Generalists and Specialists Determine the Trend and Rate of Soil Fungal Distance Decay of Similarity in a 20-ha Subtropical Forest
Abstract
:1. Introduction
2. Materials and Methods
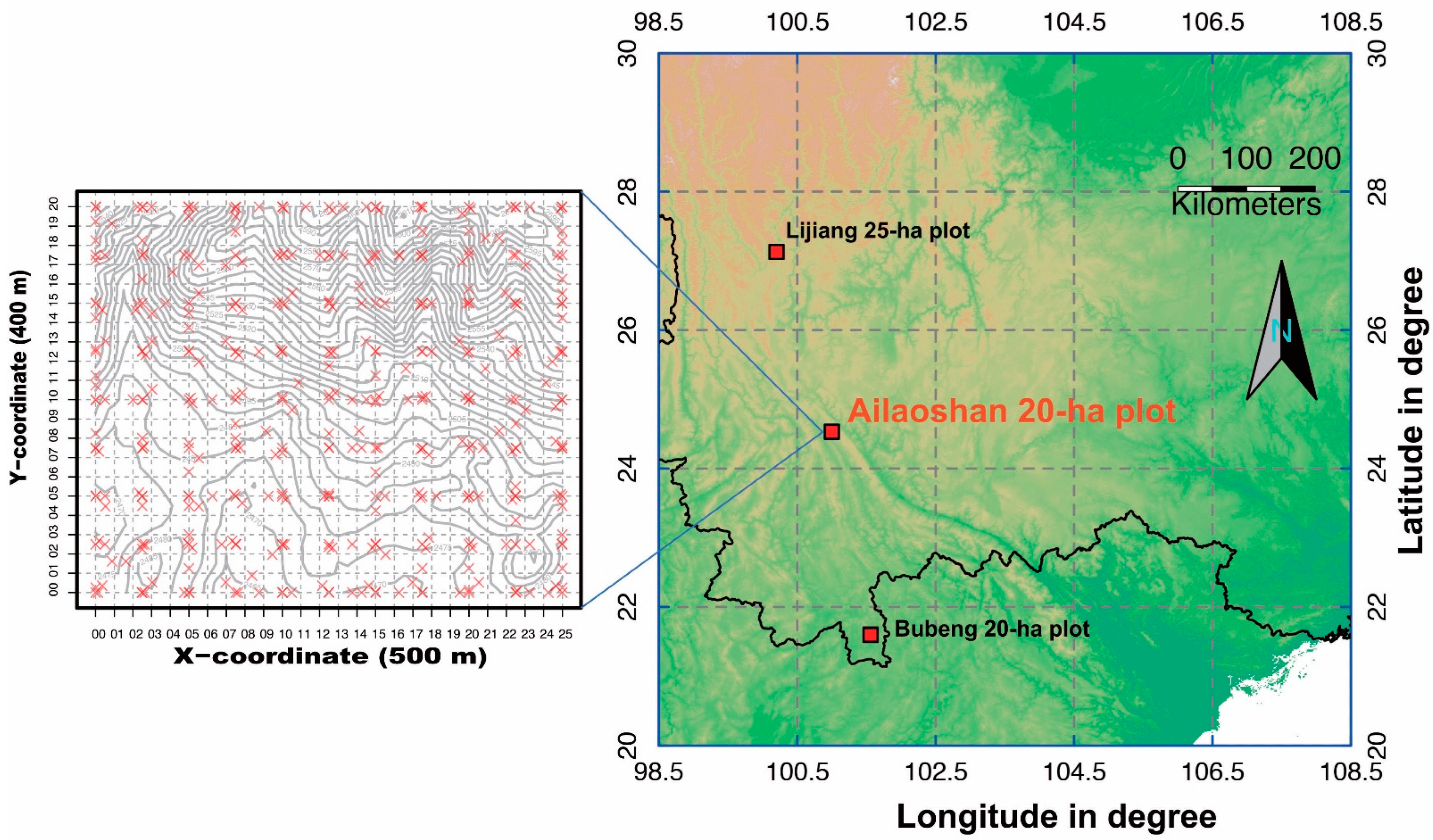
2.1. Study Site
2.2. Collection of Soil Fungal Samples
2.3. Sequencing and Sequence Data Processing
2.4. Soil Fungal Classification
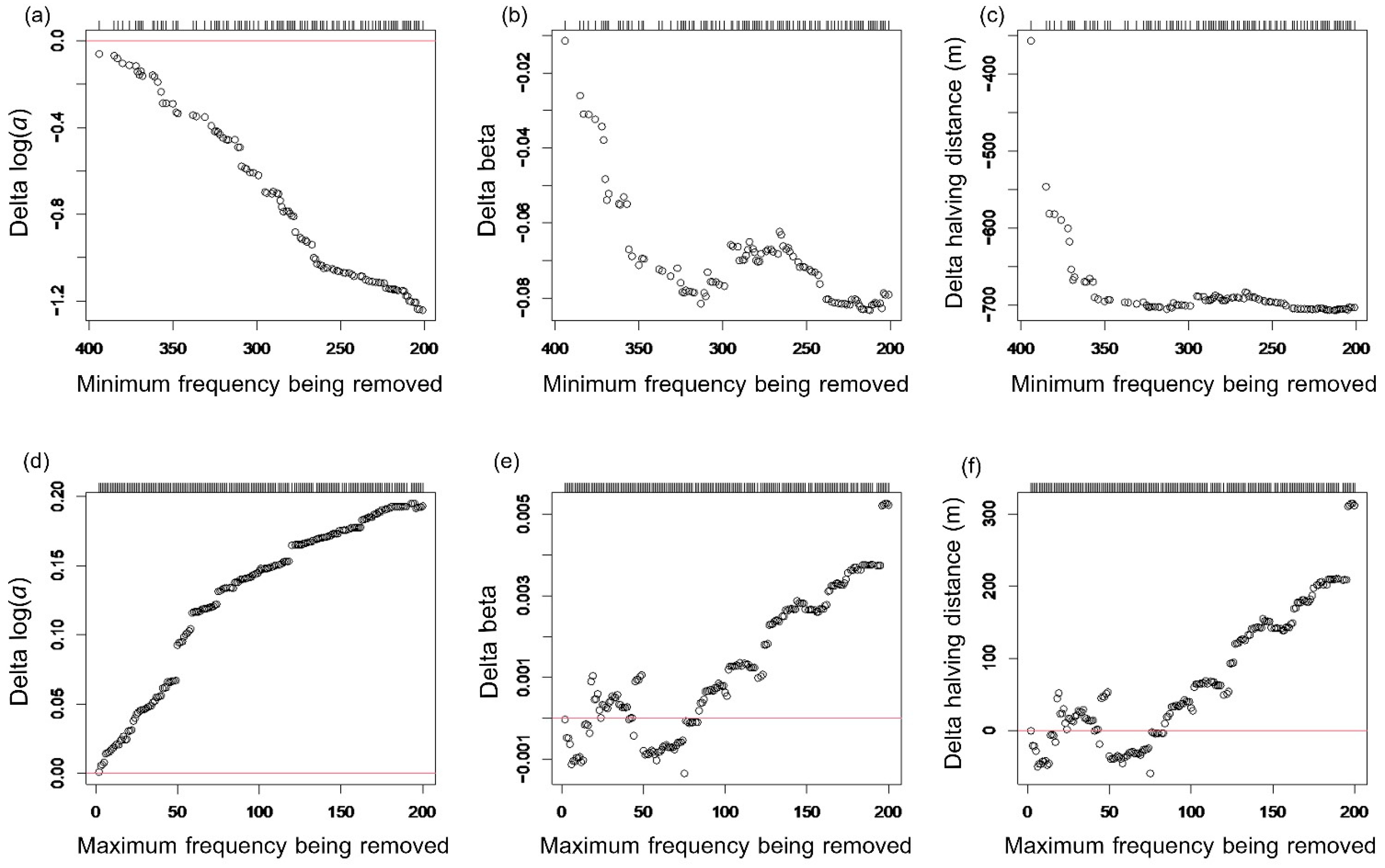
2.5. Data Analysis
3. Results
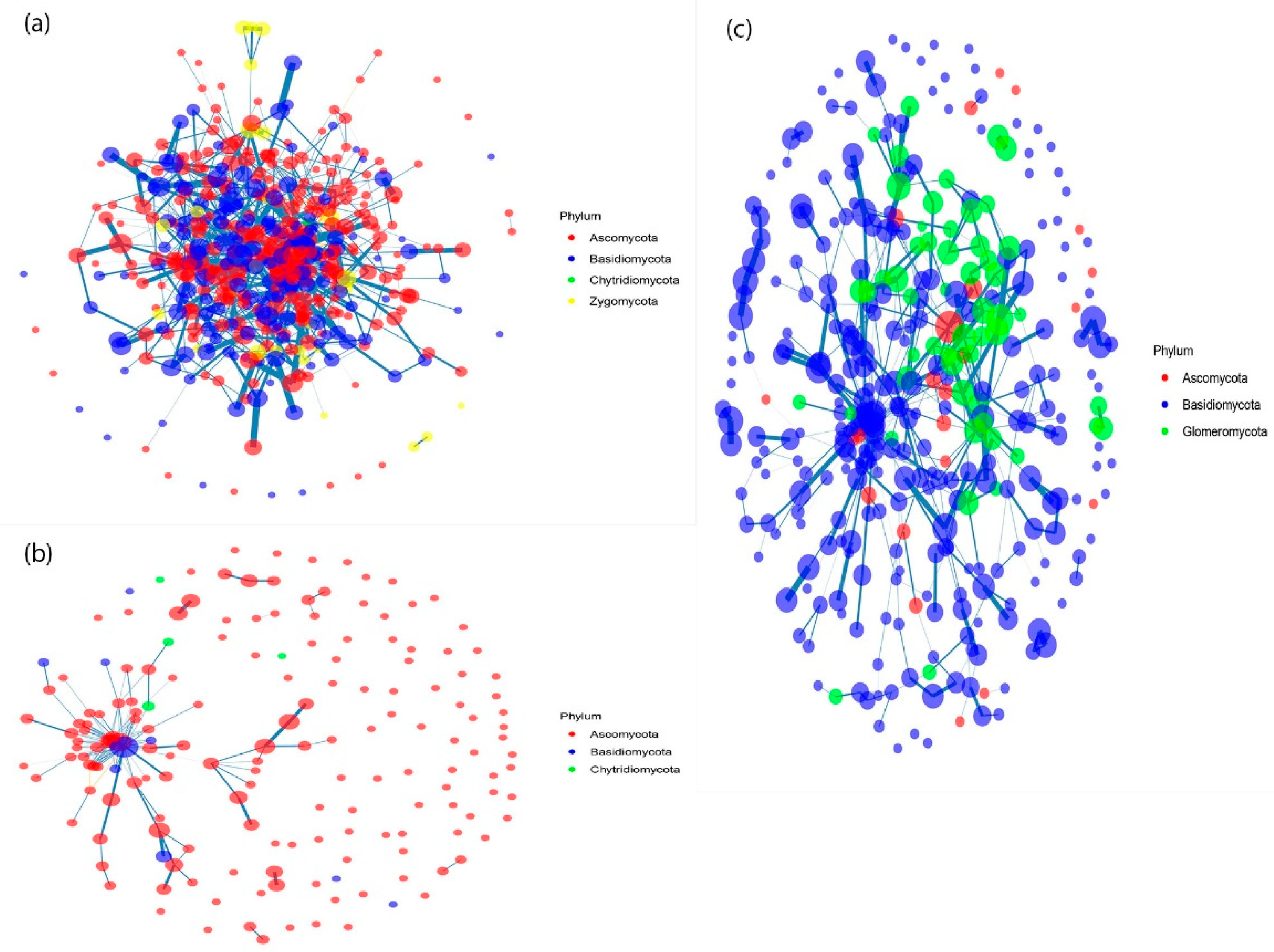
3.1. The Co-Occurrence Network of Soil Fungal Communities
3.2. Distance-Decay Relationships on Soil Fungal Communities
3.3. Beta Diversity Partitioning
4. Discussion
4.1. Soil Fungal Community Diversity and Composition
4.2. Distance–Decay Relationships of Soil Fungal Communities
4.3. Beta Diversity Partitioning
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Soininen, J.; McDonald, R.; Hillebrand, H. The distance decay of similarity in ecological communities. Ecography 2007, 30, 3–12. [Google Scholar] [CrossRef]
- Tobler, W.R. A computer movie simulating urban growth in the Detroit region. Econ. Geogr. 1970, 46 (Suppl. 1), 234–240. [Google Scholar] [CrossRef]
- Hanson, C.A.; Fuhrman, J.A.; Horner-Devine, M.C.; Martiny, J.B. Beyond biogeographic patterns: Processes shaping the microbial landscape. Nat. Rev. Microbiol. 2012, 10, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Talbot, J.M.; Bruns, T.D.; Taylor, J.W.; Smith, D.P.; Branco, S.; Glassman, S.I.; Erlandson, S.; Vilgalys, R.; Liao, H.L.; Smith, M.E.; et al. Endemism and functional convergence across the North American soil mycobiome. Proc. Natl. Acad. Sci. USA 2014, 111, 6341–6346. [Google Scholar] [CrossRef] [Green Version]
