Estimating Forest Variables for Major Commercial Timber Plantations in Northern Spain Using Sentinel-2 and Ancillary Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Data Collection and Pre-Processing
2.2.1. Field Data
2.2.2. Sentinel-2 Remote Sensing Data
Image Pre-Processing Levels and Spectral Bands
Spectral Indices
Texture Variables
2.2.3. Ancillary Data
Terrain Variables
Climatic Variables
2.3. Data Analysis, Model Fitting and Evaluation
2.3.1. Data Analysis
Analysis in Phase 1
Analysis in Phase 2
- Spectral bands.
- Spectral bands + spectral indices.
- Spectral bands + spectral indices + texture variables.
- Spectral bands + spectral indices + texture variables + terrain variables.
- Spectral bands + spectral indices + texture variables + terrain variables + climatic variables.
2.3.2. Modelling Techniques
2.3.3. Model Assessment and Evaluation
2.4. Deriving Raster Maps
3. Results
3.1. Phase 1: Best Data Configuration and Fitting Technique
3.2. Phase 2: Contribution of Each Group of Predictor Variables and Final Fitting Models
3.2.1. Contribution of Each Group of Predictor Variables
3.2.2. Model Prediction
3.3. Results of Mapping Forest Variables
4. Discussion
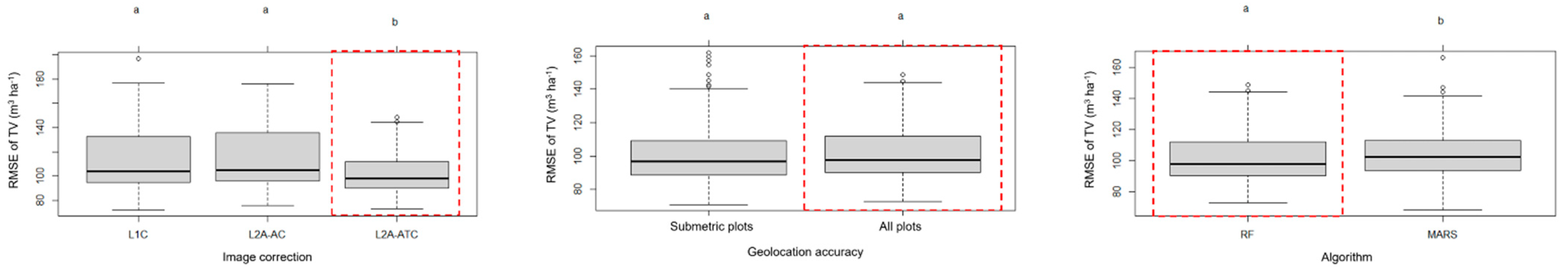
4.1. Impacts of Geolocation Accuracy, Image Correction Level and Fitting Algorithm on Total Volume Estimation
4.2. Model Accuracy and Role of Different Groups of Predictor Variables
4.3. Limitations and Future Developments
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Freer-Smith, P.; Muys, B.; Bozzano, M.; Drössler, L.; Farrelly, N.; Jactel, H.; Korhonen, J.; Minotta, G.; Nijnik, M.; Orazio, C. Plantation Forests in Europe: Challenges and Opportunities, From Science to Policy 9; European Forest Institute: Joensuu, Finland, 2019. [Google Scholar] [CrossRef]
- Dessbesell, L.; Xu, C.; Pulkki, R.; Leitch, M.; Mahmood, N. Forest biomass supply chain optimization for a biorefinery aiming to produce high-value bio-based materials and chemicals from lignin and forestry residues: A review of literature. Can. J. For. Res. 2017, 47, 277–288. [Google Scholar] [CrossRef]
- MITECO. Anuario de Estadística Forestal. Ministerio para la Transición Ecológica y el Reto Demográfico. Gobierno de España. 2022. Available online: https://www.miteco.gob.es/es/biodiversidad/estadisticas/forestal_anuarios_todos.aspx (accessed on 21 December 2023).
- Nilsson, M.; Nordkvist, K.; Jonzén, J.; Lindgren, N.; Axensten, P.; Wallerman, J.; Egberth, M.; Larsson, S.; Nilsson, L.; Eriksson, J.; et al. A nationwide forest attribute map of Sweden predicted using airborne laser scanning data and field data from the National Forest Inventory. Remote Sens. Environ. 2017, 194, 447–454. [Google Scholar] [CrossRef]
- López-Serrano, P.M.; López Sánchez, C.A.; Solís-Moreno, R.; Corral-Rivas, J.J. Geospatial Estimation of above Ground Forest Biomass in the Sierra Madre Occidental in the State of Durango, Mexico. Forests 2016, 7, 70. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Westfall, J.A. Effects of Uncertainty in Model Predictions of Individual Tree Volume on Large Area Volume Estimates. For. Sci. 2014, 60, 34–42. [Google Scholar] [CrossRef]
- Álvarez-González, J.G.; Cañellas, I.; Alberdi, I.; Gadow, K.V.; Ruiz-González, A.D. National Forest Inventory and forest observational studies in Spain: Applications to forest modeling. For. Ecol. Manag. 2014, 316, 54–64. [Google Scholar] [CrossRef]
- Moser, P.; Vibrans, A.C.; McRoberts, R.E.; Næsset, E.; Gobakken, T.; Chirici, G.; Mura, M.; Marchetti, M. Methods for variable selection in LiDAR-assisted forest inventories. For. Int. J. For. Res. 2017, 90, 112–124. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Cohen, W.B.; Næsset, E.; Stehman, S.V.; Tomppo, E.O. Using remotely sensed data to construct and assess forest attribute maps and related spatial products. Scand. J. For. Res. 2010, 25, 340–367. [Google Scholar] [CrossRef]
- Han, H.; Wan, R.; Li, B. Estimating Forest Aboveground Biomass Using Gaofen-1 Images, Sentinel-1 Images, and Machine Learning Algorithms: A Case Study of the Dabie Mountain Region, China. Remote Sens. 2022, 14, 176. [Google Scholar] [CrossRef]
- Yu, J.-W.; Yoon, Y.-W.; Baek, W.-K.; Jung, H.-S. Forest Vertical Structure Mapping Using Two-Seasonal Optic Images and LiDAR DSM Acquired from UAV Platform through Random Forest, XGBoost, and Support Vector Machine Approaches. Remote Sens. 2021, 13, 4282. [Google Scholar] [CrossRef]
- Hirschmugl, M.; Florian, L.; Carina, S. Assessing the Vertical Structure of Forests Using Airborne and Spaceborne LiDAR Data in the Austrian Alps. Remote Sens. 2023, 15, 664. [Google Scholar] [CrossRef]
- Teobaldelli, M.; Cona, F.; Saulino, L.; Migliozzi, A.; D’Urso, G.; Langella, G.; Manna, P.; Saracino, A. Detection of diversity and stand parameters in Mediterranean forests using leaf-off discrete return LiDAR data. Remote Sens. Environ. 2017, 192, 126–138. [Google Scholar] [CrossRef]
- Novo-Fernández, A.; Barrio-Anta, M.; Recondo, C.; Cámara-Obregón, A.; López-Sánchez, C.A. Integration of National Forest Inventory and Nationwide Airborne Laser Scanning Data to Improve Forest Yield Predictions in North-Western Spain. Remote Sens. 2019, 11, 1693. [Google Scholar] [CrossRef]
- CNIG. Spanish National Geographic Information Centre. ALS Data. 2022. Available online: http://centrodedescargas.cnig.es/CentroDescargas/buscadorCatalogo.do? (accessed on 22 March 2023).
