Characterisation of Methane Production Pathways in Sediment of Overwashed Mangrove Forests †
Abstract
:1. Introduction
2. Methodology
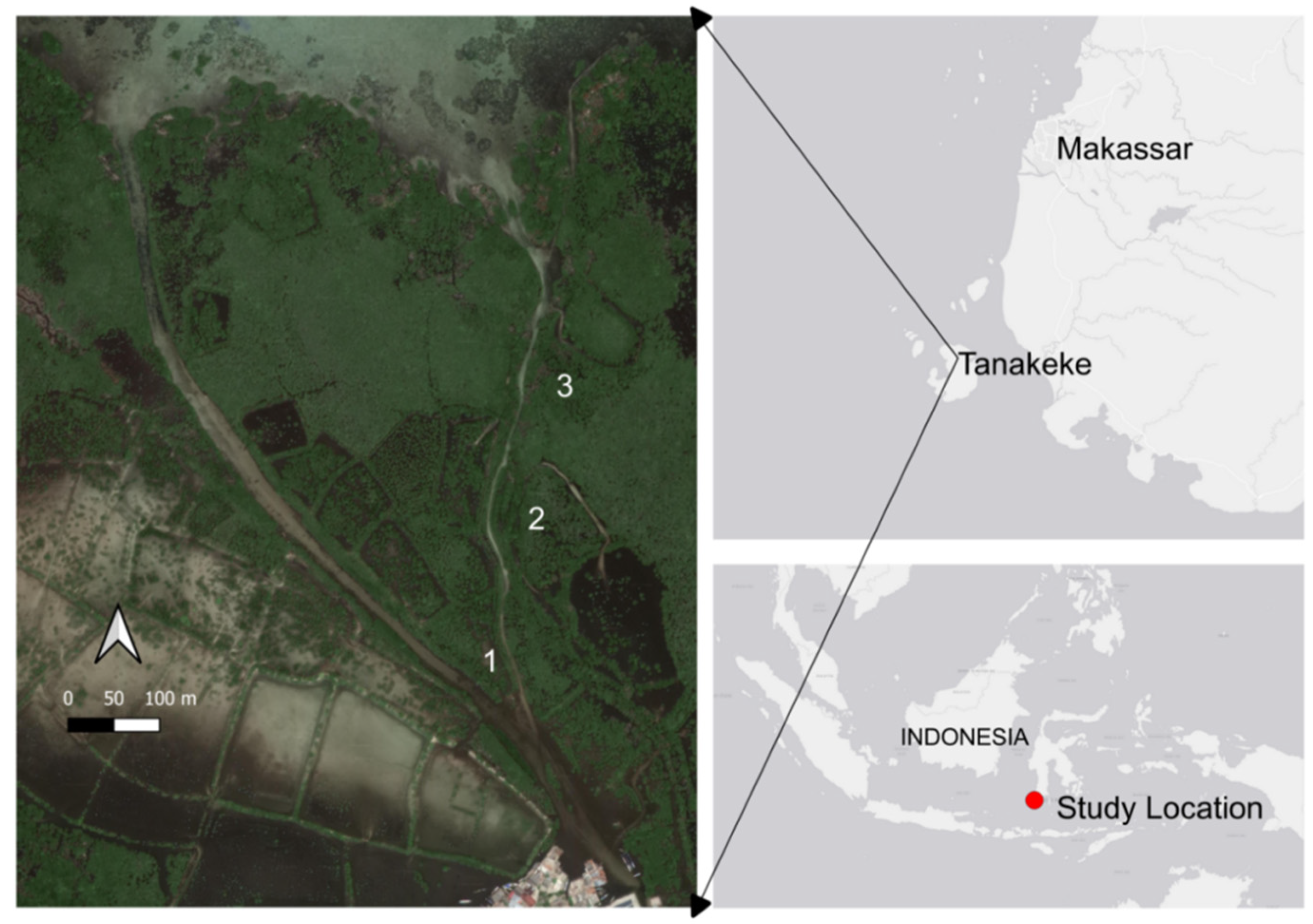
2.1. Study Location and Sampling Site
2.2. Sample Collection and Analytical Methods
2.2.1. Sample Collection
2.2.2. Microbial Enumeration
2.2.3. Sediment Slurry Experiments
3. Results
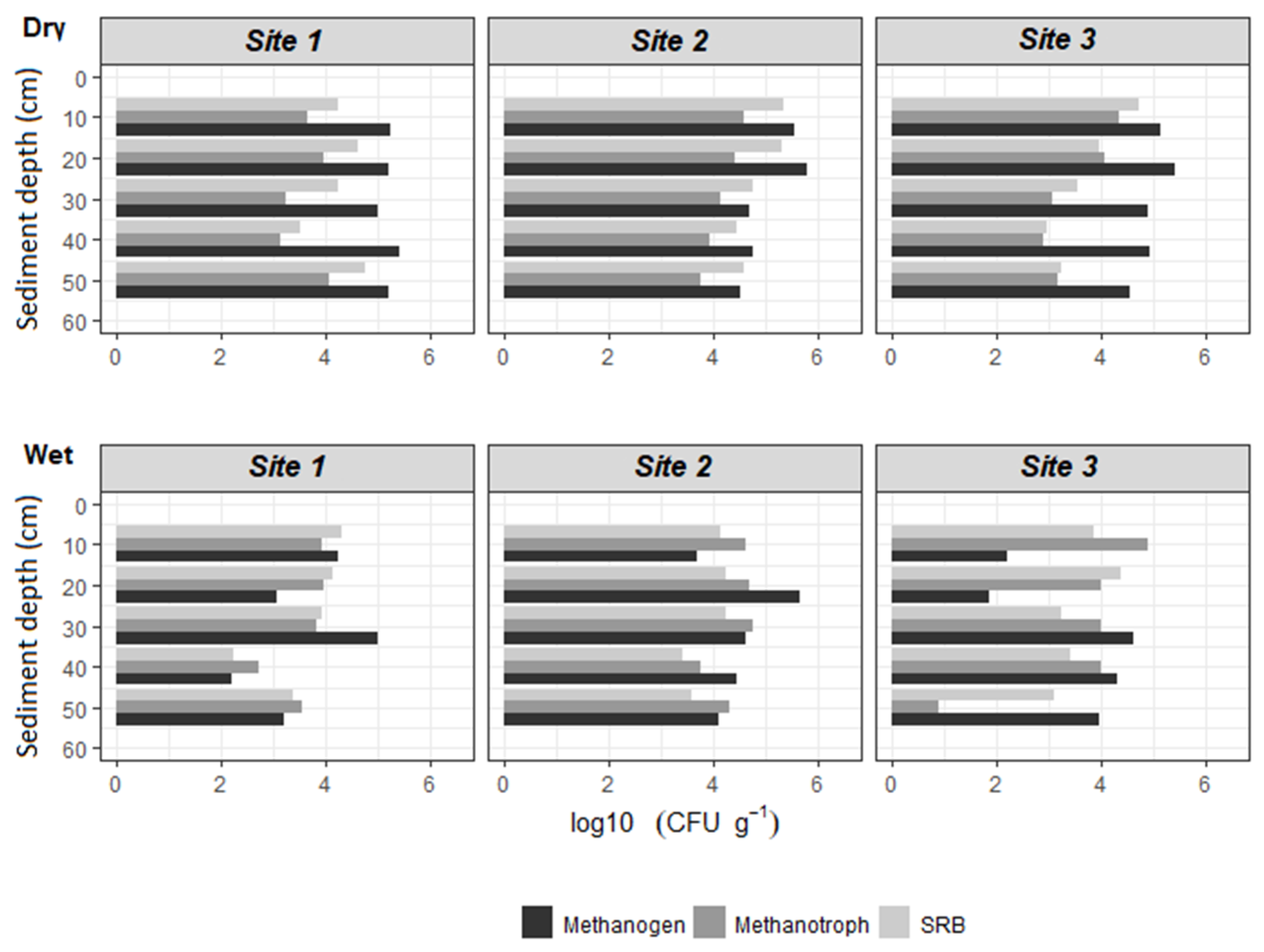
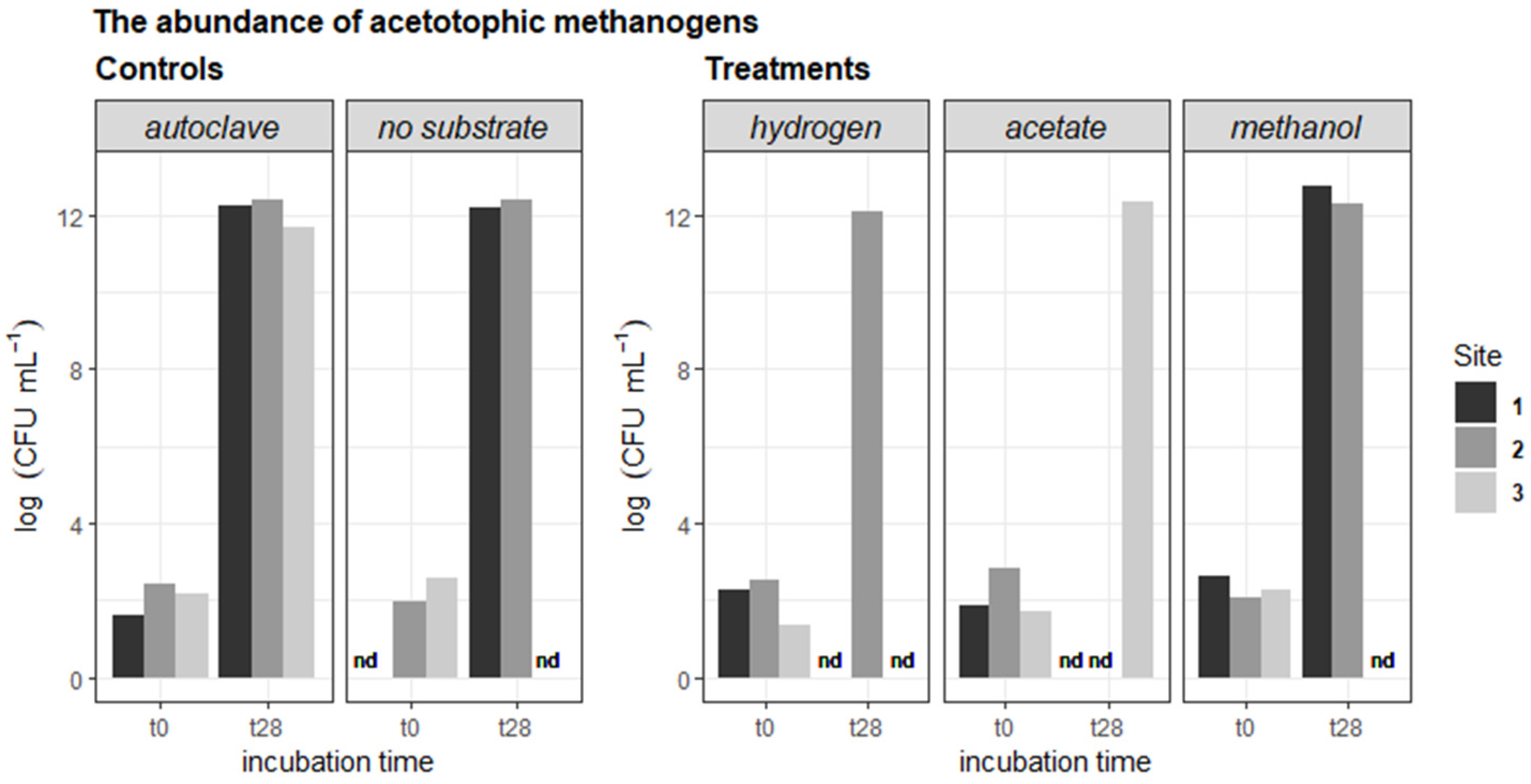
3.1. Abundances of Methanogens, Methanotrophs, and SRBs
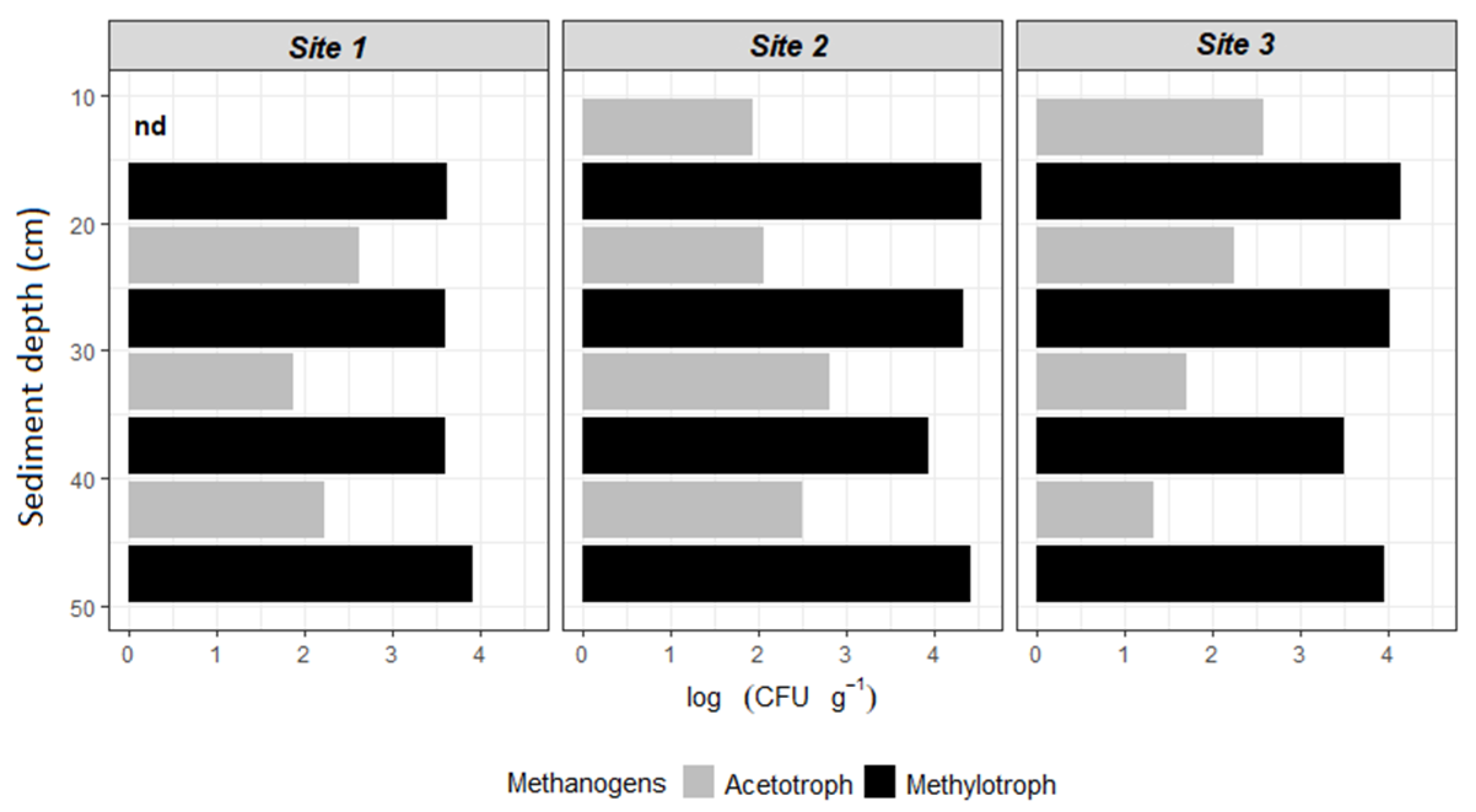
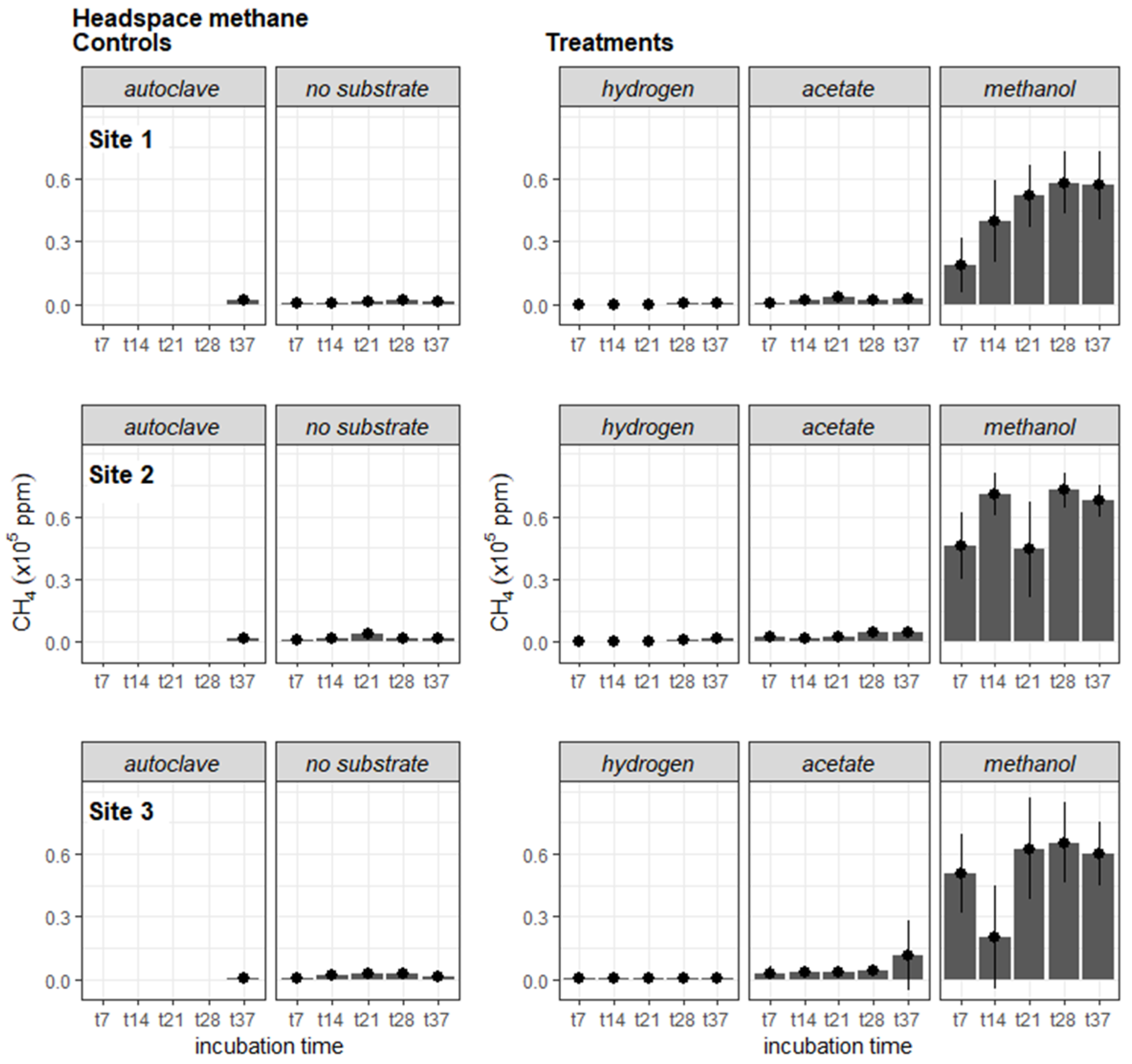
3.2. Potential CH4 Production
4. Discussion
4.1. Microbial Enumerations
4.2. Potential CH4 Production
4.3. Synthesis
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kirschke, S.; Bousquet, P.; Ciais, P.; Saunois, M.; Canadell, J.G.; Dlugokencky, E.J.; Bergamaschi, P.; Bergmann, D.; Blake, D.R.; Bruhwiler, L.; et al. Three Decades of Global Methane Sources and Sinks. Nat. Geosci. 2013, 6, 813–823. [Google Scholar] [CrossRef] [Green Version]
- Saunois, M.; Bousquet, P.; Poulter, B.; Peregon, A.; Ciais, P.; Canadell, J.G.; Dlugokencky, E.J.; Etiope, G.; Bastviken, D.; Houweling, S.; et al. The Global Methane Budget 2000–2012. Earth Syst. Sci. Data 2016, 8, 697–751. [Google Scholar] [CrossRef] [Green Version]
- Dean, J.F.; Middelburg, J.J.; Röckmann, T.; Aerts, R.; Blauw, L.G.; Egger, M.; Jetten, M.S.M.; de Jong, A.E.E.; Meisel, O.H.; Rasigraf, O.; et al. Methane Feedbacks to the Global Climate System in a Warmer World. Rev. Geophys. 2018, 56, 207–250. [Google Scholar] [CrossRef] [Green Version]
- Reay, D.S.; Smith, P.; Christensen, T.R.; James, R.H.; Clark, H. Methane and Global Environmental Change. Annu. Rev. Environ. Resour. 2018, 43, 165–192. [Google Scholar] [CrossRef]
- Zhang, Z.; Zimmermann, N.E.; Calle, L.; Hurtt, G.; Chatterjee, A.; Poulter, B. Enhanced Response of Global Wetland Methane Emissions to the 2015–2016 El Niño-Southern Oscillation Event. Environ. Res. Lett. 2018, 13, 074009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeikus, J.G.; Winfrey, M.R. Temperature Limitation of Methanogenesis in Aquatic Sediments. Appl. Environ. Microbiol. 1976, 31, 99–107. [Google Scholar] [CrossRef] [Green Version]
- Kao-Kniffin, J.; Woodcroft, B.J.; Carver, S.M.; Bockheim, J.G.; Handelsman, J.; Tyson, G.W.; Hinkel, K.M.; Mueller, C.W. Archaeal and Bacterial Communities across a Chronosequence of Drained Lake Basins in Arctic Alaska. Sci. Rep. 2015, 5, 18165. [Google Scholar] [CrossRef] [Green Version]