- Bahram, M.; Koljalg, U.; Courty, P.E.; Diedhiou, A.G.; Kjøller, R.; Polme, S.; Ryberg, M.; Veldre, V.; Tedersoo, L. The distance decay of similarity in communities of ectomycorrhizal fungi in different ecosystems and scales. J. Ecol. 2013, 101, 1335–1344. [Google Scholar] [CrossRef]
- Green, J.L.; Holmes, A.J.; Westoby, M.; Oliver, I.; Briscoe, D.; Dangerfield, M.; Gillings, M.; Beattie, A.J. Spatial scaling of microbial eukaryote diversity. Nature 2004, 432, 747–750. [Google Scholar] [CrossRef]
- Qian, H.; Ricklefs, R.E.; White, P.S. Beta diversity of angiosperms in temperate floras of eastern Asia and eastern North America. Ecol. Lett. 2005, 8, 15–22. [Google Scholar] [CrossRef]
- Hu, Y.H.; Johnson, D.J.; Mi, X.C.; Wang, X.G.; Ye, W.H.; De Li, Y.; Lian, J.Y.; Cao, M. The relative importance of space compared to topography increases from rare to common tree species across latitude. J. Biogeogr. 2018, 45, 2520–2532. [Google Scholar] [CrossRef]
- Pandit, S.N.; Kolasa, J.; Cottenie, K. Contrasts between habitat generalists and specialists: An empirical extension to the basic metacommunity framework. Ecology 2009, 90, 2253–2262. [Google Scholar] [CrossRef] [Green Version]
- Siqueira, T.; Bini, L.M.; Roque, F.O.; Marques Couceiro, S.R.; Trivinho-Strixino, S.; Cottenie, K. Common and rare species respond to similar niche processes in macroinvertebrate metacommunities. Ecography 2012, 35, 183–192. [Google Scholar] [CrossRef]
