Assessment of Variability: Chloroplast Microsatellite DNA, Defoliation, and Regeneration Potential of Old Pine Stands of Different Origins in the Context of Assisted Genotype Migration
Abstract
1. Introduction
2. Materials and Methods
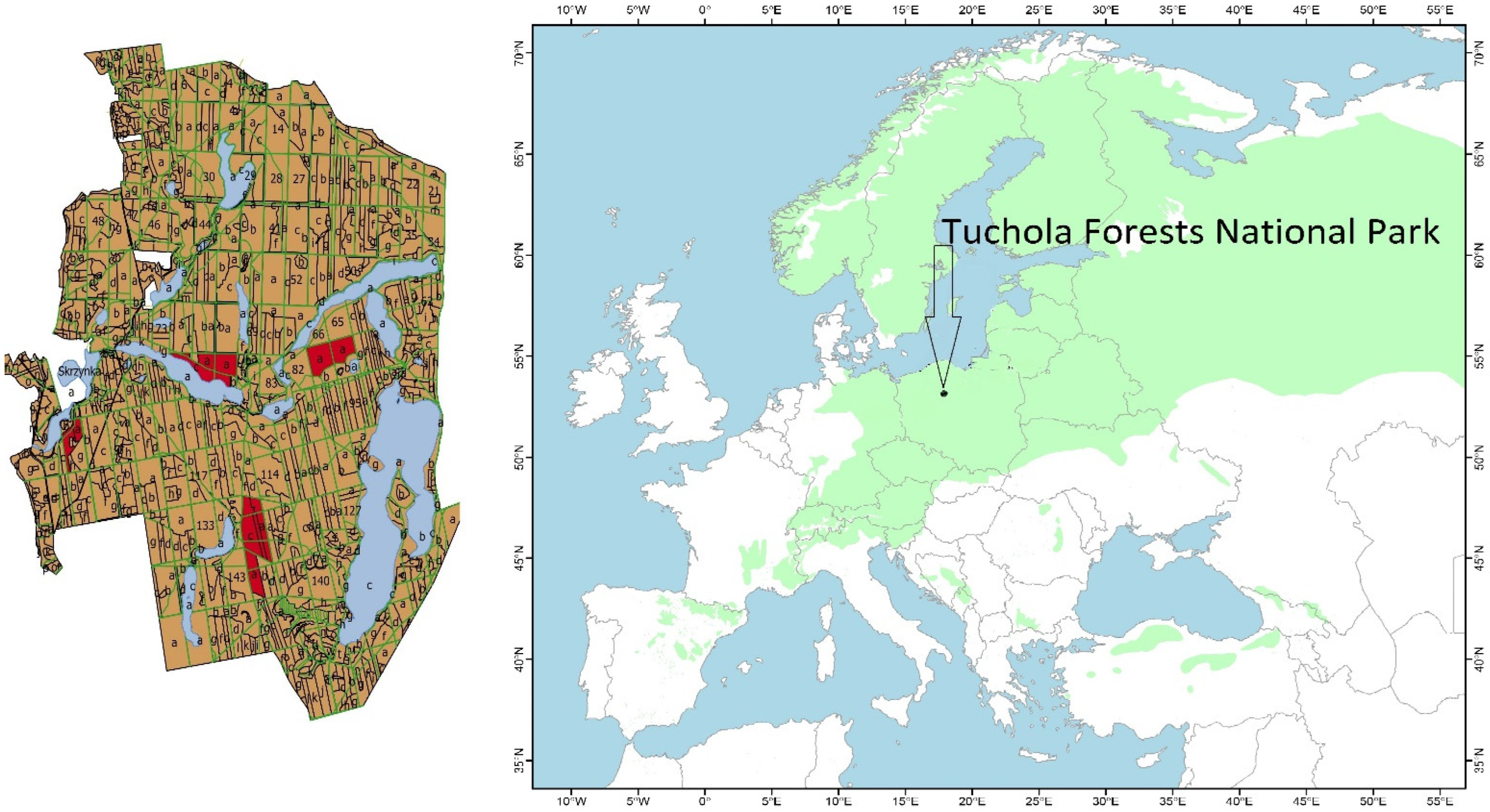
2.1. Plant Material
2.2. DNA Extraction and Microsatellite Genotyping
2.3. CpSSR Data Analysis
2.4. Defoliation Level and Regeneration Potential of the Studied Stands
2.5. Statistic Stand Health Analysis
3. Results
3.1. CpSSR Analyses
3.2. Estimated Number of Pine Seedlings and Health Statuses of Surveyed Stands
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ivetić, V.; Devetaković, J.; Nonić, M.; Stanković, D.; Šijačić-Nikolić, M. Genetic diversity and forest reproductive material—From seed source selection to planting. iFor. Biogeosci. For. 2016, 9, 801–812. [Google Scholar] [CrossRef]
- Rutkowski, P.; Diatta, J.; Konatowska, M.; Andrzejewska, A.; Tyburski, Ł.; Przybylski, P. Geochemical Referencing of Natural Forest Contamination in Poland. Forests 2020, 11, 157. [Google Scholar] [CrossRef]
- Przybylski, P.; Mohytych, V.; Rutkowski, P.; Tereba, A.; Tyburski, Ł.; Fyalkowska, K. Relationships between Some Biodiversity Indicators and Crown Damage of Pinus sylvestris L. in Natural Old Growth Pine Forests. Sustainability 2021, 13, 1239. [Google Scholar] [CrossRef]
- Interpretation Manual. EUR28. pp. 113–122. Available online: https://ec.europa.eu/environment/nature/legislation/habitatsdirective/docs/IntManual_EU28.pdf (accessed on 2 August 2021).