- He, R.; Wooller, M.J.; Pohlman, J.W.; Quensen, J.; Tiedje, J.M.; Leigh, M.B. Shifts in Identity and Activity of Methanotrophs in Arctic Lake Sediments in Response to Temperature Changes. Appl. Environ. Microbiol. 2012, 78, 4715–4723. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Tago, K.; Hayatsu, M.; Tokida, T.; Sakai, H.; Nakamura, H.; Usui, Y.; Hasegawa, T.; Asakawa, S. Effect of Elevated CO2 Concentration, Elevated Temperature and No Nitrogen Fertilization on Methanogenic Archaeal and Methane-Oxidizing Bacterial Community Structures in Paddy Soil. Microbes Environ. 2016, 31, 349–356. [Google Scholar] [CrossRef] [Green Version]
- Gamage, D.; Thompson, M.; Sutherland, M.; Hirotsu, N.; Makino, A.; Seneweera, S. New Insights into the Cellular Mechanisms of Plant Growth at Elevated Atmospheric Carbon Dioxide Concentrations: Elevated CO2 Effect on Plant Growth and Development. Plant Cell Environ. 2018, 41, 1233–1246. [Google Scholar] [CrossRef]
- Ahmadi Lahijani, M.J.; Kafi, M.; Nezami, A.; Nabati, J.; Mehrjerdi, M.Z.; Shahkoomahally, S.; Erwin, J. Variations in Assimilation Rate, Photoassimilate Translocation, and Cellular Fine Structure of Potato Cultivars (Solanum tuberosum L.) Exposed to Elevated CO2. Plant Physiol. Biochem. 2018, 130, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Chanton, J.P.; Bauer, J.E.; Glaser, P.A.; Siegel, D.I.; Kelley, C.A.; Tyler, S.C.; Romanowicz, E.H.; Lazrus, A. Radiocarbon Evidence for the Substrates Supporting Methane Formation within Northern Minnesota Peatlands. Geochim. Cosmochim. Acta 1995, 59, 3663–3668. [Google Scholar] [CrossRef]
- Kao-Kniffin, J.; Zhu, B. A Microbial Link between Elevated CO2 and Methane Emissions That Is Plant Species-Specific. Microb. Ecol. 2013, 66, 621–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, S.; Adhya, T.K. Dynamics of Methanogenesis and Methanotrophy in Tropical Paddy Soils as Influenced by Elevated CO2 and Temperature Interaction. Soil Biol. Biochem. 2012, 47, 36–45. [Google Scholar] [CrossRef]
- Kolb, S.; Carbrera, A.; Kammann, C.; Kämpfer, P.; Conrad, R.; Jäckel, U. Quantitative Impact of CO2 Enriched Atmosphere on Abundances of Methanotrophic Bacteria in a Meadow Soil. Biol. Fertil. Soils 2005, 41, 337–342. [Google Scholar] [CrossRef]
- Wei, D.; Wang, X. Recent Climatic Changes and Wetland Expansion Turned Tibet into a Net CH4 Source. Clim. Chang. 2017, 144, 657–670. [Google Scholar] [CrossRef]
- Ni, X.; Groffman, P.M. Declines in Methane Uptake in Forest Soils. Proc. Natl. Acad. Sci. USA 2018, 115, 8587–8590. [Google Scholar] [CrossRef] [Green Version]
- Grenfell, S.E.; Callaway, R.M.; Grenfell, M.C.; Bertelli, C.M.; Mendzil, A.F.; Tew, I. Will a Rising Sea Sink Some Estuarine Wetland Ecosystems? Sci. Total Environ. 2016, 554–555, 276–292. [Google Scholar] [CrossRef] [Green Version]
