRef] [PubMed]
- Lopez, A.; Liu, J. Covalent and Noncovalent Functionalization of Graphene Oxide with DNA for Smart Sensing. Adv. Intell. Syst. 2020, 2, 2000123. [Google Scholar] [CrossRef]
- Yu, W.; Sisi, L.; Haiyan, Y.; Jie, L. Progress in the Functional Modification of Graphene/Graphene Oxide: A Review. RSC Adv. 2020, 10, 15328–15345. [Google Scholar] [CrossRef]
- Chen, R.J.; Zhang, Y.; Wang, D.; Dai, H. Noncovalent Sidewall Functionalization of Single-Walled Carbon Nanotubes for Protein Immobilization. J. Am. Chem. Soc. 2001, 123, 3838–3839. [Google Scholar] [CrossRef]
- Nandanapalli, K.R.; Mudusu, D.; Lee, S. Functionalization of Graphene Layers and Advancements in Device Applications. Carbon 2019, 152, 954–985. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, Z.; Ding, Y.; Deng, D.; Zhu, X.; Wang, Z. Label-Free Electrochemical Impedance Genosensor Based on 1-Aminopyrene/Graphene Hybrids. Nanoscale 2013, 5, 5833–5840. [Google Scholar] [CrossRef] [PubMed]
- Kawata, T.; Ono, T.; Kanai, Y.; Ohno, Y.; Maehashi, K.; Inoue, K.; Matsumoto, K. Improved Sensitivity of a Graphene FET Biosensor Using Porphyrin Linkers. Jpn. J. Appl. Phys. 2018, 57, 065103. [Google Scholar] [CrossRef]
- Iliuk, A.B.; Hu, L.; Tao, W.A. Aptamer in Bioanalytical Applications. Anal. Chem. 2011, 83, 4440–4452. [Google Scholar] [CrossRef] [PubMed]
- Mascini, M.; Palchetti, I.; Tombelli, S. Nucleic Acid and Peptide Aptamers: Fundamentals and Bioanalytical Aspects. Angew. Chem. Int. Ed. Engl. 2012, 51, 1316–1332. [Google Scholar] [CrossRef] [PubMed]
- Seok Kim, Y.; Ahmad Raston, N.H.; Bock Gu, M. Aptamer-Based Nanobiosensors. Biosens. Bioelectron. 2016, 76, 2–19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lai, B.S.; Juhas, M. Recent Advances in Aptamer Discovery and Applications. Molecules 2019, 24, 941. [Google Scholar] [CrossRef] [PubMed]
- Collins, B.E.; Paulson, J.C. Cell Surface Biology Mediated by Low Affinity Multivalent Protein–Glycan Interactions. Curr. Opin. Chem. Biol. 2004, 8, 617–625. [Google Scholar] [CrossRef]
- Varki, A. Biological Roles of Glycans. Glycobiology 2017, 27, 3–49. [Google Scholar] [CrossRef]
- Suzuki, Y. Sialobiology of Influenza: Molecular Mechanism of Host Range Variation of Influenza Viruses. Biol. Pharm. Bull. 2005, 28, 399–408. [Google Scholar] [CrossRef]
- Watanabe, Y.; Ibrahim, M.S.; Suzuki, Y.; Ikuta, K. The Changing Nature of Avian Influenza A Virus (H5N1). Trends Microbiol. 2012, 20, 11–20. [Google Scholar] [CrossRef]
- Neumann, G.; Kawaoka, Y. Transmission of Influenza A Viruses. Virology 2015, 479–480, 234–246. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Ito, T.; Ibrahim, M.S.; Arai, Y.; Hotta, K.; Phuong, H.V.M.; Hang, N.L.K.; Mai, L.Q.; Soda, K.; Yamaoka, M.; et al. A Novel Immunochromatographic System for Easy-to-Use Detection of Group 1 Avian Influenza Viruses with Acquired Human-Type Receptor Binding Specificity. Biosens. Bioelectron. 2015, 65, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Ono, T.; Oe, T.; Kanai, Y.; Ikuta, T.; Ohno, Y.; Maehashi, K.; Inoue, K.; Watanabe, Y.; Nakakita, S.; Suzuki, Y.; et al. Glycan-Functionalized Graphene-FETs toward Selective Detection of Human-Infectious Avian Influenza Virus. Jpn. J. Appl. Phys. 2017, 56, 030302. [Google Scholar] [CrossRef]
- Wang, Z.; Hao, Z.; Yu, S.; De Moraes, C.G.; Suh, L.H.; Zhao, X.; Lin, Q. An Ultraflexible and Stretchable Aptameric Graphene Nanosensor for Biomarker Detection and Monitoring. Adv. Funct. Mater. 2019, 29, 1905202. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Diallo, A.K.; Dailey, J.L.; Besar, K.; Katz, H.E. Electrochemical Processes and Mechanistic Aspects of Field-Effect Sensors for Biomolecules. J. Mater. Chem. C 2015, 3, 6445–6470. [Google Scholar] [CrossRef] [PubMed]
- Kaisti, M. Detection Principles of Biological and Chemical FET Sensors. Biosens. Bioelectron. 2017, 98, 437–448. [Google Scholar] [CrossRef]