- Noss, R.F. Beyond Kyoto: Forest management in a time of rapid climate change. Conserv. Biol. 2001, 15, 578–590. [Google Scholar] [CrossRef]
- Urbański, K. Podstawy Hodowli Selekcyjnej; Wydawnictwo Świat, Biblioteczka leśniczego: Warsaw, Poland, 1999; Issue 102. [Google Scholar]
- Prus-Głowacki, W.; Urbaniak, L.; Zubrowska-Gil, M. Allozyme differentiation in some european populations of Pinus sylvestris L. Genet. Pol. 1993, 34, 159–176. [Google Scholar]
- Nowakowska, J.A. Zmienność Genetyczna Polskich Wybranych Populacji Sosny Zwyczajnej (Pinus sylvestris L.) na Podstawie Analiz Polimorfizmu DNA; dissertations and monographs; Forest Research Institute Prace IBL: Sękocin Stary, Poland, 2007. [Google Scholar]
- Przybylski, P.; Tereba, A.; Meger, J.; Szyp-Borowska, I.; Tyburski, Ł. Conservation of Genetic Diversity of Scots Pine (Pinus sylvestris L.) in a Central European National Park Based on cpDNA Studies. Diversity 2022, 14, 93. [Google Scholar] [CrossRef]
- Chałupka, W.; Matras, J.; Barzdajn, W.; Burczyk, J.; Tarasiuk, S.; Sabor, S.; Kawalczyk, J.; Fonder, W.; Grądzki, P.T.; Kacprzak, C.; et al. Program Zachowania Leśnych Zasobów Genowych i Ho-Dowli Selekcyjnej Drzew Leśnych w Polsce na Lata 2010–2035; CILP: Warsaw, Poland, 2010; p. 142. ISBN 978-83-61633-60-0. [Google Scholar]
- Reed, D.H.; Frankham, R. Correlation between Fitness and Genetic Diversity. Cons. Biol. 2003, 17, 230–237. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. Report of the 14th Regular Session of the Commision on Genetic Resources for Food and Agriculture. 2014. Available online: http://www.fao.org/docrep/meeting/028/mg468e.pdf (accessed on 2 August 2021).
