Climate Change Responses of High-Elevation Polylepis Forests
Abstract
:1. Introduction
2. Materials and Methods
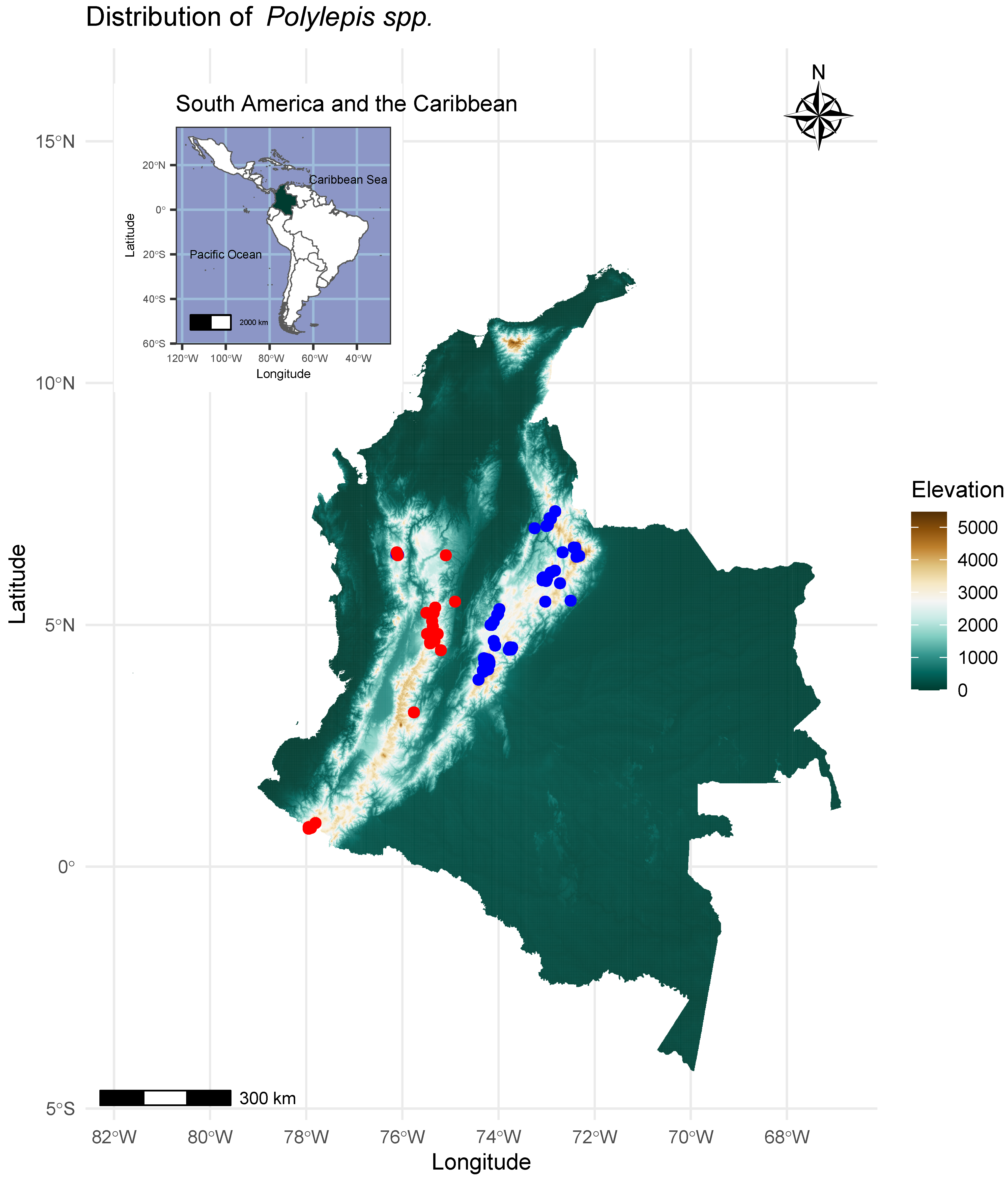
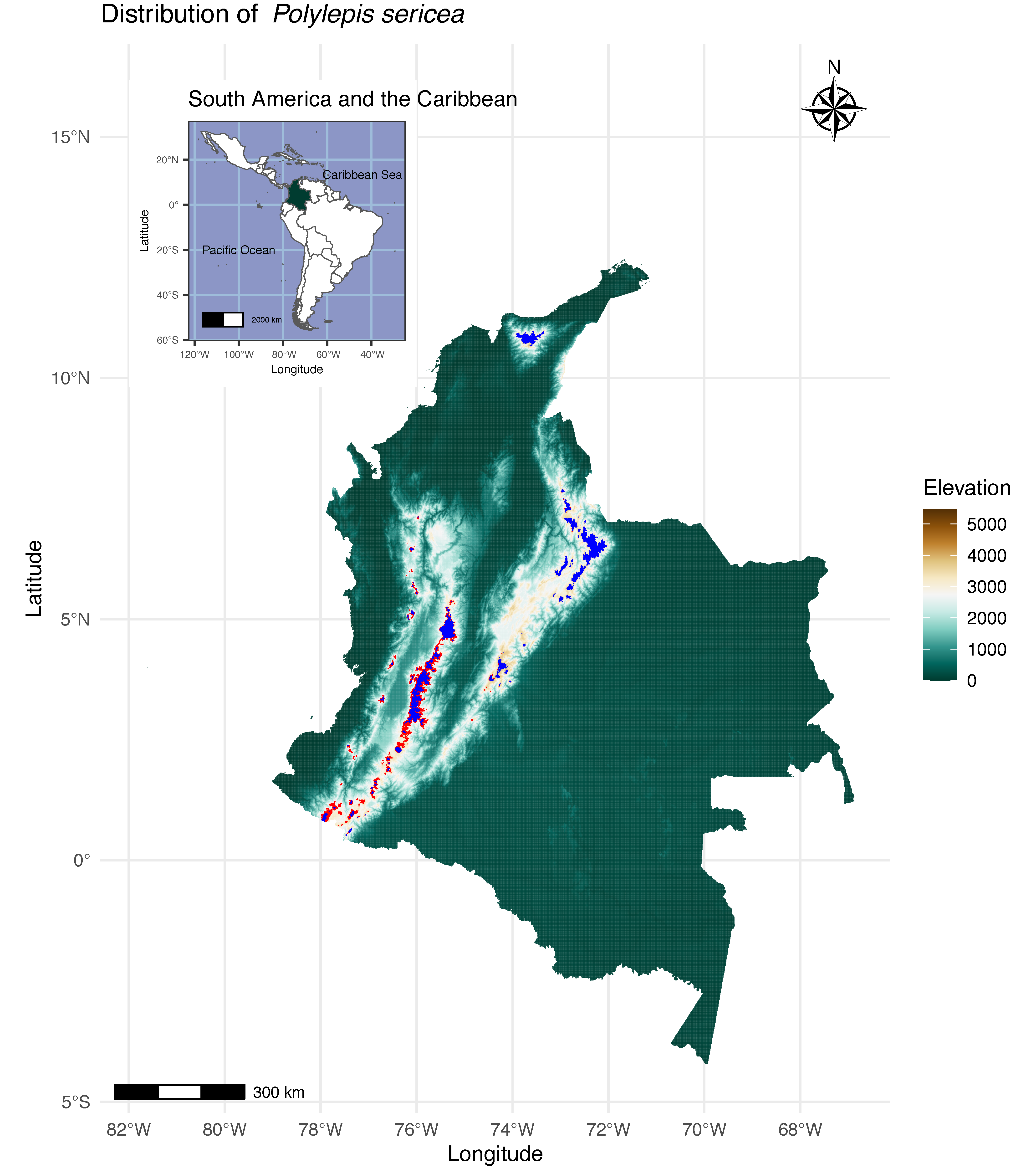