- Morlon, H.; Chuyong, G.; Condit, R.; Hubbell, S.; Kenfack, D.; Thomas, D.; Valencia, R.; Green, J.L. A general framework for the distance–decay of similarity in ecological communities. Ecol. Lett. 2008, 11, 904–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oono, R.; Rasmussen, A.; Lefèvre, E. Distance decay relationships in foliar fungal endophytes are driven by rare taxa. Environ. Microbiol. 2017, 19, 2794–2805. [Google Scholar] [CrossRef] [PubMed]
- Zinger, L.; Boetius, A.; Ramette, A. Bacterial taxa–area and distance–decay relationships in marine environments. Mol. Ecol. 2014, 23, 954–964. [Google Scholar] [CrossRef] [PubMed]
- Gloor, G.B.; Macklaim, J.M.; Pawlowsky-Glahn, V.; Egozcue, J.J. Microbiome datasets are compositional: And this is not optional. Front. Microbiol. 2017, 8, 2224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Li, W.; Dumbrell, A.J.; Liu, M.; Li, G.; Wu, M.; Jiang, C.; Li, Z. Spatial variation in soil fungal communities across paddy fields in subtropical China. mSystems 2020, 5, e00704-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [Green Version]
- Peay, K.G.; Kennedy, P.G.; Talbot, J.M. Dimensions of biodiversity in the Earth mycobiome. Nat. Rev. Microbiol. 2016, 14, 434–447. [Google Scholar] [CrossRef]
- Chen, L.; Swenson, N.G.; Ji, N.; Mi, X.; Ren, H.; Guo, L.; Ma, K. Differential soil fungus accumulation and density dependence of trees in a subtropical forest. Science 2019, 366, 124–128. [Google Scholar] [CrossRef]
- Floudas, D.; Binder, M.; Riley, R.; Barry, K.; Blanchette, R.A.; Henrissat, B.; Martinez, A.T.; Otil-lar, R.; Spatafora, J.W.; Yadav, J.S.; et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 2012, 336, 1715–1719. [Google Scholar] [CrossRef] [Green Version]
- Saikkonen, K.; Mikola, J.; Helander, M. Endophytic phyllosphere fungi and nutrient cycling in terrestrial ecosystems. Curr. Sci. 2015, 109, 121–126. [Google Scholar]