- Geselbracht, L.L.; Freeman, K.; Birch, A.P.; Brenner, J.; Gordon, D.R. Modeled Sea Level Rise Impacts on Coastal Ecosystems at Six Major Estuaries on Florida’s Gulf Coast: Implications for Adaptation Planning. PLoS ONE 2015, 10, e0132079. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Zhou, Y.; Zhuang, Q.; Prigent, C.; Liu, Y.; Teuling, A. Increasing Methane Emissions from Natural Land Ecosystems Due to Sea-Level Rise. J. Geophys. Res. Biogeosci. 2018, 123, 1756–1768. [Google Scholar] [CrossRef] [Green Version]
- Ferry, J.G. (Ed.) Methanogenesis; Springer: Boston, MA, USA, 1993. [Google Scholar] [CrossRef]
- Oremland, R.S.; Polcin, S. Methanogenesis and Sulfate Reduction: Competitive and Noncompetitive Substrates in Estuarine Sediments. Appl. Environ. Microbiol. 1982, 44, 1270–1276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyimo, T.J.; Pol, A.; Op den Camp, H.J.M. Sulfate Reduction and Methanogenesis in Sediments of Mtoni Mangrove Forest, Tanzania. AMBIO J. Hum. Environ. 2002, 31, 614–616. [Google Scholar] [CrossRef]
- Cadena, S.; García-Maldonado, J.Q.; López-Lozano, N.E.; Cervantes, F.J. Methanogenic and Sulfate-Reducing Activities in a Hypersaline Microbial Mat and Associated Microbial Diversity. Microb. Ecol. 2018, 75, 930–940. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, G.-C.; Heuer, V.B.; Lazar, C.S.; Goldhammer, T.; Wendt, J.; Samarkin, V.A.; Elvert, M.; Teske, A.P.; Joye, S.B.; Hinrichs, K.-U. Relative Importance of Methylotrophic Methanogenesis in Sediments of the Western Mediterranean Sea. Geochim. Cosmochim. Acta 2018, 224, 171–186. [Google Scholar] [CrossRef]
- Zalman, C.A.; Meade, N.; Chanton, J.; Kostka, J.E.; Bridgham, S.D.; Keller, J.K. Methylotrophic Methanogenesis in Sphagnum-Dominated Peatland Soils. Soil Biol. Biochem. 2018, 118, 156–160. [Google Scholar] [CrossRef]
- Maltby, J.; Steinle, L.; Löscher, C.R.; Bange, H.W.; Fischer, M.A.; Schmidt, M.; Treude, T. Microbial Methanogenesis in the Sulfate-Reducing Zone of Sediments in the Eckernförde Bay, SW Baltic Sea. Biogeosciences 2018, 15, 137–157. [Google Scholar] [CrossRef] [Green Version]
- King, G.M.; Klug, M.J.; Lovley, D.R. Metabolism of Acetate, Methanol, and Methylated Amines in Intertidal Sediments of Lowes Cove, Maine. Appl. Environ. Microbiol. 1983, 45, 1848–1853. [Google Scholar] [CrossRef] [Green Version]
- Lyimo, T.J.; Pol, A.; Harhangi, H.R.; Jetten, M.S.M.; Op den Camp, H.J.M. Anaerobic Oxidation of Dimethylsulfide and Methanethiol in Mangrove Sediments Is Dominated by Sulfate-Reducing Bacteria. FEMS Microbiol. Ecol. 2009, 70, 483–492. [Google Scholar] [CrossRef] [Green Version]