- Hwang, M.T.; Heiranian, M.; Kim, Y.; You, S.; Leem, J.; Taqieddin, A.; Faramarzi, V.; Jing, Y.; Park, I.; van der Zande, A.M.; et al. Ultrasensitive Detection of Nucleic Acids Using Deformed Graphene Channel Field Effect Biosensors. Nat. Commun. 2020, 11, 1543. [Google Scholar] [CrossRef]
- Kesler, V.; Murmann, B.; Soh, H.T. Going beyond the Debye Length: Overcoming Charge Screening Limitations in Next-Generation Bioelectronic Sensors. ACS Nano 2020, 14, 16194–16201. [Google Scholar] [CrossRef]
- Zheng, Z.; Zhang, H.; Zhai, T.; Xia, F. Overcome Debye Length Limitations for Biomolecule Sensing Based on Field Effective Transistors†. Chin. J. Chem. 2021, 39, 999–1008. [Google Scholar] [CrossRef]
- Vacic, A.; Criscione, J.M.; Rajan, N.K.; Stern, E.; Fahmy, T.M.; Reed, M.A. Determination of Molecular Configuration by Debye Length Modulation. J. Am. Chem. Soc. 2011, 133, 13886–13889. [Google Scholar] [CrossRef]
- Nakatsuka, N.; Yang, K.-A.; Abendroth, J.M.; Cheung, K.M.; Xu, X.; Yang, H.; Zhao, C.; Zhu, B.; Rim, Y.S.; Yang, Y.; et al. Aptamer–Field-Effect Transistors Overcome Debye Length Limitations for Small-Molecule Sensing. Science 2018, 362, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.J.; Skaletsky, E.; McPherson, A. Crystallographic Structure of an Intact IgG1 Monoclonal Antibody. J. Mol. Biol. 1998, 275, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Patolsky, F.; Zheng, G.; Hayden, O.; Lakadamyali, M.; Zhuang, X.; Lieber, C.M. Electrical Detection of Single Viruses. Proc. Natl. Acad. Sci. USA 2004, 101, 14017–14022. [Google Scholar] [CrossRef] [PubMed]
- Stern, E.; Wagner, R.; Sigworth, F.J.; Breaker, R.; Fahmy, T.M.; Reed, M.A. Importance of the Debye Screening Length on Nanowire Field Effect Transistor Sensors. Nano Lett. 2007, 7, 3405–3409. [Google Scholar] [CrossRef] [PubMed]
- Nair, P.R.; Alam, M.A. Screening-Limited Response of NanoBiosensors. Nano Lett. 2008, 8, 1281–1285. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.-K.; Jeun, M.; Jang, H.-J.; Cho, W.-J.; Lee, K.H. A Self-Amplified Transistor Immunosensor under Dual Gate Operation: Highly Sensitive Detection of Hepatitis B Surface Antigen. Nanoscale 2015, 7, 16789–16797. [Google Scholar] [CrossRef] [PubMed]
- Kwong Hong Tsang, D.; Lieberthal, T.J.; Watts, C.; Dunlop, I.E.; Ramadan, S.; del Rio Hernandez, A.E.; Klein, N. Chemically Functionalised Graphene FET Biosensor for the Label-Free Sensing of Exosomes. Sci. Rep. 2019, 9, 13946. [Google Scholar] [CrossRef]
- Ramadan, S.; Lobo, R.; Zhang, Y.; Xu, L.; Shaforost, O.; Kwong Hong Tsang, D.; Feng, J.; Yin, T.; Qiao, M.; Rajeshirke, A.; et al. Carbon-Dot-Enhanced Graphene Field-Effect Transistors for Ultrasensitive Detection of Exosomes. ACS Appl. Mater. Interfaces 2021, 13, 7854–7864. [Google Scholar] [CrossRef]
- Ohno, Y.; Maehashi, K.; Matsumoto, K. Label-Free Biosensors Based on Aptamer-Modified Graphene Field-Effect Transistors. J. Am. Chem. Soc. 2010, 132, 18012–18013. [Google Scholar] [CrossRef]
- Kim, D.-J.; Park, H.-C.; Sohn, I.Y.; Jung, J.-H.; Yoon, O.J.; Park, J.-S.; Yoon, M.-Y.; Lee, N.-E. Electrical Graphene Aptasensor for Ultra-Sensitive Detection of Anthrax Toxin with Amplified Signal Transduction. Small 2013, 9, 3352–3360. [Google Scholar] [CrossRef]
- Wang, S.; Sun, M.; Zhang, Y.; Ji, H.; Gao, J.; Song, S.; Sun, J.; Liu, H.; Zhang, Y.; Han, L. Ultrasensitive Antibiotic Perceiving Based on Aptamer-Functionalized Ultraclean Graphene Field-Effect Transistor Biosensor. Anal. Chem. 2022, 94, 14785–14793. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, S.; Ohno, Y.; Maehashi, K.; Inoue, K.; Matsumoto, K. Immunosensors Based on Graphene Field-Effect Transistors Fabricated Using Antigen-Binding Fragment. Jpn. J. Appl. Phys. 2012, 51, 06FD08. [Google Scholar] [CrossRef]