- Zhang, B.; Zhang, J.; Liu, Y.; Shi, P.; Wei, G. Co-occurrence patterns of soybean rhizosphere microbiome at a continental scale. Soil Biol. Biochem. 2018, 118, 178–186. [Google Scholar] [CrossRef]
- Li, S.; Wu, F. Diversity and co-occurrence patterns of soil bacterial and fungal communities in seven intercropping systems. Front. Microbiol. 2018, 9, 1521. [Google Scholar] [CrossRef] [PubMed]
- Bowman, E.A.; Arnold, A.E. Drivers and implications of distance decay differ for ectomycorrhizal and foliar endophytic fungi across an anciently fragmented landscape. ISME J. 2021, 15, 3437–3454. [Google Scholar] [CrossRef]
- Jiang, Y.; Liang, Y.; Li, C.; Wang, F.; Sui, Y.; Suvannang, N.; Zhou, J.; Sun, B. Crop rotations alter bacterial and fungal diversity in paddy soils across East Asia. Soil Biol. Biochem. 2016, 95, 250–261. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, N.H.; Song, Z.W.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Sterkenburg, E.; Bahr, A.; Brandström Durling, M.; Clemmensen, K.E.; Lindahl, B.D. Changes in fungal communities along a boreal forest soil fertility gradient. New Phytol. 2015, 207, 1145–1158. [Google Scholar] [CrossRef] [Green Version]
- Viana, D.S.; Chase, J.M. Spatial scale modulates the inference of metacommunity assembly processes. Ecology 2019, 100, e02576. [Google Scholar] [CrossRef] [PubMed]
- Schröter, K.; Wemheuer, B.; Pena, R.; Schöning, I.; Ehbrecht, M.; Schall, P.; Ammer, C.; Daniel, R.; Polle, A. Assembly processes of trophic guilds in the root mycobiome of temperate forests. Mol. Ecol. 2019, 28, 348–364. [Google Scholar] [CrossRef]