- Alongi, D.M.; Sasekumar, A.; Chong, V.C.; Pfitzner, J.; Trott, L.A.; Tirendi, F.; Dixon, P.; Brunskill, G.J. Sediment Accumulation and Organic Material Flux in a Managed Mangrove Ecosystem: Estimates of Land–Ocean–Atmosphere Exchange in Peninsular Malaysia. Mar. Geol. 2004, 208, 383–402. [Google Scholar] [CrossRef]
- Alongi, D.M.; Pfitzner, J.; Trott, L.A.; Tirendi, F.; Dixon, P.; Klumpp, D.W. Rapid Sediment Accumulation and Microbial Mineralization in Forests of the Mangrove Kandelia Candel in the Jiulongjiang Estuary, China. Estuar. Coast. Shelf Sci. 2005, 63, 605–618. [Google Scholar] [CrossRef]
- Welti, N.; Hayes, M.; Lockington, D. Seasonal Nitrous Oxide and Methane Emissions across a Subtropical Estuarine Salinity Gradient. Biogeochemistry 2017, 132, 55–69. [Google Scholar] [CrossRef]
- Cabezas, A.; Mitsch, W.J.; MacDonnell, C.; Zhang, L.; Bydałek, F.; Lasso, A. Methane Emissions from Mangrove Soils in Hydrologically Disturbed and Reference Mangrove Tidal Creeks in Southwest Florida. Ecol. Eng. 2018, 114, 57–65. [Google Scholar] [CrossRef]
- Ulumuddin, Y.I.; Beavis, S.; Roderick, M.; Eggins, S.; Sugoro, I.; Sukardjo, S. Potential Carbon Loss in Sediment through Methane Production during Early Development Stage of Mangrove Regeneration in Restored Mangroves. In Dynamic Sedimentary Environments of Mangrove Coasts; Elsevier: Amsterdam, The Netherlands, 2021; pp. 415–445. [Google Scholar] [CrossRef]
- Ramamurthy, T.; Raju, R.; Natarajan, R. Distribution and Ecology of Methanogenic Bacteria in Mangrove Sediments of Pitchavaram, East-Coast Of India. Indian J. Mar. Sci. 1990, 19, 269–273. [Google Scholar]
- Mohanraju, R.; Rajagopal, B.G.; Daniel, L.; Natarajan, R. Isolation and Characterization of a Methanogenic Bacterium from Mangrove Sediments. J. Mar. Biotechnol. 1997, 5, 147–152. [Google Scholar]
- Jing, H.; Cheung, S.; Zhou, Z.; Wu, C.; Nagarajan, S.; Liu, H. Spatial Variations of the Methanogenic Communities in the Sediments of Tropical Mangroves. PLoS ONE 2016, 11, e0161065. [Google Scholar] [CrossRef] [Green Version]
- McDougall, E.I. Studies on Ruminant Saliva. 1. The Composition and Output of Sheep’s Saliva. Biochem. J. 1948, 43, 99–109. [Google Scholar] [CrossRef] [Green Version]
- Hanišáková, N.; Vítězová, M.; Rittmann, S.K.-M.R. The Historical Development of Cultivation Techniques for Methanogens and Other Strict Anaerobes and Their Application in Modern Microbiology. Microorganisms 2022, 10, 412. [Google Scholar] [CrossRef]
- Luton, P.E.; Wayne, J.M.; Sharp, R.J.; Riley, P.W. The McrA Gene as an Alternative to 16S RRNA in the Phylogenetic Analysis of Methanogen Populations in Landfill b BThe GenBank Accession Numbers for the McrA Sequences Reported in This Paper Are AF414034–AF414051 (See Fig. 2) and AF414007–AF414033 (Environmental Isolates in Fig. 3). Microbiology 2002, 148, 3521–3530. [Google Scholar] [CrossRef] [Green Version]