2.1. Study Area and Data Compilation
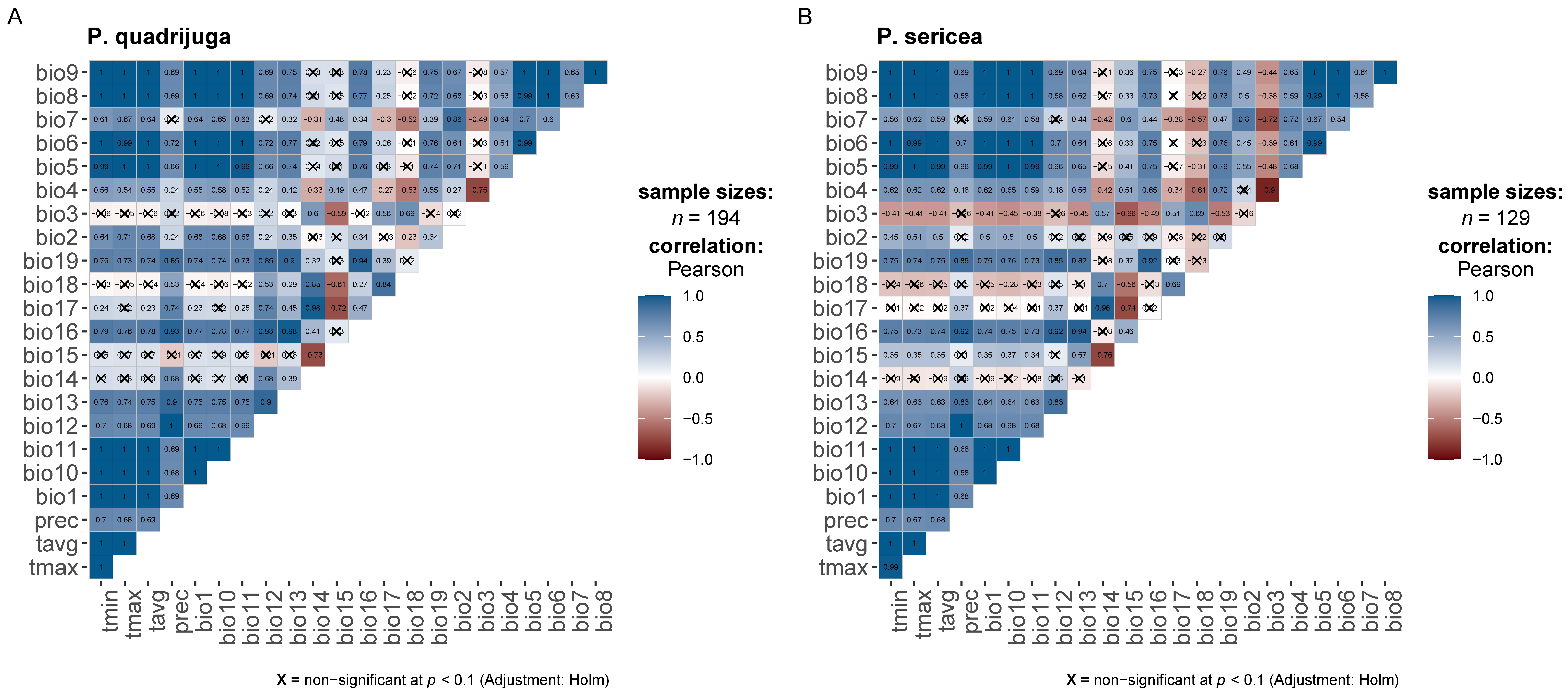
2.2. Prioritization of Bioclimatic Variables
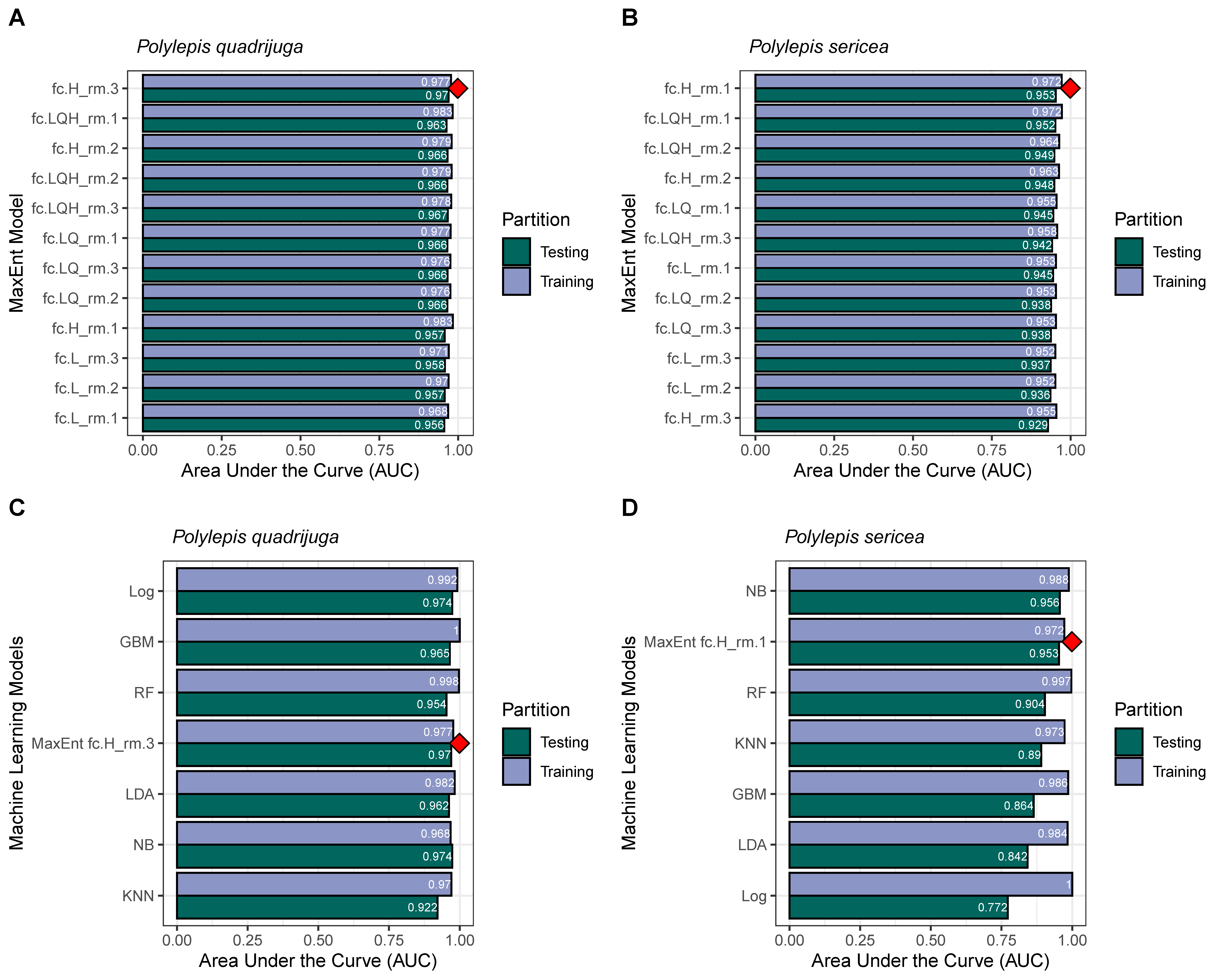