- Breidenbach, J.; Waser, L.T.; Debella-Gilo, M.; Schumacher, J.; Rahlf, J.; Hauglin, M.; Puliti, S.; Astrup, R. National mapping and estimation of forest area by dominant tree species using Sentinel-2 data. Can. J. For. Res. 2021, 51, 365–379. [Google Scholar] [CrossRef]
- Chirici, G.; Giannetti, F.; McRoberts, R.E.; Travaglini, D.; Pecchi, M.; Maselli, F.; Chiesi, M.; Corona, P. Wall-to-wall spatial prediction of growing stock volume based on Italian National Forest Inventory plots and remotely sensed data. Int. J. Appl. Earth Obs. Geoinform. 2020, 84, 101959. [Google Scholar] [CrossRef]
- López-Serrano, P.M.; López-Sánchez, C.A.; Álvarez-González, J.G.; García-Gutiérrez, J. A Comparison of Machine Learning Techniques Applied to Landsat-5 TM Spectral Data for Biomass Estimation. Can. J. Remote Sens. 2016, 42, 690–705. [Google Scholar] [CrossRef]
- Jiménez, E.; Vega, J.; Fernández-Alonso, J.; Vega-Nieva, D.; Ortiz, L.; López-Serrano, P.; López-Sánchez, C. Estimation of aboveground forest biomass in Galicia (NW Spain) by the combined use of LiDAR, LANDSAT ETM+ and National Forest Inventory data. iForest 2017, 10, 590–596. [Google Scholar] [CrossRef]
- Korhonen, L.; Hadi; Packalen, P.; Rautiainen, M. Comparison of Sentinel-2 and Landsat 8 in the estimation of boreal forest canopy cover and leaf area index. Remote Sens. Environ. 2017, 195, 259–274. [Google Scholar] [CrossRef]
- Mikeladze, G.; Gavashelishvili, A.; Akobia, I.; Metreveli, V. Estimation of forest cover change using Sentinel-2 multi-spectral imagery in Georgia (the Caucasus). iForest 2020, 13, 329–335. [Google Scholar] [CrossRef]
- Drusch, M.; Del Bello, U.; Carlier, S.; Colin, O.; Fernandez, V.; Gascon, F.; Hoersch, B.; Isola, C.; Laberinti, P.; Martimort, P.; et al. Sentinel-2: ESA’s Optical High-Resolution Mission for GMES Operational Services. Remote Sens. Environ. 2012, 120, 25–36. [Google Scholar] [CrossRef]
- Hu, Y.; Xu, X.; Wu, F.; Sun, Z.; Xia, H.; Meng, Q.; Huang, W.; Zhou, H.; Gao, J.; Li, W.; et al. Estimating Forest Stock Volume in Hunan Province, China, by Integrating In Situ Plot Data, Sentinel-2 Images, and Linear and Machine Learning Regression Models. Remote Sens. 2020, 12, 186. [Google Scholar] [CrossRef]
- Gobakken, T.; Næsset, E. Assessing effects of positioning errors and sample plot size on biophysical stand properties derived from airborne laser scanner data. Can. J. For. Res. 2009, 39, 1036–1052. [Google Scholar] [CrossRef]
- Hogland, J.; Affleck, D.L. Mitigating the Impact of Field and Image Registration Errors through Spatial Aggregation. Remote Sens. 2019, 11, 222. [Google Scholar] [CrossRef]
- López-Sánchez, C.; García-Ramírez, P.; Resl, R.; Hernández-Díaz, J.; López-Serrano, P.; Wehenkel, C. Modelling dasometric attributes of mixed and uneven-aged forests using Landsat-8 OLI spectral data in the Sierra Madre Occidental, Mexico. iForest Biogeosci. For. 2017, 10, 288–295. [Google Scholar] [CrossRef]
- García-Gutiérrez, J.; Martínez-Álvarez, F.; Troncoso, A.; Riquelme, J. A comparison of machine learning regression techniques for LiDAR-derived estimation of forest variables. Neurocomputing 2015, 167, 24–31. [Google Scholar] [CrossRef]
- Lary, D.J.; Alavi, A.H.; Gandomi, A.H.; Walker, A.L. Machine learning in geosciences and remote sensing. Geosci. Front. 2016, 7, 3–10. [Google Scholar] [CrossRef]
- EEA. Biogeographical Regions; European Environment Agency: Copenhagen, Denmark, 2016; Available online: https://www.eea.europa.eu/data-and-maps/data/biogeographical-regions-europe-3 (accessed on 21 December 2023).
- Nicolás, J.L.; Iglesias, S. Normativa de comercialización de los materiales forestales de reproducción. In Producción y Manejo de Semillas y Plantas Forestales. Tomo I. Organismo Autónomo de Parque Nacionales; Pemán, J., Navarro, R.M., Nicolás, J.L., Prada, M.A., Serrada, R., Eds.; Ministerio de Agricultura, Alimentación y Medio Ambiente: Madrid, Spain, 2012; pp. 3–41. [Google Scholar]
- MAPAMA. Spanish National Fourth Inventory Updating. Ministerio de Agricultura, Pesca y Alimentación. Gobierno de España. 2019. Available online: https://www.miteco.gob.es/es/biodiversidad/estadisticas/forestal_anuarios_todos.html/ (accessed on 21 December 2023).
- MAPAMA. Anuario de Estadística. Avance 2018. Ministerio de Agricultura. Pesca y Alimentación. Madrid. 2019. Available online: https://www.mapa.gob.es/estadistica/pags/anuario/2018/anuario/AE18.pdf (accessed on 21 December 2023).
- MARM. Inventario Forestal Nacional; Dirección General del Medio Natural y Política Forestal: Madrid, Spain, 2006. [Google Scholar]
- Fernández-Landa, A.; Navarro, J.; Condés, S.; Algeet-Abarquero, N.; Marchamalo, M. High resolution biomass mapping in tropical forests with LiDAR-derived Digital Models: Poás Volcano National Park (Costa Rica). iForest Biogeosci. For. 2017, 10, 259–266. [Google Scholar] [CrossRef]
- Gonzalez-Ferreiro, E.; Arellano-Pérez, S.; Castedo-Dorado, F.; Hevia, A.; Vega, J.A.; Vega-Nieva, D.J.; Álvarez-González, J.G.; Ruiz-González, A.D. Modelling the vertical distribution of canopy fuel load using national forest inventory and low-density airbone laser scanning data. PLoS ONE 2017, 12, e0176114. [Google Scholar] [CrossRef]
- Alberdi, I.; Cañellas, I.; Bombín, R.V. The Spanish National Forest Inventory: History, development, challenges and perspectives. Pesqui. Florest. Bras. 2017, 37, 361–368. [Google Scholar] [CrossRef]
- Castaño-Santamaría, J.; Barrio-Anta, M.; Álvarez-Álvarez, P. Potential above ground biomass production and total tree carbon sequestration in the major forest species in NW Spain. Int. For. Rev. 2013, 15, 273–289. [Google Scholar] [CrossRef]
- Mueller-Wilm. U. S2 MPC: Sen2Cor Configuration and User Manual. Ref. S2-PDGS-MPC-L2A-SUM-V2.8. 2019. Available online: http://step.esa.int/thirdparties/sen2cor/2.8.0/docs/S2-PDGS-MPC-L2A-SRN-V2.8.pdf (accessed on 16 December 2019).
- Louis, J.; L2A Team. S2 MPC: Level-2A Algorithm Theoretical Basis Document. Ref. S2-PDGS-MPC-ATBD-L2A. 2021. Available online: https://sentinels.copernicus.eu/documents/247904/446933/Sentinel-2-Level-2A-Algorithm-Theoretical-Basis-Document-ATBD.pdf/fe5bacb4-7d4c-9212-8606-6591384390c3?t=1643102691874.pdf (accessed on 29 March 2023).