- Geets, J.; Borremans, B.; Diels, L.; Springael, D.; Vangronsveld, J.; van der Lelie, D.; Vanbroekhoven, K. DsrB Gene-Based DGGE for Community and Diversity Surveys of Sulfate-Reducing Bacteria. J. Microbiol. Methods 2006, 66, 194–205. [Google Scholar] [CrossRef]
- Holmes, A.J.; Costello, A.; Lidstrom, M.E.; Murrell, J.C. Evidence That Participate Methane Monooxygenase and Ammonia Monooxygenase May Be Evolutionarily Related. FEMS Microbiol. Lett. 1995, 132, 203–208. [Google Scholar] [CrossRef]
- Oliver, D.M.; Bird, C.; Burd, E.; Wyman, M. Quantitative PCR Profiling of Escherichia coli in Livestock Feces Reveals Increased Population Resilience Relative to Culturable Counts under Temperature Extremes. Environ. Sci. Technol. 2016, 50, 9497–9505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilha, E.C.; Scariot, M.C.; Treml, D.; Pereira, T.P.; Sant′Anna, E.S.; Prudêncio, E.S.; Arisi, A.C.M. Comparison of Real-Time PCR Assay and Plate Count for Lactobacillus paracasei Enumeration in Yoghurt. Ann. Microbiol. 2016, 66, 597–606. [Google Scholar] [CrossRef]
- Anis, N.; Bonifait, L.; Quesne, S.; Baugé, L.; Chemaly, M.; Guyard-Nicodème, M. Simultaneous Detection of Salmonella Spp. and Quantification of Campylobacter Spp. in a Real-Time Duplex PCR: Myth or Reality? Pathogens 2023, 12, 338. [Google Scholar] [CrossRef]
- Dong, L.; Liu, H.; Meng, L.; Xing, M.; Lan, T.; Gu, M.; Zheng, N.; Wang, C.; Chen, H.; Wang, J. Short Communication: Quantitative PCR Coupled with Sodium Dodecyl Sulfate and Propidium Monoazide for Detection of Culturable Escherichia coli in Milk. J. Dairy Sci. 2019, 102, 6914–6919. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, G.D.; Griffin, M.J.; Mauel, M.J.; Lawrence, M.L. Validation of a Quantitative PCR Assay for the Detection of 2 Flavobacterium columnare Genomovars. J. Vet. Diagn. Investig. 2020, 32, 356–362. [Google Scholar] [CrossRef]
- Chaudhary, P.P.; Rulík, M.; Blaser, M. Is the Methanogenic Community Reflecting the Methane Emissions of River Sediments?-Comparison of Two Study Sites. MicrobiologyOpen 2017, 6, e00454. [Google Scholar] [CrossRef]
- Tong, C.; Cadillo-Quiroz, H.; Zeng, Z.H.; She, C.X.; Yang, P.; Huang, J.F. Changes of Community Structure and Abundance of Methanogens in Soils along a Freshwater–Brackish Water Gradient in Subtropical Estuarine Marshes. Geoderma 2017, 299, 101–110. [Google Scholar] [CrossRef]