- Zhao, Z.; Ma, Y.; Feng, T.; Kong, X.; Wang, Z.; Zheng, W.; Zhai, B. Assembly processes of abundant and rare microbial communities in orchard soil under a cover crop at different periods. Geoderma 2022, 406, 115543. [Google Scholar] [CrossRef]
- Jiao, S.; Lu, Y. Abundant fungi adapt to broader environmental gradients than rare fungi in agricultural fields. Glob. Chang. Biol. 2020, 26, 4506–4520. [Google Scholar] [CrossRef]
- Wen, H.D.; Lin, L.X.; Yang, J.; Hu, Y.H.; Cao, M.; Liu, Y.H.; Lu, Z.Y.; Xie, Y.N. Species composition and community structure of a 20 hm2 plot of mid-mountain moist evergreen broad-leaved forest on the Mts. Ailaoshan, Yunnan Province, China. Chin. J. Plant Ecol. 2018, 42, 419–429. (In Chinese) [Google Scholar]
- Gong, H.D.; Zhang, Y.P.; Lei, Y.B.; Liu, Y.H.; Yang, G.P.; Lu, Z.Y. Evergreen broad-leaved forest improves soil water status compared with tea tree plantation in Ailao Mountains, Southwest China. Acta Agric. Scand. 2011, 61, 384–388. [Google Scholar] [CrossRef]
- Wu, C.S.; Zhang, Y.P.; Xu, X.L.; Sha, L.Q.; You, G.Y.; Liu, Y.H.; Xie, Y.N. Influence of interactions between litter decomposition and rhizosphere activity on soil respiration and on the temperature sensitivity in a subtropical montane forest in SW China. Plant Soil 2014, 381, 215–224. [Google Scholar] [CrossRef]
- Chan, O.C.; Yang, X.; Fu, Y.; Feng, Z.; Sha, L.; Casper, P.; Zou, X. 16S rRNA gene analyses of bacterial community structures in the soils of evergreen broad-leaved forests in south-west China. FEMS Microbiol. Ecol. 2006, 58, 247–259. [Google Scholar] [CrossRef] [Green Version]