- Santini, F.; Palombo, A. Impact of Topographic Correction on PRISMA Sentinel 2 and Landsat 8 Images. Remote Sens. 2022, 14, 3903. [Google Scholar] [CrossRef]
- Delegido, J.; Verrelst, J.; Alonso, L.; Moreno, J. Evaluation of Sentinel-2 Red-Edge Bands for Empirical Estimation of Green LAI and Chlorophyll Content. Sensors 2011, 11, 7063–7081. [Google Scholar] [CrossRef] [PubMed]
- Culbert, P.D.; Pidgeon, A.M.; St.-Louis, V.; Bash, D.; Radeloff, V.C. The Impact of Phenological Variation on Texture Measures of Remotely Sensed Imagery. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2009, 2, 299–309. [Google Scholar] [CrossRef]
- Lu, D. Aboveground biomass estimation using Landsat TM data in the Brazilian Amazon. Int. J. Remote Sens. 2005, 26, 2509–2525. [Google Scholar] [CrossRef]
- Zhou, J.; Guo, R.Y.; Sun, M.; Di, T.T.; Wang, S.; Zhai, J.; Zhao, Z. The Effects of GLCM parameters on LAI estimation using texture values from Quickbird Satellite Imagery. Sci. Rep. 2017, 7, 7366. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, H.; Magdon, P.; Kleinn, C.; Flessa, H. Estimating aboveground carbon in a catchment of the Siberian forest tundra: Estimating aboveground carbon in a catchment of the Siberian forest tundra. Remote Sens. Environ. 2009, 113, 518–531. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I.H. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, 6, 610–621. [Google Scholar] [CrossRef]
- Sarker, L.R.; Nichol, J.E. Improved forest biomass estimates using ALOS AVNIR-2 texture indices. Remote Sens. Environ. 2011, 115, 968–977. [Google Scholar] [CrossRef]
- Lu, D.; Chen, Q.; Wang, G.; Liu, L.; Li, G.; Moran, E. A survey of remote sensing-based aboveground biomass estimation methods in forest ecosystems. Int. J. Digit. Earth 2014, 9, 63–105. [Google Scholar] [CrossRef]
- Liu, Y.; Gong, W.; Xing, Y.; Hu, X.; Gong, J. Estimation of the forest stand mean height and aboveground biomass in Northeast China using SAR Sentinel-1B, multispectral Sentinel-2A, and DEM imagery. ISPRS J. Photogramm. Remote Sens. 2019, 151, 277–289. [Google Scholar] [CrossRef]
- ESRI. ArcGIS Desktop: Release 10; Environmental Systems Research Institute: Redlands, CA, USA, 2011. [Google Scholar]
- Ninyerola, M.; Pons, X.; Roure, J.M. Atlas Climático Digital de la Península Ibérica. Metodología y Aplicaciones en Bioclimatología y Geobotánica; Autonomous University of Barcelona: Bellaterra, Spain, 2005; ISBN 932860-8-7. Available online: https://opengis.grumets.cat/wms/iberia/index.htm (accessed on 6 February 2020).
- Næsset, E. Predicting forest stand characteristics with airborne scanning laser using a practical two-stage procedure and field data. Remote Sens. Environ. 2002, 80, 88–99. [Google Scholar] [CrossRef]
- Straub, C.; Dees, M.; Weinacker, H.; Koch, B. Using Airborne Laser Scanner Data and CIR Orthophotos to Estimate the Stem Volume of Forest Stands. Photogramm. Fernerkund. Geoinform. 2009, 2009, 277–287. [Google Scholar] [CrossRef]
- Penner, M.; Pitt, D.G.; Woods, M.E. Parametric vs. nonparametric LiDAR models for operational forest inventory in boreal Ontario. Can. J. Remote Sens. 2013, 39, 426–443. [Google Scholar] [CrossRef]
- Prasad, A.; Iverson, L.; Liaw, A. Newer Classification and Regression Tree Techniques: Bagging and Random Forests for Ecological Prediction. Ecosystems 2006, 9, 181–199. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Latifi, H.; Nothdurft, A.; Koch, B. Non-parametric prediction and mapping of standing timber volume and biomass in a temperate forest: Application of multiple optical/LiDAR-derived predictors. For. Int. J. For. Res. 2010, 83, 395–407. [Google Scholar] [CrossRef]
- Cheng, L.; Chen, X.; De Vos, J.; Lai, X.; Witlox, F. Applying a random forest method approach to model travel mode choice behavior. Travel Behav. Soc. 2019, 14, 1–10. [Google Scholar] [CrossRef]
- Jiang, F.; Kutia, M.; Ma, K.; Chen, S.; Long, J.; Sun, H. Estimating the aboveground biomass of coniferous forest in Northeast China using spectral variables, land surface temperature and soil moisture. Sci. Total Environ. 2021, 785, 147335. [Google Scholar] [CrossRef] [PubMed]
- Immitzer, M.; Vuolo, F.; Atzberger, C. First Experience with Sentinel-2 Data for Crop and Tree Species Classifications in Central Europe. Remote Sens. 2016, 8, 166. [Google Scholar] [CrossRef]
- Gislason, P.O.; Benediktsson, J.A.; Sveinsson, J.R. Random Forests for land cover classification. Pattern Recognit. Lett. 2006, 27, 294–300. [Google Scholar] [CrossRef]
- Friedman, J.H. Multivariate Adaptive Regression Splines. Ann. Stat. 1991, 19, 1–67. Available online: https://www.jstor.org/stable/2241837 (accessed on 21 December 2023). [CrossRef]
- Alonso-Rego, C.; Arellano-Pérez, S.; Guerra-Hernández, J.; Molina-Valero, J.A.; Martínez-Calvo, A.; Pérez-Cruzado, C.; Castedo-Dorado, F.; González-Ferreiro, E.; Álvarez-González, J.G.; Ruiz-González, A.D. Estimating Stand and Fire-Related Surface and Canopy Fuel Variables in Pine Stands Using Low-Density Airborne and Single-Scan Terrestrial Laser Scanning Data. Remote Sens. 2021, 13, 5170. [Google Scholar] [CrossRef]
- Nguyen, H.Q.; Quinn, C.H.; Carrie, R.; Stringer, L.C.; Le, T.V.H.; Hackney, C.R.; Dao, V.T. Comparisons of regression and machine learning methods for estimating mangrove above-ground biomass using multiple remote sensing data in the red River Estuaries of Vietnam. Remote Sens. Appl. Soc. Environ. 2022, 26, 100725. [Google Scholar] [CrossRef]
- Castaño-Santamaría, J.; López-Sánchez, C.A.; Obeso, J.R.; Barrio-Anta, M. Development of a site form equation for predicting and mapping site quality. A case study of unmanaged beech forests in the Cantabrian range (NW Spain). For. Ecol. Manag. 2023, 528, 119512. [Google Scholar] [CrossRef]
- Fassnacht, F.; Hartig, F.; Latifi, H.; Berger, C.; Hernández, J.; Corvalán, P.; Koch, B. Importance of sample size, data type and prediction method for remote sensing-based estimations of aboveground forest biomass. Remote Sens. Environ. 2014, 154, 102–114. [Google Scholar] [CrossRef]
- R Core Team. R: A language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 18 February 2020).