- Elnathan, R.; Kwiat, M.; Pevzner, A.; Engel, Y.; Burstein, L.; Khatchtourints, A.; Lichtenstein, A.; Kantaev, R.; Patolsky, F. Biorecognition Layer Engineering: Overcoming Screening Limitations of Nanowire-Based FET Devices. Nano Lett. 2012, 12, 5245–5254. [Google Scholar] [CrossRef] [PubMed]
- Kanai, Y.; Ohmuro-Matsuyama, Y.; Tanioku, M.; Ushiba, S.; Ono, T.; Inoue, K.; Kitaguchi, T.; Kimura, M.; Ueda, H.; Matsumoto, K. Graphene Field Effect Transistor-Based Immunosensor for Ultrasensitive Noncompetitive Detection of Small Antigens. ACS Sens. 2020, 5, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Li, G.; Zhang, Y.; Zhu, M.; Wan, Y.; Shen, Y. Highly Selective and Sensitive Electrochemical Immunoassay of Cry1C Using Nanobody and π–π Stacked Graphene Oxide/Thionine Assembly. Anal. Chem. 2016, 88, 9830–9836. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; Liu, A.; Shangguan, L.; Mi, L.; Liu, X.; Liu, Y.; Zhao, Y.; Li, Y.; Wei, W.; Zhang, Y.; et al. Construction of Iron-Polymer-Graphene Nanocomposites with Low Nonspecific Adsorption and Strong Quenching Ability for Competitive Immunofluorescent Detection of Biomarkers in GM Crops. Biosens. Bioelectron. 2017, 90, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Filipiak, M.S.; Rother, M.; Andoy, N.M.; Knudsen, A.C.; Grimm, S.; Bachran, C.; Swee, L.K.; Zaumseil, J.; Tarasov, A. Highly Sensitive, Selective and Label-Free Protein Detection in Physiological Solutions Using Carbon Nanotube Transistors with Nanobody Receptors. Sens. Actuators B Chem. 2018, 255, 1507–1516. [Google Scholar] [CrossRef]
- Ferguson, B.S.; Hoggarth, D.A.; Maliniak, D.; Ploense, K.; White, R.J.; Woodward, N.; Hsieh, K.; Bonham, A.J.; Eisenstein, M.; Kippin, T.E.; et al. Real-Time, Aptamer-Based Tracking of Circulating Therapeutic Agents in Living Animals. Sci. Transl. Med. 2013, 5, 213ra165. [Google Scholar] [CrossRef]
- Gao, N.; Zhou, W.; Jiang, X.; Hong, G.; Fu, T.-M.; Lieber, C.M. General Strategy for Biodetection in High Ionic Strength Solutions Using Transistor-Based Nanoelectronic Sensors. Nano Lett. 2015, 15, 2143–2148. [Google Scholar] [CrossRef]
- Kulkarni, G.S.; Zhong, Z. Detection beyond the Debye Screening Length in a High-Frequency Nanoelectronic Biosensor. Nano Lett. 2012, 12, 719–723. [Google Scholar] [CrossRef]
- Woo, J.-M.; Kim, S.H.; Chun, H.; Kim, S.J.; Ahn, J.; Park, Y.J. Modulation of Molecular Hybridization and Charge Screening in a Carbon Nanotube Network Channel Using the Electrical Pulse Method. Lab Chip 2013, 13, 3755. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, T.; Boyle, A.; Bahreman, A.; Bao, L.; Jing, Q.; Xue, H.; Kieltyka, R.; Kros, A.; Schneider, G.F.; et al. Dielectric-Modulated Biosensing with Ultrahigh-Frequency-Operated Graphene Field-Effect Transistors. Adv. Mater. 2022, 34, 2106666. [Google Scholar] [CrossRef] [PubMed]
- Schwierz, F. Graphene Transistors. Nat. Nanotechnol. 2010, 5, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Boubanga-Tombet, S.; Knap, W.; Yadav, D.; Satou, A.; But, D.B.; Popov, V.V.; Gorbenko, I.V.; Kachorovskii, V.; Otsuji, T. Room-Temperature Amplification of Terahertz Radiation by Grating-Gate Graphene Structures. Phys. Rev. X 2020, 10, 031004. [Google Scholar] [CrossRef]
- Gao, N.; Gao, T.; Yang, X.; Dai, X.; Zhou, W.; Zhang, A.; Lieber, C.M. Specific Detection of Biomolecules in Physiological Solutions Using Graphene Transistor Biosensors. Proc. Natl. Acad. Sci. USA 2016, 113, 14633–14638. [Google Scholar] [CrossRef]
- Bliem, C.; Piccinini, E.; Knoll, W.; Azzaroni, O. Enzyme Multilayers on Graphene-Based FETs for Biosensing Applications. In Methods in Enzymology; Kumar, C.V., Ed.; Elsevier: Amsterdam, The Netherlands, 2018; Volume 609, pp. 23–46. ISBN 978-0-12-815240-9. [Google Scholar]
- Berninger, T.; Bliem, C.; Piccinini, E.; Azzaroni, O.; Knoll, W. Cascading Reaction of Arginase and Urease on a Graphene-Based FET for Ultrasensitive, Real-Time Detection of Arginine. Biosens. Bioelectron. 2018, 115, 104–110. [Google Scholar] [CrossRef]