- Gohl, D.M.; Vangay, P.; Garbe, J.; MacLean, A.; Hauge, A.; Becker, A.; Gould, T.J.; Clayton, J.B.; Johnson, T.J.; Hunter, R.; et al. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat. Biotechnol. 2016, 34, 942–949. [Google Scholar] [CrossRef] [Green Version]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Tang, G.L.; Xu, X.Y.; Nong, X.H.; Qi, S.H. Insights into deep-sea sediment fungal communities from the East Indian Ocean using targeted environmental sequencing combined with traditional cultivation. PLoS ONE 2014, 9, e109118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bengtsson-Palme, J.; Ryberg, M.; Hartmann, M.; Branco, S.; Wang, Z.; Godhe, A.; De Wit, P.; Sanchez-Garcia, M.; Ebersberger, I.; De Sousa, F.; et al. Improved software detection and extraction of ITS1 and ITS 2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013, 4, 914–919. [Google Scholar] [CrossRef]
- Al-Ghalith, G.A.; Montassier, E.; Ward, H.N.; Knights, D. NINJA-OPS: Fast accurate marker gene alignment using concatenated ribosomes. PLoS Comput. Biol. 2016, 12, e1004658. [Google Scholar] [CrossRef]
- Cregger, M.A.; Veach, A.M.; Yang, Z.K.; Crouch, M.J.; Vilgalys, R.; Tuskan, G.A.; Schadt, C.W. The Populus holobiont: Dissecting the effects of plant niches and genotype on the microbiome. Microbiome 2018, 6, 1–14. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. Int. J. Complex. 2006, 1695, 1–9. [Google Scholar]
- Nekola, J.C.; White, P.S. The distance decay of similarity in biogeography and ecology. J. Biogeogr. 1999, 26, 867–878. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2.4–3. 2017. Available online: https://cran.r-project.org/package=vegan (accessed on 8 February 2021).