- Fernández-Landa, A.; Fernández-Moya, J.; Tomé, J.L.; Algeet-Abarquero, N.; Guillén-Climent, M.L.; Vallejo, R.; Sandoval, V.; Marchamalo, M. High resolution forest inventory of pure and mixed stands at regional level combining National Forest Inventory field plots, Landsat, and low density lidar. Int. J. Remote Sens. 2018, 39, 4830–4844. [Google Scholar] [CrossRef]
- Frazer, G.; Magnussen, S.; Wulder, M.; Niemann, K. Simulated impact of sample plot size and co-registration error on the accuracy and uncertainty of LiDAR-derived estimates of forest stand biomass. Remote Sens. Environ. 2011, 115, 636–649. [Google Scholar] [CrossRef]
- Saarela, S.; Schnell, S.; Tuominen, S.; Balazs, A.; Hyyppa, J.; Grafstrom, A.; Stahl, G. Effects of positional errors in model-assisted and model-based estimation of growing stock volume. Remote Sens. Environ. 2016, 172, 101–108. [Google Scholar] [CrossRef]
- Arellano-Pérez, S.; Castedo-Dorado, F.; López-Sánchez, C.A.; González-Ferreiro, E.; Yang, Z.; Díaz-Varela, R.A.; Álvarez-González, J.G.; Vega, J.A.; Ruiz-González, A.D. Potential of Sentinel-2A Data to Model Surface and Canopy Fuel Characteristics in Relation to Crown Fire Hazard. Remote Sens. 2018, 10, 1645. [Google Scholar] [CrossRef]
- Dong, Y.; Zhang, L.; Liao, M. Improved topographic mapping in vegetated mountainous areas by high-resolution radargrammetry-assisted sar interferometry. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2020, V-3, 133–139. [Google Scholar] [CrossRef]
- Astola, H.; Häme, T.; Sirro, L.; Molinier, M.; Kilpi, J. Comparison of Sentinel-2 and Landsat 8 imagery for forest variable prediction in boreal region. Remote Sens. Environ. 2019, 223, 257–273. [Google Scholar] [CrossRef]
- Rahimzadeh-Bajgiran, P.; Hennigar, C.; Weiskittel, A.; Lamb, S. Forest Potential Productivity Mapping by Linking Remote-Sensing-Derived Metrics to Site Variables. Remote Sens. 2020, 12, 2056. [Google Scholar] [CrossRef]
- dos Reis, A.A.; Carvalho, M.C.; de Mello, J.M.; Gomide, L.R.; Filho, A.C.F.; Junior, F.W.A. Spatial prediction of basal area and volume in Eucalyptus stands using Landsat TM data: An assessment of prediction methods. N. Z. J. For. Sci. 2018, 48, 1. [Google Scholar] [CrossRef]
- Gadow, K.v.; Álvarez-González, J.G.; Zhang, C.; Pukkala, T.; Zhao, X. Sustaining Forest Ecosystems; Springer Nature: Cham, Switzerland, 2021; 419p. [Google Scholar] [CrossRef]
- Jiang, F.; Deng, M.; Tang, J.; Fu, L.; Sun, H. Integrating spaceborne LiDAR and Sentinel-2 images to estimate forest aboveground biomass in Northern China. Carbon Balance Manag. 2022, 17, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Lu, D.; Wang, G.; Wu, C.; Huang, Y.; Yu, S. Examining Spectral Reflectance Saturation in Landsat Imagery and Corresponding Solutions to Improve Forest Aboveground Biomass Estimation. Remote Sens. 2016, 8, 469. [Google Scholar] [CrossRef]
- Frampton, W.J.; Dash, J.; Watmough, G.; Milton, E.J. Evaluating the capabilities of Sentinel-2 for quantitative estimation of biophysical variables in vegetation. ISPRS J. Photogramm. Remote Sens. 2013, 82, 83–92. [Google Scholar] [CrossRef]
- Yu, T.; Pang, Y.; Liang, X.; Jia, W.; Bai, Y.; Fan, Y.; Chen, D.; Liu, X.; Deng, G.; Li, C.; et al. China’s larch stock volume estimation using Sentinel-2 and LiDAR data. Geo-Spat. Inf. Sci. 2022, 26, 392–405. [Google Scholar] [CrossRef]
- Kelsey, K.C.; Neff, J.C. Estimates of Aboveground Biomass from Texture Analysis of Landsat Imagery. Remote Sens. 2014, 6, 6407–6422. [Google Scholar] [CrossRef]
- Dube, T.; Mutanga, O.; Elfatih, M.; Abdel-Rahman, E.M.; Ismail, R.; Slotow, R. Predicting Eucalyptus spp. stand volume in Zululand, South Africa: An analysis using a stochastic gradient boosting regression ensemble with multi-source data sets. Int. J. Remote Sens. 2015, 36, 3751–3772. [Google Scholar] [CrossRef]
- Nguyen, T.T.H.; Chau, T.N.Q.; Nguyen, D.D.; Cao, T.H.; Phan, T.H.; Ho, D.B.; Ngo, T.S.; Le, Q.D.; Pham, T.A. Estimating tropical forest stand volume using Sentinel-2A imagery. In Proceedings of the 2021 Second International Conference on Intelligent Data Science Technologies and Applications (IDSTA), Tartu, Estonia, 15–16 November 2021; pp. 130–137. [Google Scholar] [CrossRef]
- Main-Knorn, M.; Moisen, G.G.; Healey, S.P.; Keeton, W.S.; Freeman, E.A.; Hostert, P. Evaluating the Remote Sensing and Inventory-Based Estimation of Biomass in the Western Carpathians. Remote Sens. 2011, 3, 1427–1446. [Google Scholar] [CrossRef]
- Canavesi, V.; Ponzoni, F.P.; Valeriano, M. Estimativa de volume de madeira em plantios de Eucalyptus spp. utilizando dados hiperespectrais e dados topográficos. Rev. Árvore 2010, 34, 539–549. [Google Scholar] [CrossRef]
- Justice, C.O.; Vermote, E.; Townshend, J.R.G.; Defries, R.; Roy, D.P.; Hall, D.K.; Salomonson, V.V.; Privette, J.L.; Riggs, G.; Strahler, A.; et al. The Moderate Resolution Imaging Spectroradiometer (MODIS): Land remote sensing for global change research. IEEE Trans. Geosci. Remote Sens. 1998, 36, 1228–1249. [Google Scholar] [CrossRef]
- Reis, A.A. Predicting Eucalyptus Stand Attributes in Minas Gerais State, Brazil. Ph.D. Thesis, Universidade Federal de Lavras, Lavras, Brazil, 2018; 188p. Available online: http://repositorio.ufla.br/bitstream/1/32173/2/TESE_Predicting%20Eucalyptus%20stand%20attributes%20in%20Minas%20Gerais%20State%2C%20Brazil%20an%20approach%20using%20machine%20learning%20algorithms%20with%20multisource%20datasets.pdf (accessed on 21 December 2023).
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Mohammadpour, P.; Viegas, D.X.; Viegas, C. Vegetation Mapping with Random Forest Using Sentinel 2 and GLCM Texture Feature—A Case Study for Lousã Region, Portugal. Remote Sens. 2022, 14, 4585. [Google Scholar] [CrossRef]
- DeVries, B.; Pratihast, A.K.; Verbesselt, J.; Kooistra, L.; Herold, M. Characterizing Forest Change Using Community-Based Monitoring Data and Landsat Time Series. PLoS ONE 2016, 11, e0147121. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Ren, C.; Zhang, B.; Wang, Z. Multi-Sensor Prediction of Stand Volume by a Hybrid Model of Support Vector Machine for Regression Kriging. Forests 2020, 11, 296. [Google Scholar] [CrossRef]
- Nichol, J.E.; Sarker, M.L.R. Efficiency of texture measurement from two optical sensors for improved biomass estimation. In Proceedings of the ISPRS TC VII Symposium—100 Years ISPRS, Vienna, Austria, 5–7 July 2010; IAPRS, Volume XXXVIII, Part 7B. Available online: https://www.isprs.org/proceedings/XXXVIII/part7/b/pdf/407_XXXVIII-part7B.pdf (accessed on 21 December 2023).
- Mauya, E.W.; Madundo, S. Modelling and Mapping Above Ground Biomass Using Sentinel 2 and Planet Scope Remotely Sensed Data in West Usambara Tropical Rainforests, Tanzania. Research Square. 2021. Available online: https://www.researchsquare.com/article/rs-942337/v1 (accessed on 21 December 2023).