- Chae, M.-S.; Yoo, Y.K.; Kim, J.; Kim, T.G.; Hwang, K.S. Graphene-Based Enzyme-Modified Field-Effect Transistor Biosensor for Monitoring Drug Effects in Alzheimer’s Disease Treatment. Sens. Actuators B Chem. 2018, 272, 448–458. [Google Scholar] [CrossRef]
- Ono, T.; Kanai, Y.; Inoue, K.; Watanabe, Y.; Nakakita, S.; Kawahara, T.; Suzuki, Y.; Matsumoto, K. Electrical Biosensing at Physiological Ionic Strength Using Graphene Field-Effect Transistor in Femtoliter Microdroplet. Nano Lett. 2019, 19, 4004–4009. [Google Scholar] [CrossRef]
- Leenaerts, O.; Partoens, B.; Peeters, F.M. Adsorption of H2O, NH3, CO, NO2, and NO on Graphene: A First-Principles Study. Phys. Rev. B 2008, 77, 125416. [Google Scholar] [CrossRef]
- Gautam, M.; Jayatissa, A.H. Graphene Based Field Effect Transistor for the Detection of Ammonia. J. Appl. Phys. 2012, 112, 064304. [Google Scholar] [CrossRef]
- Gupta Chatterjee, S.; Chatterjee, S.; Ray, A.K.; Chakraborty, A.K. Graphene–Metal Oxide Nanohybrids for Toxic Gas Sensor: A Review. Sens. Actuators B Chem. 2015, 221, 1170–1181. [Google Scholar] [CrossRef]
- Nallon, E.C.; Schnee, V.P.; Bright, C.; Polcha, M.P.; Li, Q. Chemical Discrimination with an Unmodified Graphene Chemical Sensor. ACS Sens. 2016, 1, 26–31. [Google Scholar] [CrossRef]
- Tadi, K.K.; Pal, S.; Narayanan, T.N. Fluorographene Based Ultrasensitive Ammonia Sensor. Sci. Rep. 2016, 6, 25221. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.D.; Elgammal, K.; Fan, X.; Lemme, M.C.; Delin, A.; Råsander, M.; Bergqvist, L.; Schröder, S.; Fischer, A.C.; Niklaus, F.; et al. Graphene-Based CO2 Sensing and Its Cross-Sensitivity with Humidity. RSC Adv. 2017, 7, 22329–22339. [Google Scholar] [CrossRef]
- Sugahara, T.; Hirose, Y.; Nakamura, J.; Ono, T.; Uemura, T.; Karakawa, M.; Itoh, T.; Shin, W.; Yang, Y.; Harada, N.; et al. Carrier-Type Switching with Gas Detection Using a Low-Impedance Hybrid Sensor of 2D Graphene Layer and MoOx Nanorod 3D Network. ACS Appl. Eng. Mater. 2023, 1, 1086–1092. [Google Scholar] [CrossRef]
- Rondelez, Y.; Tresset, G.; Nakashima, T.; Kato-Yamada, Y.; Fujita, H.; Takeuchi, S.; Noji, H. Highly Coupled ATP Synthesis by F1-ATPase Single Molecules. Nature 2005, 433, 773–777. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Iwai, S.; Araki, S.; Sakakihara, S.; Iino, R.; Noji, H. Large-Scale Femtoliter Droplet Array for Digital Counting of Single Biomolecules. Lab Chip 2012, 12, 4986–4991. [Google Scholar] [CrossRef]
- Witters, D.; Sun, B.; Begolo, S.; Rodriguez-Manzano, J.; Robles, W.; Ismagilov, R.F. Digital Biology and Chemistry. Lab Chip 2014, 14, 3225–3232. [Google Scholar] [CrossRef]
- Ono, T.; Noji, H. Digital Bioassay with Femtoliter Reactor Array. In Intelligent Nanosystems for Energy, Information and Biological Technologies; Sone, J., Tsuji, S., Eds.; Springer: Tokyo, Japan, 2016; pp. 107–116. ISBN 978-4-431-56429-4. [Google Scholar]
- Zhang, Y.; Noji, H. Digital Bioassays: Theory, Applications, and Perspectives. Anal. Chem. 2017, 89, 92–101. [Google Scholar] [CrossRef]
- Ono, T.; Ichiki, T.; Noji, H. Digital Enzyme Assay Using Attoliter Droplet Array. Analyst 2018, 143, 4923–4929. [Google Scholar] [CrossRef]
- Ono, T.; Akagi, T.; Ichiki, T. Anisotropic Etching of Amorphous Perfluoropolymer Films in Oxygen-Based Inductively Coupled Plasmas. J. Appl. Phys. 2009, 105, 013314. [Google Scholar] [CrossRef]
- Ono, T.; Iizuka, R.; Akagi, T.; Funatsu, T.; Ichiki, T. Damage-Free Fabrication of Perfluoropolymer Microaperture Array Device for Single-Molecule Imaging. Trans. Mater. Res. Soc. Jpn. 2011, 36, 553–556. [Google Scholar] [CrossRef][Green Version]
- Ameri, S.K.; Singh, P.K.; Sonkusale, S. Utilization of Graphene Electrode in Transparent Microwell Arrays for High Throughput Cell Trapping and Lysis. Biosens. Bioelectron. 2014, 61, 625–630. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Riaño, E.J.; Avila-Huerta, M.D.; Mancera-Zapata, D.L.; Morales-Narváez, E. Microwell Plates Coated with Graphene Oxide Enable Advantageous Real-Time Immunosensing Platform. Biosens. Bioelectron. 2020, 165, 112319. [Google Scholar] [CrossRef] [PubMed]