2.3. Potential Niche Distribution Modeling
3. Results
3.1. Prioritization of Bioclimatic Variables
3.2. Model Accuracy
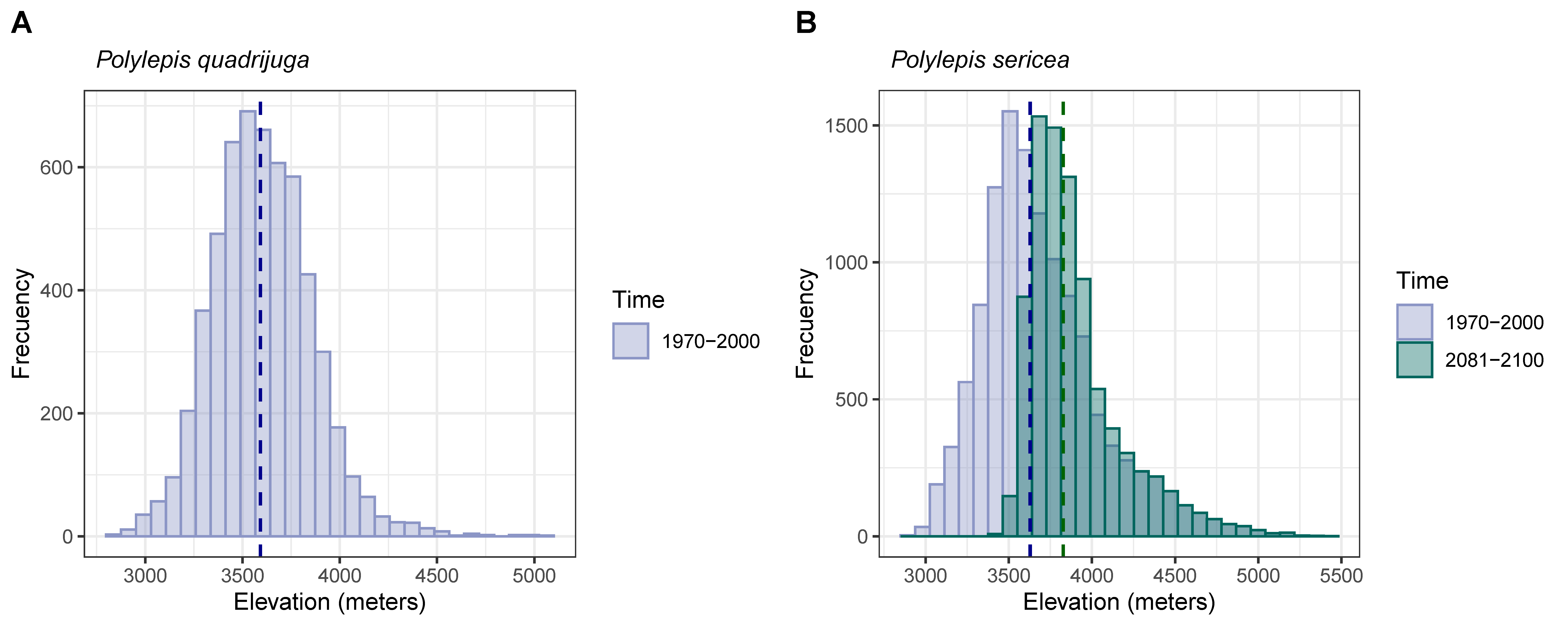
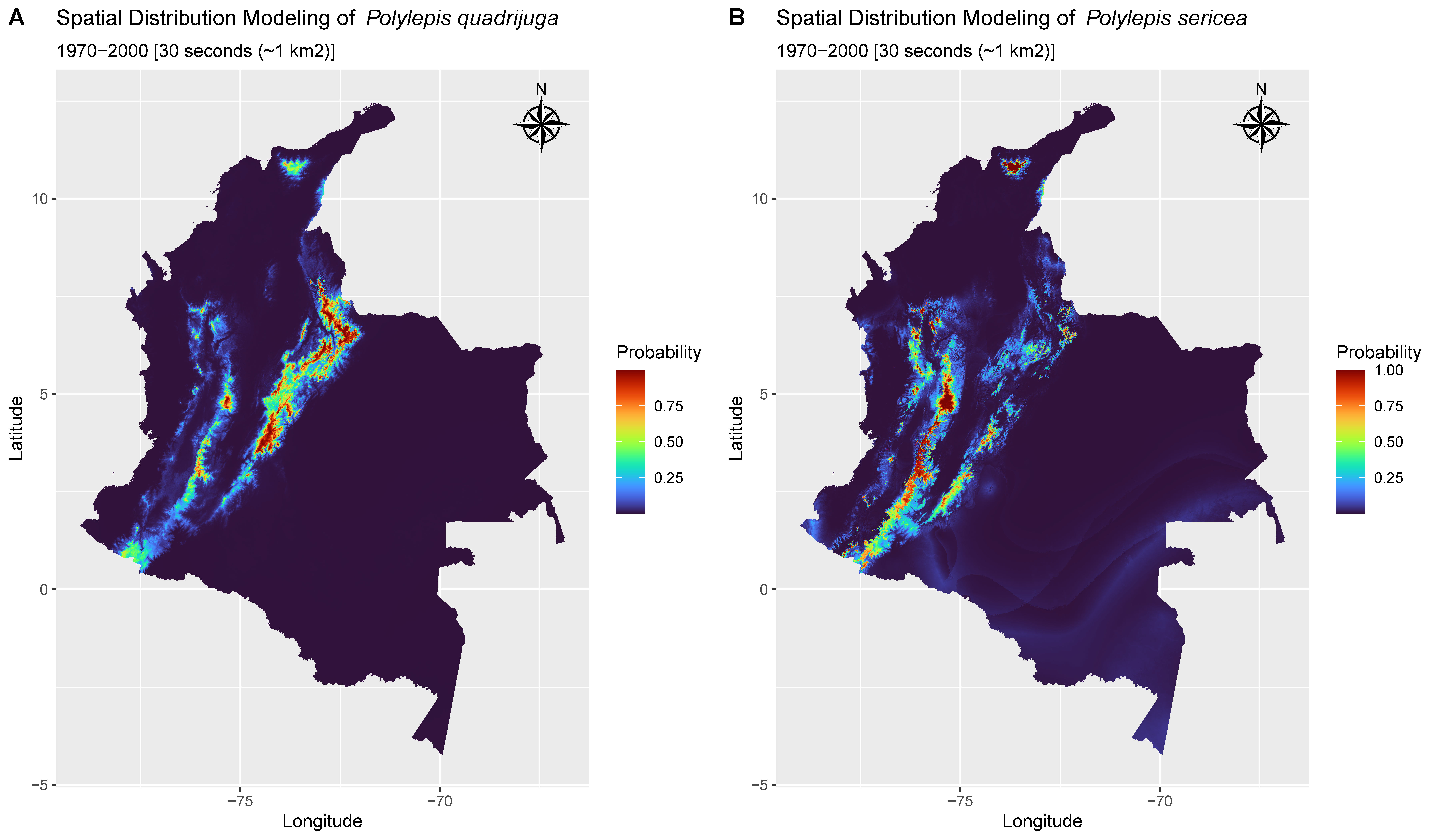
3.3. Presence Probability
4. Discussion
4.1. Range Losses Are Expected Mostly for P. quadrijuga Forests
4.2. Alternative Responses of Polylepis Forests to Climate Change
4.3. Perspectives
5. Conclusions
- Polylepis quadrijuga depicts a restricted ability to maintain its current distribution in the face of short-term climate change, with a limited potential for range shifts.