- Aboveground biomass estimation using multi-sensor data synergy and machine learning algorithms in a dense tropical forest. Appl. Geogr. 2018, 96, 29–40. [CrossRef]
- Vashum, K.T.; Jayakumar, S. Methods to Estimate Above-Ground Biomass and Carbon Stock in Natural Forests—A Review. J. Ecosyst. Ecography 2012, 2, 1–7. [Google Scholar] [CrossRef]
- Barrio-Anta, M.; Castedo-Dorado, F.; Cámara-Obregón, A.; López-Sánchez, C.A. Predicting current and future suitable habitat and productivity for Atlantic populations of maritime pine (Pinus pinaster Aiton) in Spain. Ann. For. Sci. 2020, 77, 41. [Google Scholar] [CrossRef]
- López-Serrano, P.M.; López-Sánchez, C.A.; Díaz-Varela, R.A.; Corral-Rivas, J.J.; Solis-Moreno, R.; Vargas-Larreta, B.; Alvarez-Gonzalez, J.G. Estimating biomass of mixed and uneven-aged forests using spectral data and a hybrid model combining regression trees and linear models. iForest 2015, 9, 226–234. [Google Scholar] [CrossRef]
- Alberdi, I.; Sandoval, V.; Condés, S.; Cañellas, I.; Vallejo, R. El inventario forestal nacional español, una herramienta para el conocimiento, la gestión y la conservación de los ecosistemas forestales arbolados. Ecosistemas 2016, 25, 88–97. [Google Scholar] [CrossRef]
- Hastie, T.; Friedman, J.; Tibshirani, R. The Elements of Statistical Learning; Springer Series in Statistics; Springer: New York, NY, USA, 2001; ISBN 978-1-4899-0519-2. [Google Scholar] [CrossRef]
- Lever, J.; Krzywinski, M.; Altman, N. Points of Significance: Model Selection and Overfitting. Nat. Methods 2016, 13, 703–704. [Google Scholar] [CrossRef]





| Species | No. Plots | Forest Variable | Descriptive Statistic | |||
|---|---|---|---|---|---|---|
| Mean | Min. | Max. | Std. | |||
| E. globulus | 589 | N (stems ha−1) | 833.83 | 10.19 | 2695.02 | 499.93 |
| G (m2 ha−1) | 18.30 | 0.44 | 52.25 | 0.44 | ||
| H0 (m) | 21.43 | 6.70 | 43.55 | 7.26 | ||
| TV (m3 ha−1) | 148.42 | 0.68 | 522.67 | 118.14 | ||
| AGB (Mg ha−1) | 99.44 | 0.98 | 371.55 | 81.68 | ||
| P. pinaster | 474 | N (stems ha−1) | 574.60 | 10.19 | 3176.03 | 439.15 |
| G (m2 ha−1) | 22.60 | 0.42 | 55.73 | 13.70 | ||
| H0 (m) | 16.67 | 3.40 | 31.78 | 6.34 | ||
| TV (m3 ha−1) | 164.05 | 0.88 | 460.72 | 119.37 | ||
| AGB (Mg ha−1) | 92.26 | 0.80 | 298.64 | 68.25 | ||
| P. radiata | 408 | N (stems ha−1) | 453.66 | 25.46 | 1773.48 | 294.07 |
| G (m2 ha−1) | 27.82 | 0.67 | 66.62 | 13.54 | ||
| H0 (m) | 22.55 | 5.70 | 39.55 | 6.28 | ||
| TV (m3 ha−1) | 246.23 | 2.25 | 699.31 | 147.64 | ||
| AGB (Mg ha−1) | 127.43 | 1.59 | 356.93 | 75.38 | ||
| Satellite/Granule | Acquisition Date | Solar Zenith (°) | Solar Azimuth (°) |
|---|---|---|---|
| S2A/29TMH | 11 August 2018 | 30.86 | 148.82 |
| S2A/29TNG | 19 June 2018 | 22.94 | 138.83 |
| S2A/29TNH | 11 August 2018 | 30.42 | 150.94 |
| S2A/29TNJ | 11 August 2018 | 31.22 | 151.58 |
| S2A/29TPG | 19 June 2018 | 22.36 | 141.25 |
| S2B/29TPH | 14 June 2018 | 23.10 | 143.23 |
| S2B/29TPJ | 24 June 2018 | 23.95 | 143.16 |
| S2B/29TQH | 24 June 2018 | 22.67 | 144.43 |
| S2B/29TQJ | 24 June 2018 | 23.41 | 145.61 |
| S2A/30TUN | 5 August 2018 | 29.46 | 146.64 |
| S2A/30TUP | 5 August 2018 | 30.25 | 147.34 |
| S2A/30TVN | 5 August 2018 | 29.00 | 148.81 |
| S2A/30TVP | 5 August 2018 | 29.80 | 149.51 |
| S2B/30TWN | 27 August 2018 | 35.52 | 153.22 |
| S2B/30TWP | 27 August 2018 | 36.34 | 153.70 |
| Band | Symbol | Spectral Region | Wavelength (µm) | Spatial Resolution (m) |
|---|---|---|---|---|
| Band 2 | B2 | Blue | 0.46–0.52 | 10 |
| Band 3 | B3 | Green | 0.54–0.58 | 10 |
| Band 4 | B4 | Red | 0.65–0.68 | 10 |
| Band 5 | B5 | Red-Edge-1 (RE1) | 0.70–0.71 | 20 |
| Band 6 | B6 | Red-Edge-2 (RE2) | 0.73–0.75 | 20 |
| Band 7 | B7 | Red-Edge-3 (RE3) | 0.76–0.78 | 20 |
| Band 8 | B8 | Near-Infrared (NIR) | 0.78–0.90 | 10 |
| Band 8A | B8A | Narrow NIR (nNIR) | 0.85–0.87 | 20 |
| Band 11 | B11 | Shortwave infrared (SWIR-1) | 1.56–1.65 | 20 |
| Band 12 | B12 | Shortwave infrared (SWIR-2) | 2.10–2.28 | 20 |
| Group | Variable Name |
|---|---|
| Spectral bands | Band 2—Blue (B2), Band 3—Green (B3), Band 4—Red (B4), Band 5—Vegetation Red-Edge-1 (B5), Band 6—Vegetation Red-Edge-2 (B6), Band 7—Vegetation Red-Edge-3 (B7), Band 8—NIR (B8), Band 8A—Narrow NIR (B8A), Band 11—SWIR-1 (B11), Band 12—SWIR-2 (B12). |
| Spectral indices | Anthocyanin Reflectance Index (ARI), Chlorophyll Red-Edge (CRE), Enhanced Vegetation Index (EVI), Enhanced Vegetation Index 2 (EVI2), Green Normalized Difference Vegetation Index (GNDVI), Modified Anthocyanin Reflectance Index (MARI), Modified Chlorophyll Absorption in Reflectance Index (MCARI), Modified Soil Adjusted Vegetation Index (MSAVI), Modified Soil Adjusted Vegetation Index (MSI), Normalized Burn Ratio (NBR), Normalized Burn Ratio 2 (NBR2), Normalized Difference Moisture Index (NDMI), Normalized Difference Vegetation Index (NDVI), Pigment-Specific Simple Ratio (PSSR), Soil Adjusted Vegetation Index (SAVI), Tasseled Cap Angle (TCA), Tasseled Cap Brightness (TCB), Tasseled Cap Greenness (TCG), Tasseled Cap Wetness (TCW). |
| Texture | Angular Second Moment (SEC), Contrast (CON), Correlation (COR), Dissimilarity (DIS), Energy (ENE), Entropy (ENT), Homogeneity (HOM), Max (MAX), Mean (MEN), Standard Deviation (STD). |
| Terrain | Aspect (ASP), Aspect/Slope Ratio (ASR), Curvature (CU), Elevation (ELV), Heat Load Index (HLI), Plan Curvature (PLC), Profile curvature (PFC), Slope (SLP), Terrain Shape Index (TSI), Wetness Index (WI). |