- Semerikov, V.L.; Semerikova, S.A.; Dymshakova, O.S.; Zatsepina, K.G.; Tarakanov, V.V.; Tikhonova, I.V.; Ekart, A.K.; Vidyakin, A.I.; Jamiyansuren, S.; Rogovtsev, R.V. Microsatellite loci polymorphism ofchloroplast DNA of the pine tree (Pinus sylvestris L.) in Asia and Eastern Europe. Genetika 2014, 50, 660–669. [Google Scholar]
- Pazouki, L.; Shanjani, P.S.; Fields, P.D.; Martins, K.; Suhhorutsenko, M.; Viinalass, H.; Niinemets, U. Large within-population genetic diversity of the widespread conifer Pinus sylvestris at its soil fertility limit characterized by nuclear and chloroplast microsatellite markers. Eur. J. For. Res. 2016, 135, 161–177. [Google Scholar] [CrossRef]
- Wojnicka-Półtorak, A.; Celiński, K.; Chudzińska, E. Genetic Diversity among Age Classes of a Pinus sylvestris (L.) Population from the Białowieza Primeval Forest, Poland. Forests 2017, 8, 227. [Google Scholar] [CrossRef]
- Blumenröther, M.; Bachmann, M.; Müller-Starck, G. Genetic characters and diameter growth of provenances of Scots pine (Pinus sylvestris L.). Silvae Genet. 2001, 50, 212–222. [Google Scholar]
- Prus-Głowacki, W.; Sukovata, L.; Lewandowska-Wosik, A.; Nowak-Bzowy, R. Shikimate dehydrogenase (E.C. 1.1.1. 25 ShDH) alleles as potential markers for flowering phenology in Pinus sylvestris. Dendrobiology 2015, 73, 153–162. [Google Scholar] [CrossRef][Green Version]
- Bell, D.L.; Sultan, S.E. Dynamic phenotypic plasticity for root growth in Polygonum: A comparative study. Am. J. Bot. 1999, 86, 807–819. [Google Scholar] [CrossRef] [PubMed]
- Gulyaeva, E.N.; Tarelkina, T.V.; Galibina, N.A. Functional characteristics of EST-SSR markers available for Scots pine. Math. Biol. Bioinform. 2022, 17, 82–155. [Google Scholar] [CrossRef]
- O’Neill, G.A.; Ukrainetz, N.K.; Carlson, M.R.; Cartwright, C.V.; Jaquish, B.C.; King, J.N.; Krakowski, J.; Russell, J.H.; Stoehr, M.U.; Xie, C.; et al. Assisted Migration to Address Climate Change in British Columbia: Recommendations for Interim Seed Transfer Standards; Technical Report 048; Ministry of Forests and Range, Research Branch: Victoria, BC, Canada, 2008. [Google Scholar]
- Vendramin, G.G.; Lelli, L.; Rossi, P.; Morgante, M. A set of primers for the amplificationof chloroplast microsatellites in Pinaceae. Mol. Ecol. 1996, 5, 595–598. [Google Scholar] [CrossRef] [PubMed]
- Provan, J.; Soranzo, N.; Wilson, N.J.; McNicol, J.W.; Forrest, G.I.; Cottrell, J.; Powell, W. Gene-poolvariation in Caledonian and European Scots pine (Pinus sylvestris L.) revealed by chloroplastsimple-sequence repeats. Proc. R. Soc. Lond. Ser. B Biol. Sci. 1998, 265, 1697–1705. [Google Scholar] [CrossRef]
- Nowakowska, J.A.; Oszako, T.; Tereba, A.; Konecka, A. Forest tree species traced with a DNA-based proof for illegal logging case in Poland. In Evolutionary Biology: Biodiversification from Genotype to Phenotype, 1st ed.; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Eliades, N.-G.; Eliades, D.G. Haplotype Analysis: Software for Analysis of Haplotypes Data. In Forest Genetics and Forest Tree Breeding; Georg-Augst University: Goettingen, Germany, 2009. [Google Scholar]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver. 3.5, A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P. Genealex 6.5, Genetic Analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Lorenz, M. International co-operative programme on assessment and monitoring of air pollution effects on forests–ICP forests. Water Air Soil Pollut. 1995, 85, 1221–1226. [Google Scholar] [CrossRef]
- Tyszkiewicz, S. Nasiennictwo Leśne; Forest Research Institute: Warsaw, Poland, 1949. [Google Scholar]
- Przybylski, P.; Konatowska, M.; Jastrzębowski, S.; Tereba, A.; Mohytych, V.; Tyburski, Ł.; Rutkowski, P. The Possibility of Regenerating a Pine Stand through Natural Regeneration. Forests 2021, 12, 1055. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 29 March 2020).