- Mcnulty, C.M.; Wise, R. Rapid Diagnosis of Campylobacter-Associated Gastritis. Lancet 1985, 325, 1443–1444. [Google Scholar] [CrossRef] [PubMed]
- Mobley, H.L.T.; Island, M.D.; Hausinger, R.P. Molecular Biology of Microbial Ureases. Microbiol. Rev. 1995, 59, 451–480. [Google Scholar] [CrossRef]
- Uemura, N.; Okamoto, S.; Yamamoto, S.; Matsumura, N.; Yamaguchi, S.; Yamakido, M.; Taniyama, K.; Sasaki, N.; Schlemper, R.J. Helicobacter Pylori Infection and the Development of Gastric Cancer. N. Engl. J. Med. 2001, 345, 784–789. [Google Scholar] [CrossRef]
- Liu, F.; Choi, K.S.; Park, T.J.; Lee, S.Y.; Seo, T.S. Graphene-Based Electrochemical Biosensor for Pathogenic Virus Detection. BioChip J. 2011, 5, 123–128. [Google Scholar] [CrossRef]
- Benvidi, A.; Rajabzadeh, N.; Mazloum-Ardakani, M.; Heidari, M.M.; Mulchandani, A. Simple and Label-Free Electrochemical Impedance Amelogenin Gene Hybridization Biosensing Based on Reduced Graphene Oxide. Biosens. Bioelectron. 2014, 58, 145–152. [Google Scholar] [CrossRef]
- Liu, B.; Huang, P.J.; Kelly, E.Y.; Liu, J. Graphene Oxide Surface Blocking Agents Can Increase the DNA Biosensor Sensitivity. Biotechnol. J. 2016, 11, 780–787. [Google Scholar] [CrossRef]
- Wang, F.; Horikawa, S.; Hu, J.; Wikle, H.; Chen, I.-H.; Du, S.; Liu, Y.; Chin, B. Detection of Salmonella Typhimurium on Spinach Using Phage-Based Magnetoelastic Biosensors. Sensors 2017, 17, 386. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Naranjo, J.; Aguilar, O. Suppressing Non-Specific Binding of Proteins onto Electrode Surfaces in the Development of Electrochemical Immunosensors. Biosensors 2019, 9, 15. [Google Scholar] [CrossRef] [PubMed]
- Lichtenberg, J.Y.; Ling, Y.; Kim, S. Non-Specific Adsorption Reduction Methods in Biosensing. Sensors 2019, 19, 2488. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, K.; Sun, H.; Zhao, S.; Chen, X.; Qian, D.; Mao, H.; Zhao, J. Novel Graphene Biosensor Based on the Functionalization of Multifunctional Nano-Bovine Serum Albumin for the Highly Sensitive Detection of Cancer Biomarkers. Nano-Micro Lett. 2019, 11, 20. [Google Scholar] [CrossRef] [PubMed]
- Bungon, T.; Haslam, C.; Damiati, S.; O’Driscoll, B.; Whitley, T.; Davey, P.; Siligardi, G.; Charmet, J.; Awan, S.A. Graphene FET Sensors for Alzheimer’s Disease Protein Biomarker Clusterin Detection. Front. Mol. Biosci. 2021, 8, 651232. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Kang, H.; Naylor, C.H.; Streller, F.; Ducos, P.; Serrano, M.D.; Ping, J.; Zauberman, J.; Rajesh; Carpick, R.W.; et al. Scalable Production of Sensor Arrays Based on High-Mobility Hybrid Graphene Field Effect Transistors. ACS Appl. Mater. Interfaces 2016, 8, 27546–27552. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Khan, N.I.; Tsavalas, J.G.; Song, E. Selective Detection of Lysozyme Biomarker Utilizing Large Area Chemical Vapor Deposition-Grown Graphene-Based Field-Effect Transistor. Front. Bioeng. Biotechnol. 2018, 6, 29. [Google Scholar] [CrossRef]
- Chen, H.; Chen, P.; Huang, J.; Selegård, R.; Platt, M.; Palaniappan, A.; Aili, D.; Tok, A.I.Y.; Liedberg, B. Detection of Matrilysin Activity Using Polypeptide Functionalized Reduced Graphene Oxide Field-Effect Transistor Sensor. Anal. Chem. 2016, 88, 2994–2998. [Google Scholar] [CrossRef]
- Wang, X.; Zhu, Y.; Olsen, T.R.; Sun, N.; Zhang, W.; Pei, R.; Lin, Q. A Graphene Aptasensor for Biomarker Detection in Human Serum. Electrochim. Acta 2018, 290, 356–363. [Google Scholar] [CrossRef]
- Rafiee, J.; Mi, X.; Gullapalli, H.; Thomas, A.V.; Yavari, F.; Shi, Y.; Ajayan, P.M.; Koratkar, N.A. Wetting Transparency of Graphene. Nat. Mater. 2012, 11, 217–222. [Google Scholar] [CrossRef]