| Climatic | Average Temperature (TM), Maximum Temperature (TMAX), Minimum Temperature (TMIN), Precipitation (PT), Radiation (RA). |
| Species | E. globulus | P. pinaster | P. radiata | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Image Correction | L1C | L2A-AC | L2A-ATC | L1C | L2A-AC | L2A-ATC | L1C | L2A-AC | L2A-ATC |
| Total plots | 589 | 589 | 589 | 474 | 474 | 474 | 408 | 408 | 408 |
| Outliers | 13 + 32 | 13 + 32 | 13 + 32 | 36 + 27 | 36 + 26 | 36 + 23 | 4 + 20 | 4 + 20 | 4 + 23 |
| % Outliers | 7.64 | 7.64 | 7.64 | 13.29 | 13.08 | 12.44 | 5.88 | 5.88 | 6.61 |
| Species | Image Correction | Geolocation Accuracy | No. Plot | MARS | RF | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| R2 | Bias | RMSE | RMSE% | R2 | Bias | RMSE | RMSE% | ||||
| E. globulus | L1C | All plots | 544 | 0.36 | −0.40 | 96.98 | 64.18% | 0.35 | −1.35 | 97.66 | 64.63% |
| Sub-meter plots | 457 | 0.34 | −0.99 | 100.86 | 66.20% | 0.34 | −1.04 | 100.70 | 66.09% | ||
| L2A-AC | All plots | 544 | 0.33 | 0.72 | 98.43 | 65.61% | 0.29 | −0.91 | 100.73 | 67.15% | |
| Sub-meter plots | 458 | 0.31 | −0.07 | 102.37 | 67.91% | 0.29 | −0.75 | 103.50 | 68.66% | ||
| L2A-ATC | All plots | 544 | 0.37 | 0.19 | 94.53 | 63.69% | 0.42 | −0.89 | 90.76 | 61.15% | |
| Sub-meter plots | 457 | 0.36 | −0.32 | 97.49 | 65.45% | 0.43 | −0.59 | 91.11 | 61.17% | ||
| P. pinaster | L1C | All plots | 411 | 0.37 | −0.44 | 95.89 | 58.04% | 0.33 | −1.05 | 98.22 | 59.45% |
| Sub-meter plots | 351 | 0.38 | −1.23 | 97.04 | 60.01% | 0.37 | 1.10 | 97.09 | 60.03% | ||
| L2A-AC | All plots | 412 | 0.38 | 0.32 | 95.48 | 57.94% | 0.36 | −2.13 | 96.79 | 58.74% | |
| Sub-meter plots | 353 | 0.39 | 0.40 | 96.24 | 59.55% | 0.40 | −2.28 | 94.84 | 58.69% | ||
| L2A-ATC | All plots | 415 | 0.32 | −0.42 | 99.72 | 60.79% | 0.38 | −1.18 | 94.97 | 57.89% | |
| Sub-meter plots | 354 | 0.37 | 0.22 | 98.04 | 60.78% | 0.40 | −1.29 | 94.55 | 58.62% | ||
| P. radiata | L1C | All plots | 384 | 0.24 | 0.76 | 132.90 | 52.96% | 0.12 | −1.10 | 142.91 | 56.95% |
| Sub-meter plots | 172 | 0.24 | −0.72 | 125.08 | 57.87% | 0.09 | −3.80 | 138.99 | 64.31% | ||
| L2A-AC | All plots | 384 | 0.27 | 0.21 | 132.02 | 52.61% | 0.14 | −2.60 | 145.22 | 57.87% | |
| Sub-meter plots | 171 | 0.29 | 0.98 | 120.15 | 55.59% | 0.11 | −1.89 | 134.58 | 62.26% | ||
| L2A-ATC | All plots | 381 | 0.36 | 0.54 | 119.82 | 48.66% | 0.36 | 0.37 | 118.45 | 48.10% | |
| Sub-meter plots | 172 | 0.29 | 1.72 | 115.69 | 54.08% | 0.26 | −0.73 | 116.10 | 54.27% | ||
| Species | E. globulus | P. pinaster | P. radiata | |
|---|---|---|---|---|
| Image correction | L1C | 64.63% | 59.45% | 56.95% |
| L2A-AC | 67.15% (−2.51%) | 58.74% (+0.72%) | 57.87 (−0.92%) | |
| L2A-ATC | 61.15% (+3.49%) | 57.89% (+1.56%) | 48.10 (+8.50%) | |
| Plot variable | Average slope (%) | 28.06 | 23.76 | 35.93 |
| Average aspect (°) | 179.14 | 179.21 | 179.60 | |
| % Plots with slope > 20% | 67.91 | 52.95 | 75.74 |
| Type | Dependent Variable | Statistic | Eucalyptus globulus | Pinus pinaster | Pinus radiata | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group of Predictor Variables | Group of Predictor Variables | Group of Predictor Variables | |||||||||||||||
| (1) | (2) | (3) | (4) | (5) | (1) | (2) | (3) | (4) | (5) | (1) | (2) | (3) | (4) | (5) | |||
| Density | Number of stems, N (stems ha−1) | R2 | 0.24 | +4.17% | +8.33% | +8.33% | +8.33% | 0.15 | +6.67% | +26.67% | +40.00% | +53.33% | 0.1 | +20.00% | +60.00% | +80.00% | +50.00% |
| Bias | 4.64 | +34.91% | +65.52% | +17.24% | +62.72% | −4.91 | +61.30% | +102.85% | +92.26% | +178.00% | −6.02 | −9.30% | +20.43% | +13.79% | +33.06% | ||
| RMSE | 438.49 | -0.88% | −1.34% | −1.48% | −1.20% | 412.04 | −2.70% | −4.18% | −5.30% | −6.63% | 283.12 | −1.60% | −3.78% | −5.09% | −3.87% | ||
| Basal área, G (m2 ha−1) | R2 | 0.40 | +10.00% | +12.50% | +15.00% | +15.00% | 0.41 | 0.00% | +12.20% | +12.20% | +12.20% | 0.33 | 0.00% | +9.09% | +18.18% | +18.18% | |
| Bias | −0.05 | −20.00% | +20.00% | +40.00% | +80.00% | −0.08 | −25.00% | −50.00% | −50.00% | +25.00% | −0.01 | −300.00% | −500.00% | +100.00% | −500.00% | ||
| RMSE | 9.50 | −3.26% | −3.68% | −4.63% | −4.42% | 10.59 | +0.38% | −4.53% | −4.25% | −4.72% | 11.18 | 0.00% | −2.15% | −5.10% | −5.10% | ||
| Size | Dominant height, H0 (m) | R2 | 0.26 | +3.85% | +11.54% | +23.08% | +26.92% | 0.26 | +7.69% | +7.69% | +34.62% | +42.31% | 0.28 | +14.29% | +21.43% | +32.14% | +28.57% |
| Bias | 0.04 | −75.00% | −100.00% | −200.00% | −150.00% | −0.04 | +50.00% | +50.00% | +100.00% | +25.00% | −0.01 | +300.00% | −300.00% | −400.00% | −500.00% | ||
| RMSE | 6.31 | −0.63% | −2.06% | −4.28% | −4.75% | 5.49 | −1.28% | −1.28% | −6.38% | −7.47% | 5.36 | −2.61% | −4.66% | −6.16% | −5.97% | ||
| Yield | Total volume with bark, TV (m3 ha−1) | R2 | 0.41 | +7.32% | +9.76% | +12.20% | +12.20% | 0.38 | +2.63% | +7.89% | +10.53% | +18.42% | 0.37 | +2.70% | +8.11% | +18.92% | +16.22% |
| Bias | −0.62 | +24.19% | +122.58% | +35.48% | +35.48% | −1.15 | −45.22% | +20.00% | +37.39% | −11.30% | 0.11 | +254.55% | +618.18% | −27.27% | +218.18% | ||
| RMSE | 91.21 | −3.03% | −3.66% | −4.14% | −4.14% | 94.53 | −1.03% | −2.84% | −3.09% | −5.11% | 117.94 | −1.09% | −2.42% | −5.27% | −4.58% | ||
| Aboveground Biomass, AGB (Mg ha−1)(Mg/ha) | R2 | 0.41 | +4.88% | +2.44% | +4.88% | +4.88% | 0.36 | +2.78% | +8.33% | +11.11% | +13.89% | 0.35 | +5.71% | +8.57% | +20.00% | +20.00% | |
| Bias | −0.79 | +16.46% | +40.51% | +31.65% | +20.25% | −1.03 | +2.91% | −27.18% | −35.92% | −35.92% | −0.41 | −82.93% | −168.29% | −119.51% | −48.78% | ||