- On the other hand, P. sericea forests could persist in their current ranges despite moderate vulnerability in the trailing edge.
- Contrasting species responses via range shifts to climate change may be due to higher niche specialization of P. quadrijuga, a strategy likely favored by divergent selection in the environmentally and topographically complex Eastern Cordillera, while a generalist response of P. sericea is expected from a species that faced less heterogenous landscape as the one found in the Central Cordillera of the northern Andes.
- The current analytical pipeline is applicable to other montane forest tree species in the tropics, and would benefit by the inclusion of additional variables concerning dispersal dynamics, germination probabilities, explicit eco-physiological indices, as well as high-resolution environmental, edaphic, and biotic masks.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cortés, A.J.; Restrepo-Montoya, M.; Bedoya-Canas, L.E. Modern Strategies to Assess and Breed Forest Tree Adaptation to Changing Climate. Front. Plant Sci. 2020, 11, 583323. [Google Scholar] [CrossRef] [PubMed]
- Mina, M.; Bugmann, H.; Cordonnier, T.; Irauschek, F.; Klopcic, M.; Pardos, M.; Cailleret, M.; Brando, P. Future ecosystem services from European mountain forests under climate change. J. Appl. Ecol. 2016, 54, 389–401. [Google Scholar] [CrossRef]
- Lenoir, J.; Gegout, J.C.; Marquet, P.A.; de Ruffray, P.; Brisse, H. A significant upward shift in plant species optimum elevation during the 20th century. Science 2008, 320, 1768–1771. [Google Scholar] [CrossRef] [PubMed]
- Steinbauer, M.J.; Grytnes, J.A.; Jurasinski, G.; Kulonen, A.; Lenoir, J.; Pauli, H.; Rixen, C.; Winkler, M.; Bardy-Durchhalter, M.; Barni, E.; et al. Accelerated increase in plant species richness on mountain summits is linked to warming. Nature 2018, 556, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Lenoir, J.; Gégout, J.-C.; Guisan, A.; Vittoz, P.; Wohlgemuth, T.; Zimmermann, N.E.; Dullinger, S.; Pauli, H.; Willner, W.; Svenning, J.-C. Going against the flow: Potential mechanisms for unexpected downslope range shifts in a warming climate. Ecography 2010, 33, 295–303. [Google Scholar] [CrossRef]
- Jump, A.S.; Penuelas, J. Running to stand still: Adaptation and the response of plants to rapid climate change. Ecol. Lett. 2005, 8, 1010–1020. [Google Scholar] [CrossRef] [PubMed]
- Nicotra, A.B.; Segal, D.L.; Hoyle, G.L.; Schrey, A.W.; Verhoeven, K.J.; Richards, C.L. Adaptive plasticity and epigenetic variation in response to warming in an Alpine plant. Ecol. Evol. 2015, 5, 634–647. [Google Scholar] [CrossRef] [PubMed]
- Nicotra, A.B.; Atkin, O.K.; Bonser, S.P.; Davidson, A.M.; Finnegan, E.J.; Mathesius, U.; Poot, P.; Purugganan, M.D.; Richards, C.L.; Valladares, F.; et al. Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 2010, 15, 684–692. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.A.; Sgro, C.M. Climate change and evolutionary adaptation. Nature 2011, 470, 479–485. [Google Scholar] [CrossRef]
- Berry, P.M.; Dawson, T.P.; Harrison, P.A.; Pearson, R.; Butt, N. The sensitivity and vulnerability of terrestrial habitats and species in Britain and Ireland to climate change. J. Nat. Conserv. 2003, 11, 15–23. [Google Scholar] [CrossRef]