- Bao, Q.-L.; Xiao, K.-Q.; Chen, Z.; Yao, H.-Y.; Zhu, Y.-G. Methane Production and Methanogenic Archaeal Communities in Two Types of Paddy Soil Amended with Different Amounts of Rice Straw. FEMS Microbiol. Ecol. 2014, 88, 372–385. [Google Scholar] [CrossRef]
- Zeleke, J.; Sheng, Q.; Wang, J.-G.; Huang, M.-Y.; Xia, F.; Wu, J.-H.; Quan, Z.-X. Effects of Spartina Alterniflora Invasion on the Communities of Methanogens and Sulfate-Reducing Bacteria in Estuarine Marsh Sediments. Front. Microbiol. 2013, 4, 243. [Google Scholar] [CrossRef] [Green Version]
- Gao, G.F.; Li, P.F.; Shen, Z.J.; Qin, Y.Y.; Zhang, X.M.; Ghoto, K.; Zhu, X.Y.; Zheng, H.L. Exotic Spartina Alterniflora Invasion Increases CH4 While Reduces CO2 Emissions from Mangrove Wetland Soils in Southeastern China. Sci. Rep. 2018, 8, 9243. [Google Scholar] [CrossRef] [Green Version]
- Wilms, R.; Sass, H.; Köpke, B.; Cypionka, H.; Engelen, B. Methane and Sulfate Profiles within the Subsurface of a Tidal Flat Are Reflected by the Distribution of Sulfate-Reducing Bacteria and Methanogenic Archaea: Sulfate Reducers and Methanogens in Tidal Flats. FEMS Microbiol. Ecol. 2007, 59, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, N.; Wang, W.; Li, B.; Xie, S.; Liu, Y. Vertical Profiles of Sediment Methanogenic Potential and Communities in Two Plateau Freshwater Lakes. Biogeosciences 2017, 14, 341–351. [Google Scholar] [CrossRef] [Green Version]
- Cameron, C.; Hutley, L.B.; Friess, D.A.; Brown, B. Community Structure Dynamics and Carbon Stock Change of Rehabilitated Mangrove Forests in Sulawesi, Indonesia. Ecol. Appl. 2019, 29, e01810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murdiyarso, D.; Purbopuspito, J.; Kauffman, J.B.; Warren, M.W.; Sasmito, S.D.; Donato, D.C.; Manuri, S.; Krisnawati, H.; Taberima, S.; Kurnianto, S. The Potential of Indonesian Mangrove Forests for Global Climate Change Mitigation. Nat. Clim. Chang. 2015, 5, 1089–1092. [Google Scholar] [CrossRef]
- Reshmi, R.R.; Deepa Nair, K.; Zachariah, E.J.; Vincent, S.G.T. Methanogenesis: Seasonal Changes in Human Impacted Regions of Ashtamudi Estuary (Kerala, South India). Estuar. Coast. Shelf Sci. 2015, 156, 144–154. [Google Scholar] [CrossRef]
- Yuan, J.; Ding, W.; Liu, D.; Kang, H.; Xiang, J.; Lin, Y. Shifts in Methanogen Community Structure and Function across a Coastal Marsh Transect: Effects of Exotic Spartina Alterniflora Invasion. Sci. Rep. 2016, 6, 18777. [Google Scholar] [CrossRef] [PubMed]
- Torres-Alvarado, M.d.R.; Fernández, F.J.; Ramírez Vives, F.; Varona-Cordero, F. Dynamics of the Methanogenic Archaea in Tropical Estuarine Sediments. Archaea 2013, 2013, 582646. [Google Scholar] [CrossRef] [Green Version]