| RMSE | 63.13 | −2.47% | −1.39% | −2.14% | −2.08% | 54.88 | −0.80% | −2.53% | −3.37% | −3.81% | 61.15 | −1.77% | −2.29% | −5.10% | −4.73% | ||
| Eucalyptus globulus | Pinus pinaster | Pinus radiata | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | G | H0 | TV | AGB | Avg. | N | G | H0 | TV | AGB | Avg. | N | G | H0 | TV | AGB | Avg. | |||
| Independent variables | Group | (1) | 0.24 (2) | 0.47 (4) | 0.18 (2) | 0.26 (2) | 0.48 (6) | 0.33 | 0.29 (2) | 0.53 (3) | 0.25 (3) | 0.51 (4) | 0.43 (3) | 0.40 | 0.41 (5) | 0.35 (4) | 0.32 (4) | 0.44 (4) | 0.39 (3) | 0.38 |
| (2) | 0.55 (6) | 0.36 (5) | 0.47 (9) | 0.56 (6) | 0.52 (4) | 0.50 | 0.34 (5) | 0.20 (2) | 0.23 (5) | 0.13 (2) | 0.23 (3) | 0.23 | 0.28 (3) | 0.31 (3) | 0.29 (4) | 0.24 (3) | 0.22 (3) | 0.26 | ||
| (3) | 0.07 (1) | 0.11 (2) | 0.07 (2) | 0.06 (1) | - | 0.06 | 0.09 (3) | 0.27 (3) | 0.15 (3) | 0.14 (2) | 0.16 (2) | 0.16 | 0.14 (1) | 0.16 (3) | 0.16 (3) | 0.10 (2) | 0.13 (4) | 0.13 | ||
| (4) | 0.14 (2) | 0.06 (1) | 0.17 (4) | 0.13 (2) | - | 0.10 | 0.20 (3) | - | 0.23 (3) | 0.07 (1) | 0.08 (1) | 0.12 | 0.17 (2) | 0.17 (3) | 0.23 (4) | 0.22 (4) | 0.27 (5) | 0.21 | ||
| (5) | - | - | 0.11 (3) | - | - | 0.02 | 0.08 (1) | - | 0.15 (2) | 0.16 (2) | 0.10 (1) | 0.10 | - | - | - | - | - | 0.00 | ||
| No. of variables | 11 | 12 | 20 | 11 | 10 | 14 | 8 | 16 | 11 | 10 | 11 | 13 | 15 | 13 | 15 | |||||
| Goodness-of-fit statistics | R2 | 0.26 | 0.46 | 0.33 | 0.46 | 0.43 | 0.23 | 0.46 | 0.37 | 0.45 | 0.41 | 0.18 | 0.39 | 0.37 | 0.44 | 0.42 | ||||
| Bias | −5.44 | −0.07 | −0.02 | −0.84 | −0.92 | −13.65 | −0.10 | −0.05 | −1.02 | −0.66 | −6.85 | −0.02 | 0.03 | 0.08 | 0.08 | |||||
| Bias% | −0.007 | −0.004 | −0.001 | −0.006 | −0.009 | −0.024 | −0.005 | −0.003 | −0.006 | −0.007 | −0.015 | −0.001 | 0.001 | 0.000 | 0.001 | |||||
| RMSE | 432.63 | 9.06 | 6.01 | 87.43 | 61.57 | 384.72 | 10.01 | 5.08 | 89.7 | 52.79 | 268.7 | 10.61 | 5.03 | 111.73 | 58.03 | |||||
| RMSE% | 51.8 | 49.5 | 28.0 | 58.9 | 61.9 | 67.0% | 44.6 | 30.5 | 54.7 | 57.2 | 59.2 | 38.1 | 22.3 | 45.4 | 45.5 | |||||
| Type | Indep. Variable | Eucalyptus globulus | Pinus pinaster | Pinus radiata | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | G | H0 | TV | AGB | Sum. | N | G | H0 | TV | AGB | Sum. | N | G | H0 | TV | AGB | Sum. | ||
| Spectral bands | B2 | - | - | - | - | 0.06 | 0.06 | - | - | - | - | - | - | 0.09 | 0.06 | - | - | - | 0.15 |
| B3 | - | - | - | - | - | - | - | - | - | - | - | - | 0.07 | 0.09 | - | 0.10 | 0.16 | 0.42 | |
| B4 | - | - | - | - | - | - | - | - | 0.11 | 0.08 | - | 0.19 | 0.07 | 0.08 | 0.06 | 0.10 | 0.09 | 0.40 | |
| B5 | 0.11 | - | 0.08 | - | 0.10 | 0.29 | - | - | 0.08 | 0.09 | - | 0.17 | - | - | 0.12 | - | - | 0.12 | |
| B6 | 0.13 | 0.11 | - | - | 0.06 | 0.30 | - | 0.07 | - | - | 0.04 | 0.11 | - | - | 0.08 | - | - | 0.08 | |
| B7 | - | 0.05 | - | 0.06 | 0.04 | 0.15 | - | - | - | - | - | - | - | - | - | - | - | - | |
| B8 | - | - | - | - | - | - | - | 0.07 | - | - | - | 0.07 | - | 0.12 | - | 0.10 | 0.14 | 0.36 | |
| B8A | - | 0.06 | - | - | 0.04 | 0.10 | - | - | - | - | 0.04 | 0.04 | - | - | 0.06 | - | - | 0.06 | |
| B11 | - | 0.26 | 0.10 | 0.20 | 0.19 | 0.75 | 0.20 | 0.39 | 0.06 | 0.19 | 0.34 | 1.18 | 0.11 | - | - | 0.14 | - | 0.25 | |
| B12 | - | - | - | - | - | - | 0.08 | - | - | 0.14 | - | 0.22 | 0.07 | - | - | - | - | 0.07 | |
| Spectral indices | ARI | 0.10 | 0.10 | 0.06 | 0.11 | 0.08 | 0.45 | - | - | - | - | - | - | 0.15 | - | - | - | - | 0.15 |
| CRE | - | - | - | - | - | - | - | - | - | - | - | - | 0.06 | - | - | - | - | 0.06 | |
| EVI | - | 0.08 | 0.05 | 0.09 | 0.08 | 0.30 | 0.06 | 0.10 | - | - | - | 0.16 | - | 0.08 | 0.10 | 0.11 | 0.13 | 0.42 | |
| EVI2 | - | - | - | - | 0.04 | 0.04 | - | - | - | - | - | - | - | - | - | - | - | - | |
| GNDVI | - | 0.06 | - | - | - | 0.06 | 0.08 | - | 0.04 | 0.06 | - | 0.18 | - | 0.06 | 0.05 | 0.05 | 0.05 | 0.21 | |
| MARI | - | - | - | - | 0.05 | 0.05 | - | 0.10 | 0.04 | 0.07 | 0.10 | 0.31 | - | - | - | - | - | - | |
| MCARI | - | - | 0.05 | 0.07 | 0.06 | 0.18 | - | - | - | - | - | - | - | - | - | - | - | - | |
| MSAVI | - | 0.07 | - | 0.06 | - | 0.13 | - | - | - | - | 0.05 | 0.05 | - | - | - | - | 0.04 | 0.04 | |
| MSI | 0.08 | - | - | - | - | 0.08 | 0.05 | - | - | - | - | 0.05 | - | - | 0.04 | - | - | 0.04 | |
| NBR | - | - | - | - | - | 0.07 | - | - | 0.05 | - | - | 0.05 | - | - | - | - | - | - | |
| NBR2 | - | - | 0.03 | - | - | 0.03 | 0.10 | - | 0.04 | - | - | 0.14 | 0.06 | - | - | - | - | 0.06 | |
| NDMI | - | - | 0.08 | - | - | 0.08 | 0.05 | - | 0.06 | - | - | 0.11 | - | - | - | - | - | - | |
| NDVI | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| PSSR | 0.08 | - | 0.03 | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | |
| SAVI | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| TCA | 0.07 | 0.06 | 0.04 | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | |
| TCB | 0.14 | - | 0.07 | - | - | 0.21 | - | - | - | - | 0.08 | 0.08 | - | 0.17 | - | - | - | 0.17 | |
| TCG | - | - | 0.06 | 0.06 | 0.05 | 0.17 | - | - | - | - | - | - | - | - | - | 0.07 | - | 0.07 | |
| TCW | - | - | 0.09 | 0.17 | 0.16 | 0.42 | - | - | - | - | - | - | - | - | 0.09 | - | - | 0.09 | |
| Texture | SEC | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.03 | 0.03 |
| CON | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.05 | - | 0.04 | 0.09 | |
| COR | - | 0.05 | - | - | - | 0.05 | - | - | - | - | - | - | 0.14 | 0.06 | - | - | - | 0.20 | |