- McVicar, T.R.; Körner, C. On the use of elevation, altitude, and height in the ecological and climatological literature. Oecologia 2013, 171, 335–337. [Google Scholar] [CrossRef] [PubMed]
- Madriñán, S.; Cortés, A.J.; Richardson, J.E. Páramo is the world’s fastest evolving and coolest biodiversity hotspot. Front. Genet. 2013, 4, 192. [Google Scholar] [CrossRef] [PubMed]
- Pouchon, C.; Fernández, A.; Nassar, J.M.; Boyer, F.d.r.; Aubert, S.; Lavergne, S.b.; Mavárez, J.S. Phylogenomic Analysis of the Explosive Adaptive Radiation of the Espeletia complex (Asteraceae) in the Tropical Andes. Syst. Biol. 2018, 67, 1041–1060. [Google Scholar] [CrossRef]
- Valencia, J.B.; Mesa, J.; León, J.G.; Madriñán, S.; Cortés, A.J. Climate Vulnerability Assessment of the Espeletia Complex on Páramo Sky Islands in the Northern Andes. Front. Ecol. Evol. 2020, 8, 565708. [Google Scholar] [CrossRef]
- Peyre, G.; Lenoir, J.; Karger, D.N.; Gomez, M.; Gonzalez, A.; Broennimann, O.; Guisan, A.; Jiménez-Alfaro, B. The fate of páramo plant assemblages in the sky islands of the northern Andes. J. Veg. Sci. 2020, 31, 967–980. [Google Scholar] [CrossRef]
- Mavárez, J.; Bézy, S.; Goeury, T.; Fernández, A.; Aubert, S. Current and future distributions of Espeletiinae (Asteraceae) in the Venezuelan Andes based on statistical downscaling of climatic variables and niche modelling. Plant Ecol. Divers. 2018, 12, 633–647. [Google Scholar] [CrossRef]
- Anthelme, F.; Carrasquer, I.; Ceballos, J.L.; Peyre, G. Novel plant communities after glacial retreat in Colombia: (many) losses and (few) gains. Alp. Bot. 2022, 132, 211–222. [Google Scholar] [CrossRef]
- Luteyn, J.L. Páramos: A Checklist of Plant Diversity, Geographic Distribution and Botanical Literature; The New York Botanical Garden Press: New York, NY, USA, 1999. [Google Scholar]
- Hughes, C.E.; Atchison, G.W. The ubiquity of alpine plant radiations: From the Andes to the Hengduan Mountains. New Phytol. 2015, 207, 275–282. [Google Scholar] [CrossRef]
- Cortés, A.J.; Garzón, L.N.; Valencia, J.B.; Madriñán, S. On the Causes of Rapid Diversification in the Páramos: Isolation by Ecology and Genomic Divergence in Espeletia. Front. Plant Sci. 2018, 9, 408949. [Google Scholar] [CrossRef]
- Sklenář, P.; Kučerová, A.; Macková, J.; Romoleroux, K. Temperature Microclimates of Plants in a Tropical Alpine Environment: How Much does Growth Form Matter? Arct. Antarct. Alp. Res. 2016, 48, 61–78. [Google Scholar] [CrossRef]
- Sklenář, P.; Kučerová, A.; Macek, P.; Macková, J. The frost-resistance mechanism in páramo plants is related to geographic origin. N. Z. J. Bot. 2012, 50, 391–400. [Google Scholar] [CrossRef]
- Sklenář, P.; KucErová, A.; Macek, P.; Macková, J. Does plant height determine the freezing resistance in the páramo plants? Austral Ecol. 2010, 35, 929–934. [Google Scholar] [CrossRef]
- Monasterio, M.; Sarmiento, L. Adaptive radiation of Espeletia in the cold andean tropics. Trends Ecol. Evol. 1991, 6, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Flantua, S.G.A.; Payne, D.; Borregaard, M.K.; Beierkuhnlein, C.; Steinbauer, M.J.; Dullinger, S.; Essl, F.; Irl, S.D.H.; Kienle, D.; Kreft, H.; et al. Snapshot isolation and isolation history challenge the analogy between mountains and islands used to understand endemism. Glob. Ecol. Biogeogr. 2020, 29, 1651–1673. [Google Scholar] [CrossRef]
- Sklenář, P.; Hedberg, I.; Cleef, A.M. Island biogeography of tropical alpine floras. J. Biogeogr. 2014, 41, 287–297. [Google Scholar] [CrossRef]