- Millar, R.B.; Anderson, M.J.; Tolimieri, N. Much ado about nothings: Using zero similarity points in distance-decay curves. Ecology 2011, 92, 1717–1722. [Google Scholar] [CrossRef] [Green Version]
- Baselga, A. Partitioning the turnover and nestedness components of beta diversity. Glob. Ecol. Biogeogr. 2010, 19, 134–143. [Google Scholar] [CrossRef]
- Baselga, A.; Orme, C.D.L. betapart: An R package for the study of beta diversity. Methods Ecol. Evol. 2012, 3, 808–812. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017; Available online: https://www.r-project.org/ (accessed on 26 July 2022).
- Mougi, A.; Kondoh, M. Diversity of Interaction Types and Ecological Community Stability. Science 2012, 337, 349–351. [Google Scholar] [CrossRef]
- Deacon, L.J.; Pryce-Miller, E.J.; Frankland, J.C.; Bainbridge, B.W.; Moore, P.D.; Robinson, C.H. Diversity and function of decomposer fungi from a grassland soil. Soil Biol. Biochem. 2006, 38, 7–20. [Google Scholar] [CrossRef]
- Shi, L.L.; Mortimer, P.E.; Slik, J.F.; Zou, X.M.; Xu, J.; Feng, W.T.; Qiao, L. Variation in forest soil fungal diversity along a latitudinal gradient. Fungal Divers. 2014, 64, 305–315. [Google Scholar] [CrossRef]
- Xiong, J.; Peng, F.; Sun, H.; Xue, X.; Chu, H. Divergent Responses of Soil Fungi Functional Groups to Short-term Warming. Microb. Ecol. 2014, 68, 708–715. [Google Scholar] [CrossRef]
- Tatsumi, C.; Azuma, W.A.; Ogawa, Y.; Komada, N. Nitrogen Availability and Microbial Communities of Canopy Soils in a Large Cercidiphyllum japonicum Tree of a Cool-Temperate Old Growth Forest. Microb. Ecol. 2021, 82, 919–931. [Google Scholar] [CrossRef]
- Cairney, J.W. Basidiomycete mycelia in forest soils: Dimensions, dynamics and roles in nutrient distribution. Mycol. Res. 2005, 109, 7–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konstantinidis, G.; Kaounas, V. Elaphomyces muricatus and Fischerula macrospora, two interesting hypogeous fungi from Greece. Ascomycete 2012, 4, 95–98. [Google Scholar]
- Zsigmond, A.R.; Kántor, I.; May, Z.; Urák, I.; Héberger, K. Elemental composition of Russula cyanoxantha along an urbanizationgradient in Cluj-Napoca (Romania). Chemosphere 2020, 238, 124566. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Logares, R.; Huang, B.; Hsieh, C.H. Abundant and rare picoeukaryotic sub-communities present contrasting patterns in the epipelagic waters of marginal seas in the northwestern Pacific Ocean. Environ. Microbiol. 2017, 19, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Lu, Y. Soil pH and temperature regulate assembly processes of abundant and rare bacterial communities in agricultural ecosystems. Environ. Microbiol. 2020, 22, 1052–1065. [Google Scholar] [CrossRef]
- Liang, Y.; Xiao, X.; Nuccio, E.E.; Yuan, M.; Zhang, N.; Xue, K.; Cohan, F.M.; Zhou, J.; Sun, B. Differentiation strategies of soil rare and abundant microbial taxa in response to changing climatic regimes. Environ. Microbiol. 2020, 22, 1327–1340. [Google Scholar] [CrossRef] [PubMed]
- Szekely, A.J.; Langenheder, S. The importance of species sorting differs between habitat generalists and specialists in bacterial communities. FEMS Microbiol. Ecol. 2014, 87, 102–112. [Google Scholar] [CrossRef]