| DIS | - | - | - | - | - | - | 0.03 | 0.08 | - | - | - | 0.11 | - | - | 0.05 | 0.06 | 0.03 | 0.14 | |
| ENE | - | - | - | - | - | - | 0.03 | - | - | - | - | 0.03 | - | 0.05 | - | 0.04 | - | 0.09 | |
| ENT | - | - | - | - | - | - | 0.03 | - | 0.04 | - | - | 0.07 | - | - | - | - | - | - | |
| HOM | 0.07 | - | - | - | - | 0.07 | - | - | 0.04 | - | - | 0.04 | - | 0.05 | - | - | 0.02 | 0.07 | |
| MAX | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| MEN | - | 0.06 | 0.04 | 0.06 | - | 0.16 | - | 0.10 | 0.06 | 0.07 | 0.09 | 0.32 | - | - | - | - | - | - | |
| SDT | - | - | 0.04 | - | - | 0.04 | - | 0.09 | - | 0.07 | 0.08 | 0.24 | - | - | 0.05 | - | - | 0.05 | |
| Terrain | ASP | - | 0.06 | 0.04 | 0.06 | - | 0.16 | - | - | - | - | - | - | - | 0.07 | 0.05 | 0.06 | 0.06 | 0.24 |
| ASR | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |||||
| CU | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.06 | - | - | 0.06 | |
| ELV | - | - | 0.04 | - | - | 0.04 | - | - | 0.14 | 0.07 | - | 0.21 | - | - | - | 0.05 | 0.06 | 0.11 | |
| HLI | 0.07 | - | - | 0.07 | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | |
| PLC | 0.07 | - | - | - | - | 0.07 | 0.06 | - | - | - | 0.08 | 0.14 | - | 0.05 | - | - | - | 0.05 | |
| PFC | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.04 | 0.04 | |
| SLP | - | - | 0.04 | - | - | 0.04 | 0.06 | - | 0.04 | - | - | 0.10 | 0.08 | - | 0.07 | 0.05 | 0.05 | 0.25 | |
| TSI | - | - | 0.04 | - | - | 0.04 | - | - | - | - | - | - | - | - | 0.05 | - | - | 0.05 | |
| WI | - | - | - | - | - | - | 0.08 | - | 0.05 | - | - | 0.13 | 0.09 | 0.06 | - | 0.06 | 0.06 | 0.27 | |
| Climatic | TM | - | - | 0.04 | - | - | 0.04 | - | - | - | 0.09 | - | 0.09 | - | - | - | - | - | - |
| TMAX | - | - | 0.04 | - | - | 0.04 | 0.08 | - | 0.06 | 0.07 | 0.10 | 0.31 | - | - | - | - | - | - | |
| TMIN | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| PT | - | - | 0.03 | - | - | 0.03 | - | - | 0.09 | - | - | 0.09 | - | - | - | - | - | - | |
| RA | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| Region | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Galicia | Asturias | Cantabria | Basque Country | ||||||
| Avg. (Sd) | Total | Avg. (Sd) | Total | Avg. (Sd) | Total | Avg. (Sd) | Total | ||
| E. globulus | N | 816.42 (120.11) | 108,162,865.38 | 782.00 (131.34) | 30,174,276.15 | 790.58 (129.26) | 26,771,979.95 | 868.86 (180.21) | 8,625,895.65 |
| G | 18.21 (3.95) | 2,412,527.76 | 15.79 (3.34) | 609,186.45 | 16.89 (4.07) | 572,048.59 | 20.12 (6.37) | 199,773.22 | |
| H0 | 21.66 (2.03) | 2,870,200.23 | 20.17 (1.49) | 778,171.45 | 21.25 (1.88) | 719,627.29 | 22.23 (2.81) | 220,672.33 | |
| TV | 152.15 (38.17) | 20,157,482.01 | 126.57 (29.66) | 4,883,988.79 | 141.38 (38.21) | 4,787,726.89 | 170.94 (61.01) | 1,697,084.24 | |
| AGB | 103.48 (26.00) | 13,709,146.49 | 86.45 (19.72) | 3,335,870.38 | 95.80 (25.94) | 3,244,040.50 | 117.41 (41.94) | 1,165,589.52 | |
| P. pinaster | N | 663.05 (129.72) | 111,420.57 | 605.84 (92.03) | 8,788,913,94 | 809.37 (271.92) | 173,420.57 | 794.72 (185.78) | 4,177,647.49 |
| G | 21.19 (4.86) | 3,569,806.40 | 22.90 (4.05) | 322,160.32 | 25.96 (9.04) | 5,561.59 | 27.65 (5.65) | 145,354.34 | |
| H0 | 15.88 (2.54) | 2,675,987.48 | 15.95 (1.62) | 231,398.86 | 16.19 (2.66) | 3,468.13 | 17.69 (1.57) | 92,982.27 | |
| TV | 156.43 (41.86) | 26,353,790.70 | 164.17 (32.25) | 2,381,631.85 | 197.49 (78.15) | 42,316.14 | 208.83 (43.58) | 1,097,773.53 | |
| AGB | 87.27 (24.04) | 14,701,927.30 | 113.07 (26.48) | 1,640,230.48 | 66.97 (16.88) | 14,349.93 | 103.61 (26.08) | 544,669.58 | |
| P. radiata | N | 513.70 (87.71) | 30,609,531.69 | 521.11 (76.32) | 9,831,803.62 | 466.87 (76.92) | 3,032,877.78 | 476.96 (64.09) | 54,692,959.82 |
| G | 26.05 (4.10) | 1,552,099.38 | 26.99 (4.23) | 509,313.68 | 26.32 (4.93) | 171,007.17 | 27.80 (5.46) | 3,187,960.25 | |
| H0 | 20.52 (1.47) | 1,222,504.17 | 20.44 (1.83) | 385,672.55 | 21.70 (2.17) | 140,998.67 | 23.24 (2.18) | 2,664,458.16 | |
| TV | 215.59 (38.68) | 12,846,469.77 | 222.08 (41.58) | 4,189,903.77 | 230.08 (50.87) | 1,494,652.27 | 259.30 (61.23) | 29,733,439.08 | |
| AGB | 114.94 (20.58) | 6,848,897.27 | 116.21 (21.72) | 2,192,591.69 | 114.32 (27.88) | 742,616.38 | 132.73 (30.24) | 15,219,668.09 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Novo-Fernández, A.; López-Sánchez, C.A.; Cámara-Obregón, A.; Barrio-Anta, M.; Teijido-Murias, I. Estimating Forest Variables for Major Commercial Timber Plantations in Northern Spain Using Sentinel-2 and Ancillary Data. Forests 2024, 15, 99. https://doi.org/10.3390/f15010099
Novo-Fernández A, López-Sánchez CA, Cámara-Obregón A, Barrio-Anta M, Teijido-Murias I. Estimating Forest Variables for Major Commercial Timber Plantations in Northern Spain Using Sentinel-2 and Ancillary Data. Forests. 2024; 15(1):99. https://doi.org/10.3390/f15010099
Chicago/Turabian StyleNovo-Fernández, Alís, Carlos A. López-Sánchez, Asunción Cámara-Obregón, Marcos Barrio-Anta, and Iyán Teijido-Murias. 2024. "Estimating Forest Variables for Major Commercial Timber Plantations in Northern Spain Using Sentinel-2 and Ancillary Data" Forests 15, no. 1: 99. https://doi.org/10.3390/f15010099
APA StyleNovo-Fernández, A., López-Sánchez, C. A., Cámara-Obregón, A., Barrio-Anta, M., & Teijido-Murias, I. (2024). Estimating Forest Variables for Major Commercial Timber Plantations in Northern Spain Using Sentinel-2 and Ancillary Data. Forests, 15(1), 99. https://doi.org/10.3390/f15010099