- Zhang, X.; Wan, S.; Pu, J.; Wang, L.; Liu, X. Highly Hydrophobic and Adhesive Performance of Graphene Films. J. Mater. Chem. 2011, 21, 12251–12258. [Google Scholar] [CrossRef]
- Yu, Y.; Li, Y.-T.; Jin, D.; Yang, F.; Wu, D.; Xiao, M.-M.; Zhang, H.; Zhang, Z.-Y.; Zhang, G.-J. Electrical and Label-Free Quantification of Exosomes with a Reduced Graphene Oxide Field Effect Transistor Biosensor. Anal. Chem. 2019, 91, 10679–10686. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Kwon, D.; Choi, W.; Jung, G.Y.; Au, A.K.; Folch, A.; Jeon, S. 3D-Printed Microfluidic Device for the Detection of Pathogenic Bacteria Using Size-Based Separation in Helical Channel with Trapezoid Cross-Section. Sci. Rep. 2015, 5, 7717. [Google Scholar] [CrossRef] [PubMed]
- Chircov, C.; Bîrcă, A.C.; Grumezescu, A.M.; Andronescu, E. Biosensors-on-Chip: An Up-to-Date Review. Molecules 2020, 25, 6013. [Google Scholar] [CrossRef]
- Zhang, Q.; Rawal, G.; Qian, J.; Ibrahim, H.; Zhang, J.; Dong, L.; Lu, M. An Integrated Magneto-Opto-Fluidic Biosensor for Rapid on-Chip Assay of Respiratory Viruses of Livestock. Lab Chip 2022, 22, 3236–3244. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.; Shukla, S.; Bajpai, V.K.; Han, Y.-K.; Huh, Y.S.; Ghosh, A.; Gandhi, S. Microfluidic-Based Graphene Field Effect Transistor for Femtomolar Detection of Chlorpyrifos. Sci. Rep. 2019, 9, 276. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wang, X.; Li, Z.; Zhang, S.; Xing, F. Recent Advances in the Fabrication and Application of Graphene Microfluidic Sensors. Micromachines 2020, 11, 1059. [Google Scholar] [CrossRef] [PubMed]
- Xing, F.; Meng, G.-X.; Zhang, Q.; Pan, L.-T.; Wang, P.; Liu, Z.-B.; Jiang, W.-S.; Chen, Y.; Tian, J.-G. Ultrasensitive Flow Sensing of a Single Cell Using Graphene-Based Optical Sensors. Nano Lett. 2014, 14, 3563–3569. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Lu, C.-C.; Yeh, C.-H.; Jin, C.; Suenaga, K.; Chiu, P.-W. Graphene Annealing: How Clean Can It Be? Nano Lett. 2012, 12, 414–419. [Google Scholar] [CrossRef]
- Jung, J.H.; Sohn, I.Y.; Kim, D.J.; Kim, B.Y.; Jang, M.; Lee, N.-E. Enhancement of Protein Detection Performance in Field-Effect Transistors with Polymer Residue-Free Graphene Channel. Carbon 2013, 62, 312–321. [Google Scholar] [CrossRef]
- Jang, M.; Quang Trung, T.; Jung, J.-H.; Kim, B.-Y.; Lee, N.-E. Improved Performance and Stability of Field-Effect Transistors with Polymeric Residue-Free Graphene Channel Transferred by Gold Layer. Phys. Chem. Chem. Phys. 2014, 16, 4098–4105. [Google Scholar] [CrossRef] [PubMed]
- Tien, D.H.; Park, J.-Y.; Kim, K.B.; Lee, N.; Seo, Y. Characterization of Graphene-Based FET Fabricated Using a Shadow Mask. Sci. Rep. 2016, 6, 25050. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, B.; Li, S.; Li, S.; Yin, J. Ways to Eliminate PMMA Residues on Graphene—Superclean Graphene. Carbon 2021, 173, 609–636. [Google Scholar] [CrossRef]
- Laaksonen, P.; Kainlauri, M.; Laaksonen, T.; Shchepetov, A.; Jiang, H.; Ahopelto, J.; Linder, M.B. Interfacial Engineering by Proteins: Exfoliation and Functionalization of Graphene by Hydrophobins. Angew. Chem. Int. Ed. Engl. 2010, 49, 4946–4949. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Teweldebrhan, D.; Ashraf, K.; Liu, G.; Jing, X.; Yan, Z.; Li, R.; Ozkan, M.; Lake, R.K.; Balandin, A.A.; et al. Gating of Single-Layer Graphene with Single-Stranded Deoxyribonucleic Acids. Small 2010, 6, 1150–1155. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.; Zhu, Y.; Wang, X.; Rotti, P.G.; DiMarco, C.; Tyler, S.R.; Zhao, X.; Engelhardt, J.F.; Hone, J.; Lin, Q. Real-Time Monitoring of Insulin Using a Graphene Field-Effect Transistor Aptameric Nanosensor. ACS Appl. Mater. Interfaces 2017, 9, 27504–27511. [Google Scholar] [CrossRef]
- Chen, F.; Qing, Q.; Xia, J.; Li, J.; Tao, N. Electrochemical Gate-Controlled Charge Transport in Graphene in Ionic Liquid and Aqueous Solution. J. Am. Chem. Soc. 2009, 131, 9908–9909. [Google Scholar] [CrossRef]