- Silva, L.J.; Laercio, S. Duncan Test, Tukey Test and Scott-Knott Test. R package Version 1.0-1. Available online: https://CRAN.Rproject.org/package=laercio (accessed on 29 March 2020).
- Wei, T.; Simko, V. R Package “Corrplot”: Visualization of a Correlation Matrix (Version 0.84). Available online: https://github.com/taiyun/corrplot (accessed on 29 March 2020).
- Vu, V.Q. Ggbiplot: A Ggplot2 Based Biplot. R Package Version 0.55. Available online: http://github.com/vqv/ggbiplot (accessed on 29 March 2020).
- Millar, C.I.; Stephenson, N.L.; Stephens, S.L. Climate change and forests of the future: Managing in the face of uncertainty. Ecol. Appl. 2007, 17, 2145–2151. [Google Scholar] [CrossRef] [PubMed]
- Hoegh−Guldberg, O.; Hughes, L.; McIntyre, S.; Lindenmayer, D.B.; Parmesan, C.; Possingham, H.P.; Thomas, C.D. Assisted colonization and rapid climate change. Science 2008, 321, 345–346. [Google Scholar] [CrossRef] [PubMed]
- Kosinska, J.; Lewandowski, A.; Chalupka, W. Genetic variability of Scots pine maternal populations and their progenies. Silva Fenn. 2007, 41, 5–12. [Google Scholar] [CrossRef]
- Bush, R.M.; Smouse, P.E. The impact of electrophoretic on life history traits in Pinus taeda. Evolution 1991, 45, 481–498. [Google Scholar] [CrossRef]
- Müller-Starck, G. Genetic differences between ‘tolerant’ and ‘sensitive’ beeches (Fagus sylvatica L.) in an environmentally stressed adult forest stand. Silvae Genetica 1985, 34, 241–247. [Google Scholar]
- Cheng, Z.-M.; Shi, N.-Q.; Herman, D.E.; Capps, T.K. Building in resistance to Dutch elm disease. J. For. 1997, 95, 24–27. [Google Scholar]
- Rajora, O.P.; Rahman, M.H.; Buchert, G.P.; Dancik, B.P. Microsatellite DNA analysis of genetic effects of harvesting in old-growth eastern white pine (Pinus strobus) in Ontario. Mol. Ecol. 2000, 9, 339–348. [Google Scholar] [CrossRef]
- Available online: www.Gios.gov.pl/monlas/raporty.html (accessed on 24 September 2022).