- Mohanraju, R.; Natarajan, R. Methanogenic Bacteria in Mangrove Sediments. Hydrobiologia 1992, 247, 187–193. [Google Scholar] [CrossRef]
- Peters, V.; Conrad, R. Methanogenic and Other Strictly Anaerobic Bacteria in Desert Soil and Other Oxic Soils. Appl. Environ. Microbiol. 1995, 61, 1673–1676. [Google Scholar] [CrossRef] [Green Version]
- Otte, J.M.; Blackwell, N.; Soos, V.; Rughöft, S.; Maisch, M.; Kappler, A.; Kleindienst, S.; Schmidt, C. Sterilization Impacts on Marine Sediment—Are We Able to Inactivate Microorganisms in Environmental Samples? FEMS Microbiol. Ecol. 2018, 94, fiy189. [Google Scholar] [CrossRef]
- Setlow, P. Spores of Bacillus Subtilis: Their Resistance to and Killing by Radiation, Heat and Chemicals. J. Appl. Microbiol. 2006, 101, 514–525. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, L.A.; Roussel, E.G.; Weightman, A.J.; Webster, G.; Hubert, C.R.; Bell, E.; Head, I.; Sass, H.; Parkes, R.J. Survival of Desulfotomaculum Spores from Estuarine Sediments after Serial Autoclaving and High-Temperature Exposure. ISME J. 2015, 9, 922–933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, J.F.; Shah, N.N.; Nelson, C.M.; Ludlow, J.M.; Clark, D.S. Pressure and Temperature Effects on Growth and Methane Production of the Extreme Thermophile Methanococcus jannaschii. Appl. Environ. Microbiol. 1988, 54, 3039–3042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takai, K.; Nakamura, K.; Toki, T.; Tsunogai, U.; Miyazaki, M.; Miyazaki, J.; Hirayama, H.; Nakagawa, S.; Nunoura, T.; Horikoshi, K. Cell Proliferation at 122 °C and Isotopically Heavy CH 4 Production by a Hyperthermophilic Methanogen under High-Pressure Cultivation. Proc. Natl. Acad. Sci. USA 2008, 105, 10949–10954. [Google Scholar] [CrossRef] [Green Version]
- Chuang, P.-C.; Young, M.B.; Dale, A.W.; Miller, L.G.; Herrera-Silveira, J.A.; Paytan, A. Methane and Sulfate Dynamics in Sediments from Mangrove-Dominated Tropical Coastal Lagoons, Yucatán, Mexico. Biogeosciences 2016, 13, 2981–3001. [Google Scholar] [CrossRef] [Green Version]
- Xiao, K.-Q.; Beulig, F.; Kjeldsen, K.U.; Jørgensen, B.B.; Risgaard-Petersen, N. Concurrent Methane Production and Oxidation in Surface Sediment from Aarhus Bay, Denmark. Front. Microbiol. 2017, 8, 1198. [Google Scholar] [CrossRef] [Green Version]
- Bebout, B.M.; Hoehler, T.M.; Thamdrup, B.; Albert, D.; Carpenter, S.P.; Hogan, M.; Turk, K.; Des Marais, D.J. Methane Production by Microbial Mats under Low Sulphate Concentrations. Geobiology 2004, 2, 87–96. [Google Scholar] [CrossRef]
- Avrahamov, N.; Antler, G.; Yechieli, Y.; Gavrieli, I.; Joye, S.B.; Saxton, M.; Turchyn, A.V.; Sivan, O. Anaerobic Oxidation of Methane by Sulfate in Hypersaline Groundwater of the Dead Sea Aquifer. Geobiology 2014, 12, 511–528. [Google Scholar] [CrossRef] [Green Version]