- Merkel, T.C.; Bondar, V.I.; Nagai, K.; Freeman, B.D.; Pinnau, I. Gas Sorption, Diffusion, and Permeation in Poly(Dimethylsiloxane). J. Polym. Sci. B Polym. Phys. 2000, 38, 415–434. [Google Scholar] [CrossRef]
- Makhloufi, C.; Lasseuguette, E.; Remigy, J.C.; Belaissaoui, B.; Roizard, D.; Favre, E. Ammonia Based CO2 Capture Process Using Hollow Fiber Membrane Contactors. J. Membr. Sci. 2014, 455, 236–246. [Google Scholar] [CrossRef]
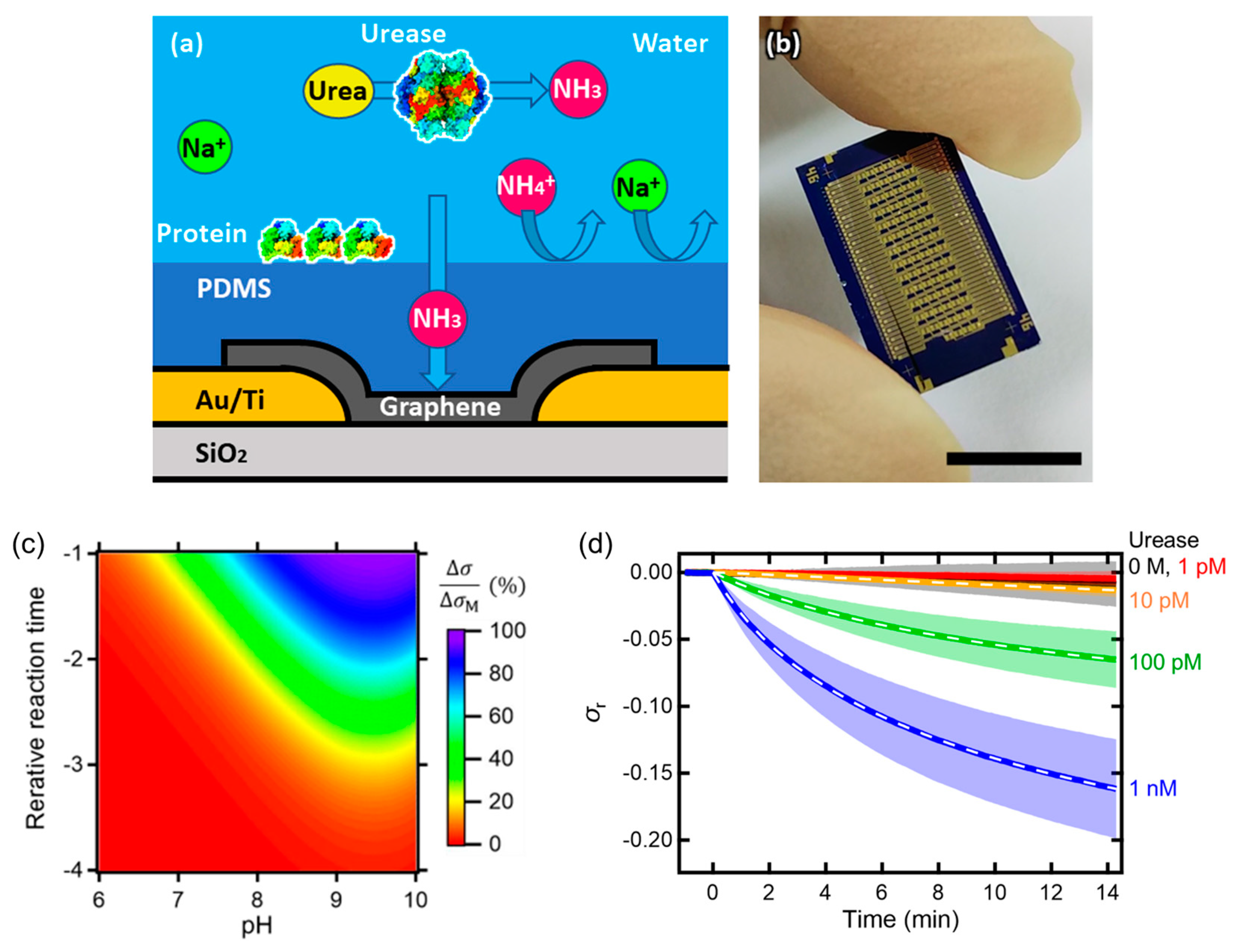
- Ono, T.; Kannaka, M.; Kanai, Y.; Miyakawa, N.; Shinagawa, A.; Nakakita, S.; Watanabe, Y.; Ushiba, S.; Tani, S.; Suzuki, Y.; et al. Elastomer-Coated Graphene Biosensor and Its Response to Enzymatic Reactions. Jpn. J. Appl. Phys. 2023, 62, 067002. [Google Scholar] [CrossRef]
- Huang, X.; Leng, T.; Zhu, M.; Zhang, X.; Chen, J.; Chang, K.; Aqeeli, M.; Geim, A.K.; Novoselov, K.S.; Hu, Z. Highly Flexible and Conductive Printed Graphene for Wireless Wearable Communications Applications. Sci. Rep. 2015, 5, 18298. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Tao, L.; Liu, F.; Ji, L.; Hu, Y.; Cheng, M.M.-C.; Chen, P.-Y.; Akinwande, D. Chemical-Sensitive Graphene Modulator with a Memory Effect for Internet-of-Things Applications. Microsyst. Nanoeng. 2016, 2, 16018. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Kim, J.; Won, S.M.; Ma, Y.; Kang, D.; Xie, Z.; Lee, K.-T.; Chung, H.U.; Banks, A.; Min, S.; et al. Battery-Free, Wireless Sensors for Full-Body Pressure and Temperature Mapping. Sci. Transl. Med. 2018, 10, eaan4950. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-J.; Saviers, K.R.; Fisher, T.S.; Irazoqui, P.P. Continuous Glucose Monitoring with a Flexible Biosensor and Wireless Data Acquisition System. Sens. Actuators B Chem. 2018, 275, 237–243. [Google Scholar] [CrossRef]
- Hwang, M.T.; Wang, Z.; Ping, J.; Ban, D.K.; Shiah, Z.C.; Antonschmidt, L.; Lee, J.; Liu, Y.; Karkisaval, A.G.; Johnson, A.T.C.; et al. DNA Nanotweezers and Graphene Transistor Enable Label-Free Genotyping. Adv. Mater. 2018, 30, 1802440. [Google Scholar] [CrossRef]
- Zhang, X.; Jing, Q.; Ao, S.; Schneider, G.F.; Kireev, D.; Zhang, Z.; Fu, W. Ultrasensitive Field-Effect Biosensors Enabled by the Unique Electronic Properties of Graphene. Small 2020, 16, 1902820. [Google Scholar] [CrossRef] [PubMed]
- Laliberte, K.E.; Scott, P.; Khan, N.I.; Mahmud, M.S.; Song, E. A Wearable Graphene Transistor-Based Biosensor for Monitoring IL-6 Biomarker. Microelectron. Eng. 2022, 262, 111835. [Google Scholar] [CrossRef]
- Sharma, S.; Lernoud, C.-A.; Fain, B.; Othmen, R.; Bouchiat, V.; Yvert, B.; Hébert, C. Graphene Solution-Gated Field-Effect Transistor for Ultrasound-Based Wireless and Battery-Free Biosensing. Adv. Mater. Technol. 2023, 8, 2300163. [Google Scholar] [CrossRef]
- Liu, X.; Wang, J.-Y.; Mao, X.-B.; Ning, Y.; Zhang, G.-J. Single-Shot Analytical Assay Based on Graphene-Oxide-Modified Surface Acoustic Wave Biosensor for Detection of Single-Nucleotide Polymorphisms. Anal. Chem. 2015, 87, 9352–9359. [Google Scholar] [CrossRef]
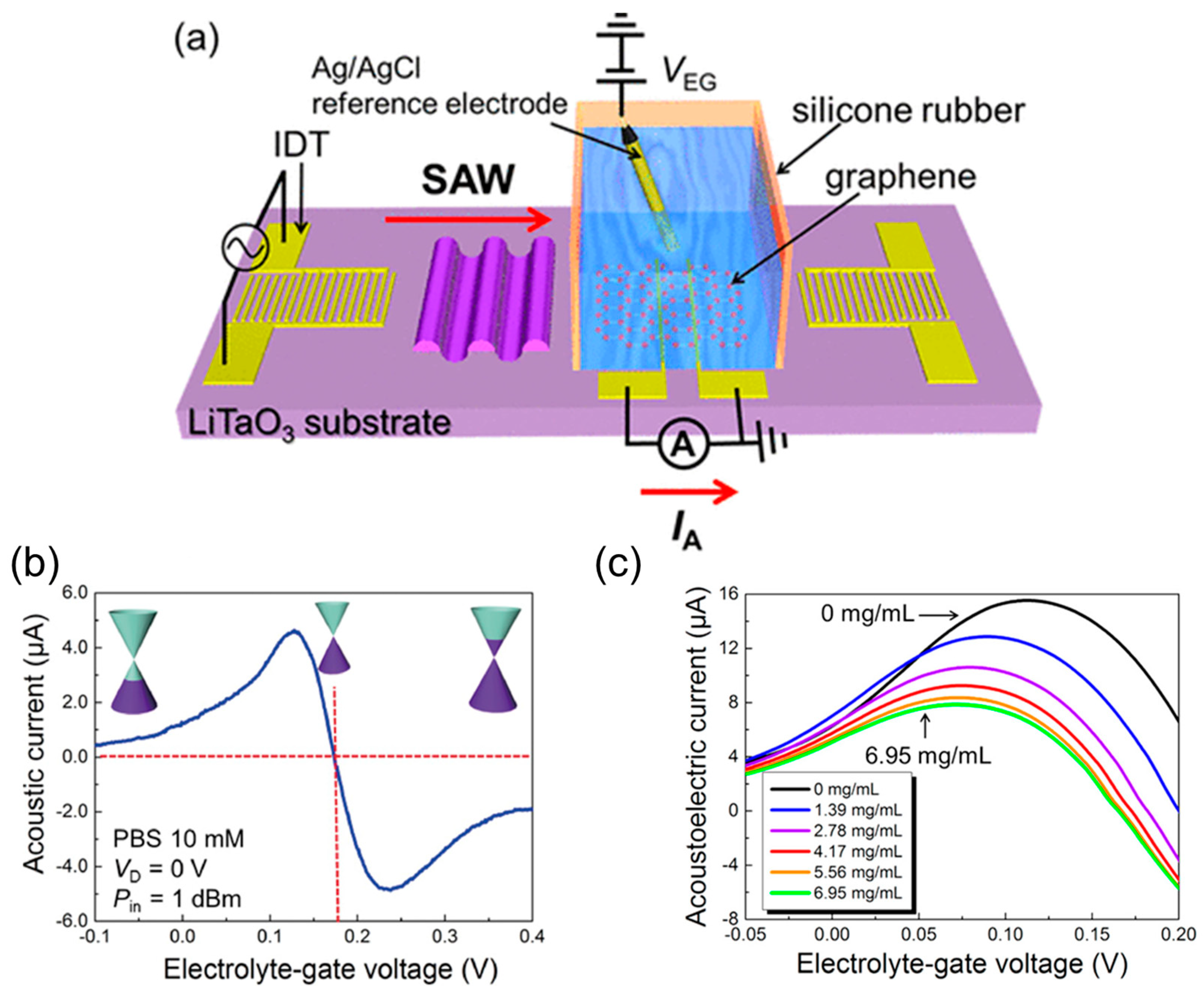
- Okuda, S.; Ono, T.; Kanai, Y.; Ikuta, T.; Shimatani, M.; Ogawa, S.; Maehashi, K.; Inoue, K.; Matsumoto, K. Graphene Surface Acoustic Wave Sensor for Simultaneous Detection of Charge and Mass. ACS Sens. 2018, 3, 200–204. [Google Scholar] [CrossRef]
- Ji, J.; Pang, Y.; Li, D.; Huang, Z.; Zhang, Z.; Xue, N.; Xu, Y.; Mu, X. An Aptamer-Based Shear Horizontal Surface Acoustic Wave Biosensor with a CVD-Grown Single-Layered Graphene Film for High-Sensitivity Detection of a Label-Free Endotoxin. Microsyst. Nanoeng. 2020, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Okuda, S.; Ikuta, T.; Kanai, Y.; Ono, T.; Ogawa, S.; Fujisawa, D.; Shimatani, M.; Inoue, K.; Maehashi, K.; Matsumoto, K. Acoustic Carrier Transportation Induced by Surface Acoustic Waves in Graphene in Solution. Appl. Phys. Express 2016, 9, 045104. [Google Scholar] [CrossRef]