- Aleksandrowicz-Trzcińska, M.; Drozdowski, S.; Studnicki, M.; Żybura, H. Effects of Site Preparation Methods on the Establishment and Natural-Regeneration Traits of Scots Pines (Pinus sylvestris L.) in Northeastern Poland. Forests 2018, 9, 717. [Google Scholar] [CrossRef]
- Grzesiuk, S. Wpływ chemizacji rolnictwa na fizjologiczne właściwości nasion. Biul. IHAR 1973, 5–6, 9–14. [Google Scholar]



| Location | Gacno | Plesno | Mielnica | Kociol |
|---|---|---|---|---|
| Acronyms | GAC | PLE | MIE | KOC |
| Coordinates | N53.792663 E17.569418 | N53.814655 E17.554436 | N53.800613 E17.518334 | N53.824116 E17.591752 |
| Age * of the dominant P. sylvestris | 130–140 (avg.: 135) | 119–132 (avg.: 125) | 150–190 (avg.: 170) | 110–120 (avg.: 115) |
| Forest habitat type | fresh coniferous forest | fresh coniferous forest | fresh mixed coniferous | fresh coniferous forest |
| Population | N | A | Ph | Rh | He | D2sh |
|---|---|---|---|---|---|---|
| KOC | 50 | 47 | 27 | 44.238 | 0.998 | 3.785 |
| GAC | 48 | 31 | 18 | 30.000 | 0.977 | 2.096 |
| PLE | 50 | 36 | 21 | 33.915 | 0.982 | 2.763 |
| MIE | 50 | 38 | 23 | 35.796 | 0.986 | 3.243 |
| Mean | 49.5 | 38 | 22.250 | 35.987 | 0.986 | 2.971 |
| Source of Variation | Sum of Squares | Variance Components | Percent of Variation |
|---|---|---|---|
| Among populations | 50.930 | 0.15009 | 9.43186 |
| Among individuals within populations | 530.357 | 1.44119 | 90.56814 |
| Within individuals | 0.000 | 0.00000 | 0.0000 |
| Total | 581.287 | 1.59127 |
| KOC | GAC | PLE | MIE | |
|---|---|---|---|---|
| KOC | - | 0.00594 | 0.00000 | 0.00000 |
| GAC | 0.04242 | - | 0.00000 | 0.00000 |
| PLE | 0.07938 | 0.15318 | - | 0.21842 |
| MIE | 0.12080 | 0.20449 | 0.01577 | - |
| Population | Number of Seedlings in the Stand | Number of Cones in the Stand |
|---|---|---|
| GAC | 71 | 2120 |
| PLE | 22 | 1516 |
| MIE | 67 | 2328 |
| KOC | 2 | 1388 |
| Df | Sum Sq | Mean Sq | Fvalue | p-Value | |
|---|---|---|---|---|---|
| Stands | 3 | 653 | 218 | 2.725 | 0.045 |
| Number of vintages of needles | 2 | 17,132 | 17,132 | 214.609 | 0.000 |
| Biosocial position | 1 | 360 | 360 | 4.514 | 0.034 |
| Residuals | 192 | 15,327 | 80 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Przybylski, P. Assessment of Variability: Chloroplast Microsatellite DNA, Defoliation, and Regeneration Potential of Old Pine Stands of Different Origins in the Context of Assisted Genotype Migration. Forests 2022, 13, 1829. https://doi.org/10.3390/f13111829
Przybylski P. Assessment of Variability: Chloroplast Microsatellite DNA, Defoliation, and Regeneration Potential of Old Pine Stands of Different Origins in the Context of Assisted Genotype Migration. Forests. 2022; 13(11):1829. https://doi.org/10.3390/f13111829
Chicago/Turabian StylePrzybylski, Paweł. 2022. "Assessment of Variability: Chloroplast Microsatellite DNA, Defoliation, and Regeneration Potential of Old Pine Stands of Different Origins in the Context of Assisted Genotype Migration" Forests 13, no. 11: 1829. https://doi.org/10.3390/f13111829
APA StylePrzybylski, P. (2022). Assessment of Variability: Chloroplast Microsatellite DNA, Defoliation, and Regeneration Potential of Old Pine Stands of Different Origins in the Context of Assisted Genotype Migration. Forests, 13(11), 1829. https://doi.org/10.3390/f13111829