- Xu, Q.; Vandenkoornhuyse, P.; Li, L.; Guo, J.; Zhu, C.; Guo, S.; Ling, N.; Shen, Q. Microbial generalists and specialists differently contribute to the community diversity in farmland soils. J. Adv. Res. 2021; in press. [Google Scholar]
- Liu, L.; Yang, J.; Yu, Z.; Wilkinson, D.M. The biogeography of abundant and rare bacterioplankton in the lakes and reservoirs of China. ISME J. 2015, 9, 2068–2077. [Google Scholar] [CrossRef]
- Liao, J.; Cao, X.; Zhao, L.; Wang, J.; Gao, Z.; Wang, M.C.; Huang, Y. The importance of neutral and niche processes for bacterial community assembly differs between habitat generalists and specialists. FEMS Microbiol. Ecol. 2016, 92, fiw174. [Google Scholar] [CrossRef] [Green Version]
- Rosenzweig, M.L. Species Diversity in Space and Time; Cambridge University Press (CUP): Cambridge, UK, 1995. [Google Scholar]
- Wang, J.; Zhang, T.; Li, L.; Li, J.; Feng, Y.; Lu, Q. The patterns and drivers of bacterial and fun-gal β-diversity in a typical dryland ecosystem of northwest China. Front. Microbiol. 2017, 8, 2126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genevieve, L.; Pierre-Luc, C.; Roxanne, G.-T.; Amélie, M.; Danny, B.; Vincent, M.; Hugo, G. Estimation of fungal diversity and identification of major abiotic drivers influencing fungal richness and communities in northern temperate and boreal Quebec forests. Forests 2019, 10, 1096. [Google Scholar] [CrossRef] [Green Version]
- Buckley, L.B.; Jetz, W. Linking global turnover of species and environments. Proc. Natl. Acad. Sci. USA 2008, 105, 17836–17841. [Google Scholar] [CrossRef] [Green Version]
- Podani, J.; Schmera, D. A new conceptual and methodological framework for exploring and ex-plaining pattern in presence–absence data. Oikos 2011, 120, 1625–1638. [Google Scholar] [CrossRef]
- Legendre, P. Interpreting the replacement and richness difference components of beta diversity. Glob. Ecol. Biogeogr. 2014, 23, 1324–1334. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaithaisong, P.; Alcantara, M.J.M.; Song, L.; Hu, Y.-H. Generalists and Specialists Determine the Trend and Rate of Soil Fungal Distance Decay of Similarity in a 20-ha Subtropical Forest. Forests 2022, 13, 1188. https://doi.org/10.3390/f13081188
Chaithaisong P, Alcantara MJM, Song L, Hu Y-H. Generalists and Specialists Determine the Trend and Rate of Soil Fungal Distance Decay of Similarity in a 20-ha Subtropical Forest. Forests. 2022; 13(8):1188. https://doi.org/10.3390/f13081188
Chicago/Turabian StyleChaithaisong, Pachchara, Mark Jun M. Alcantara, Liang Song, and Yue-Hua Hu. 2022. "Generalists and Specialists Determine the Trend and Rate of Soil Fungal Distance Decay of Similarity in a 20-ha Subtropical Forest" Forests 13, no. 8: 1188. https://doi.org/10.3390/f13081188
APA StyleChaithaisong, P., Alcantara, M. J. M., Song, L., & Hu, Y.-H. (2022). Generalists and Specialists Determine the Trend and Rate of Soil Fungal Distance Decay of Similarity in a 20-ha Subtropical Forest. Forests, 13(8), 1188. https://doi.org/10.3390/f13081188