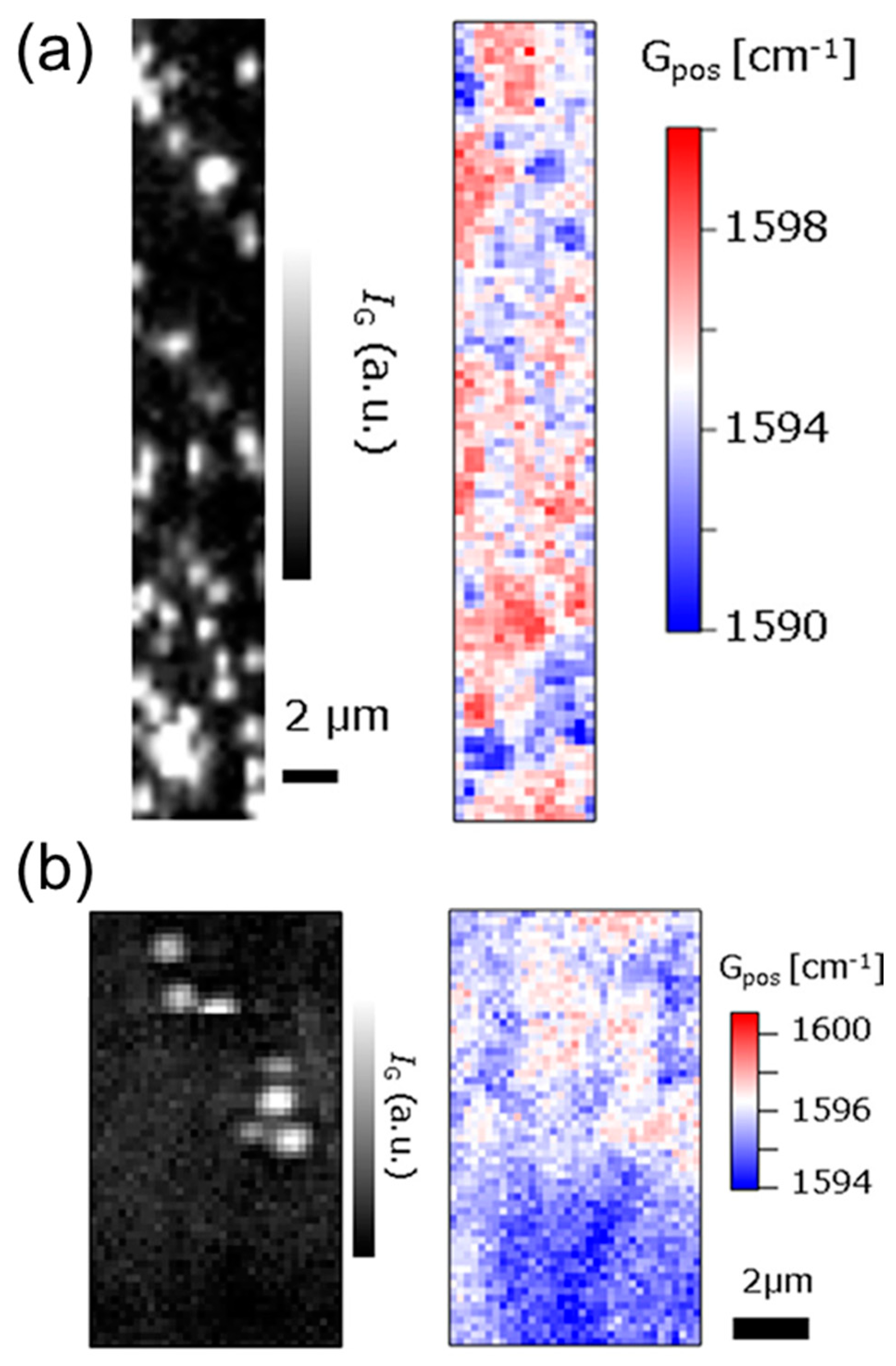
- Das, A.; Pisana, S.; Chakraborty, B.; Piscanec, S.; Saha, S.K.; Waghmare, U.V.; Novoselov, K.S.; Krishnamurthy, H.R.; Geim, A.K.; Ferrari, A.C.; et al. Monitoring Dopants by Raman Scattering in an Electrochemically Top-Gated Graphene Transistor. Nat. Nanotechnol. 2008, 3, 210–215. [Google Scholar] [CrossRef]
- Ryu, S.; Liu, L.; Berciaud, S.; Yu, Y.-J.; Liu, H.; Kim, P.; Flynn, G.W.; Brus, L.E. Atmospheric Oxygen Binding and Hole Doping in Deformed Graphene on a SiO2 Substrate. Nano Lett. 2010, 10, 4944–4951. [Google Scholar] [CrossRef] [PubMed]
- Ushiba, S.; Ono, T.; Kanai, Y.; Inoue, K.; Kimura, M.; Matsumoto, K. Graphene as an Imaging Platform of Charged Molecules. ACS Omega 2018, 3, 3137–3142. [Google Scholar] [CrossRef]
- Silver, A.; Kitadai, H.; Liu, H.; Granzier-Nakajima, T.; Terrones, M.; Ling, X.; Huang, S. Chemical and Bio Sensing Using Graphene-Enhanced Raman Spectroscopy. Nanomaterials 2019, 9, 516. [Google Scholar] [CrossRef]
- Khalil, I.; Julkapli, N.M.; Yehye, W.A.; Basirun, W.J.; Bhargava, S.K. Graphene–Gold Nanoparticles Hybrid—Synthesis, Functionalization, and Application in a Electrochemical and Surface-Enhanced Raman Scattering Biosensor. Materials 2016, 9, 406. [Google Scholar] [CrossRef]
- Morales-Narváez, E.; Merkoçi, A. Graphene Oxide as an Optical Biosensing Platform. Adv. Mater. 2012, 24, 3298–3308. [Google Scholar] [CrossRef]
- Zhu, C.; Du, D.; Lin, Y. Graphene and Graphene-like 2D Materials for Optical Biosensing and Bioimaging: A Review. 2D Mater. 2015, 2, 032004. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, X.; Zhou, X.; Zhang, Y. Review on the Graphene Based Optical Fiber Chemical and Biological Sensors. Sens. Actuators B Chem. 2016, 231, 324–340. [Google Scholar] [CrossRef]
- Jiang, W.-S.; Xin, W.; Xun, S.; Chen, S.-N.; Gao, X.-G.; Liu, Z.-B.; Tian, J.-G. Reduced Graphene Oxide-Based Optical Sensor for Detecting Specific Protein. Sens. Actuators B Chem. 2017, 249, 142–148. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, S.; Xu, T.; Zhang, T.; Mo, Y.; Liu, J.; Yan, L.; Xing, F. Ultra-Sensitive and Ultra-Fast Detection of Whole Unlabeled Living Cancer Cell Responses to Paclitaxel with a Graphene-Based Biosensor. Sens. Actuators B Chem. 2018, 263, 417–425. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, W.; Xing, F. Graphene Optical Biosensors. Int. J. Mol. Sci. 2019, 20, 2461. [Google Scholar] [CrossRef]
- Ushiba, S.; Nakano, T.; Miyakawa, N.; Shinagawa, A.; Ono, T.; Kanai, Y.; Tani, S.; Kimura, M.; Matsumoto, K. Robust Graphene Field-Effect Transistor Biosensors via Hydrophobization of SiO2 Substrates. Appl. Phys. Express 2022, 15, 115002. [Google Scholar] [CrossRef]
- Miyakawa, N.; Shinagawa, A.; Nakano, T.; Ushiba, S.; Ono, T.; Kanai, Y.; Tani, S.; Kimura, M.; Matsumoto, K. Semi-Quantitative Graphene Chemiresistor Enzyme Immunoassay for Simple and Sensitive Antigen Detection. Microchem. J. 2024, 196, 109594. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ono, T.; Okuda, S.; Ushiba, S.; Kanai, Y.; Matsumoto, K. Challenges for Field-Effect-Transistor-Based Graphene Biosensors. Materials 2024, 17, 333. https://doi.org/10.3390/ma17020333
Ono T, Okuda S, Ushiba S, Kanai Y, Matsumoto K. Challenges for Field-Effect-Transistor-Based Graphene Biosensors. Materials. 2024; 17(2):333. https://doi.org/10.3390/ma17020333
Chicago/Turabian StyleOno, Takao, Satoshi Okuda, Shota Ushiba, Yasushi Kanai, and Kazuhiko Matsumoto. 2024. "Challenges for Field-Effect-Transistor-Based Graphene Biosensors" Materials 17, no. 2: 333. https://doi.org/10.3390/ma17020333
APA StyleOno, T., Okuda, S., Ushiba, S., Kanai, Y., & Matsumoto, K. (2024). Challenges for Field-Effect-Transistor-Based Graphene Biosensors. Materials, 17(2), 333. https://doi.org/10.3390/ma17020333