| Oligo Name | Micro-Organisms | Sequence | References |
|---|---|---|---|
| mcrA | methanogens | Forward: M13F (5′- TGTAAAACGACGGCCAGTGGTGGTGTMGGATTCACACARTAYGCWACAGC-3′) Reverse: M13R (5′-CA- GGAAACAGCTATGACCTTCATTGCRTAGTTWGGR- TAGTT-3′) | [40] |
| dsrB | SRBs | Forward: DSRp2060F (5′-CAACATCGTYCAYACCCAGGG- 3′) Reverse: DSR4R (5′- GTGTAGCAGTTACCGCA-3′) | [41] |
| pmoA | methanotrophs | Forward: A189 (5′-GGNGACTGGGACTTCTGG-3′) Reverse: A682 (5′-GAASGCNGAGAAGAASGC-3′) | [42] |
| No. | Bacteria | Slope | Regression Model Y = Cq and X = log10 CFU | R2 | References |
|---|---|---|---|---|---|
| 1 | Lactobacillus paracasei FNU | −3.24 | Y = −3.24 X + 43.25 | 0.97 | [44] |
| 2 | Campylobacter spp. | −3.3 | 1.00 | [45] | |
| 3 | Salmonella spp. | −3.4 | 1.00 | [45] | |
| 4 | E. coli | −3.3 | Y = −3.31 X + 34.18 | 0.99 | [46] |
| 5 | Flavobacterium columnare ATCC 49512 | −2.8 | Y = −2.8 X + 41.38 | 0.97 | [47] |
| 6 | Flavobacterium columnare ATCC 94-081 | −2.9 | Y = −2.9 X + 40.51 | 0.99 | [47] |
| 7 | Escherichia coli strain InaCC B5 | −3.5 | Y = −3.52 X + 44.53 | 0.99 | This study |
| Treatments | Volume, Duration, and Concentration | Experiment Length (Days) | Replicates | Number of Sites | Depth Levels | |
|---|---|---|---|---|---|---|
| Controls | No additional substrates | N2 headspace | 37 | 2 | 3 | 5 |
| Autoclaved | N2 headspace | 37 | 2 | 3 | 5 | |
| Treatments (methanogenic substrates) | Methanol | 30 µL, 10 mM | 37 | 2 | 3 | 5 |
| Acetate | 30 µL, 10 mM | 37 | 2 | 3 | 5 | |
| H2 gas | 100%, 1 min | 37 | 2 | 3 | 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ulumuddin, Y.I.; Sugoro, I.; Beavis, S.; Roderick, M.; Eggins, S.; Muarif, M.R. Characterisation of Methane Production Pathways in Sediment of Overwashed Mangrove Forests. Forests 2023, 14, 564. https://doi.org/10.3390/f14030564
Ulumuddin YI, Sugoro I, Beavis S, Roderick M, Eggins S, Muarif MR. Characterisation of Methane Production Pathways in Sediment of Overwashed Mangrove Forests. Forests. 2023; 14(3):564. https://doi.org/10.3390/f14030564
Chicago/Turabian StyleUlumuddin, Yaya Ihya, Irawan Sugoro, Sara Beavis, Michael Roderick, Stephen Eggins, and Muhammad Rizky Muarif. 2023. "Characterisation of Methane Production Pathways in Sediment of Overwashed Mangrove Forests" Forests 14, no. 3: 564. https://doi.org/10.3390/f14030564
APA StyleUlumuddin, Y. I., Sugoro, I., Beavis, S., Roderick, M., Eggins, S., & Muarif, M. R. (2023). Characterisation of Methane Production Pathways in Sediment of Overwashed Mangrove Forests. Forests, 14(3), 564. https://doi.org/10.3390/f14030564