- Vargas, O.M.; Ortiz, E.M.; Simpson, B.B. Conflicting phylogenomic signals reveal a pattern of reticulate evolution in a recent high-Andean diversification (Asteraceae: Astereae: Diplostephium). New Phytol. 2017, 214, 1736–1750. [Google Scholar] [CrossRef] [PubMed]
- Flantua, S.G.A.; O’Dea, A.; Onstein, R.E.; Giraldo, C.; Hooghiemstra, H. The flickering connectivity system of the north Andean páramos. J. Biogeogr. 2019, 46, 1808–1825. [Google Scholar] [CrossRef]
- Papadopoulou, A.; Knowles, L.L. Genomic tests of the species-pump hypothesis: Recent island connectivity cycles drive population divergence but not speciation in Caribbean crickets across the Virgin Islands. Evolution 2015, 69, 1501–1517. [Google Scholar] [CrossRef] [PubMed]
- Vargas, O.M.; Madriñán, S.; Simpson, B.B. Allopatric speciation is more prevalent than parapatric ecological divergence in tropical montane system. PeerJ 2019, 11, e15479. [Google Scholar] [CrossRef]
- Muellner-Riehl, A.N. Mountains as Evolutionary Arenas: Patterns, Emerging Approaches, Paradigm Shifts, and Their Implications for Plant Phylogeographic Research in the Tibeto-Himalayan Region. Front. Plant Sci. 2019, 10, 195. [Google Scholar] [CrossRef]
- Ronikier, M. Biogeography of high-mountain plants in the Carpathians: An emerging phylogeographical perspective. Taxon. 2011, 6, 373–389. [Google Scholar] [CrossRef]
- Hazzi, N.A.; Moreno, J.S.; Ortiz-Movliav, C.; Palacio, R.D. Biogeographic regions and events of isolation and diversification of the endemic biota of the tropical Andes. Proc. Natl. Acad. Sci. USA 2018, 115, 7985–7990. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.H.; Delzon, S.; Urli, M.; Samalens, J.-C.; Lamy, J.-B.; Lischke, H.; Sin, F.; Zimmermann, N.E.; Porté, A.J. Field Evidence of Colonisation by Holm Oak, at the Northern Margin of Its Distribution Range, during the Anthropocene Period. PLoS ONE 2013, 8, e80443. [Google Scholar] [CrossRef] [PubMed]
- Castillo, D.D.; Carrasco, J.C.; Quevedo, L.A.; Ricaurte, C.B.; Gavilanes, A.V.; Borz, S.A. Diversity, composition and structure of Andean high forest in Ecuador, South America. Bull. Transilv. Univ. Bras.-Ser. II For. 2017, 10, 1–16. [Google Scholar]
- Rangel-Churio, O.; Arellano-Peña, H. Bosques de Polylepis: Un tipo de vegetación condenado a la extinción. In Colombia Diversidad Biotica X: Cambio Global (Natural) y Climático (Antrópico) en el Páramo Colombiano; Rangel-Churio, O., Ed.; Instituto de Ciencias Naturales: Bogotá Colombia, 2010; pp. 443–478. [Google Scholar]
- Segoia-Salcedo, M.C.; Domic, A.; Boza, T.E.; Kessler, M. Taxonomic review of the genus Polylepis: Implications for ecological studies, conservation and restoration. Ecol. Austral 2018, 18, 1. [Google Scholar]
- Caballero-Villalobos, L.; Fajardo-Gutiérrez, F.; Calbi, M.; Silva-Arias, G.A. Climate Change Can Drive a Significant Loss of Suitable Habitat for Polylepis quadrijuga, a Treeline Species in the Sky Islands of the Northern Andes. Front. Ecol. Evol. 2021, 9, 661550. [Google Scholar] [CrossRef]
- Gradstein, S.R.; León-Yánez, S. Liverwort diversity in Polylepis pauta forests of Ecuador under different climatic conditions. Neotrop. Biodivers. 2020, 6, 138–146. [Google Scholar] [CrossRef]
- Zutta, B.R.; Rundel, P.W. Modeled shifts in Polylepis species ranges in the Andes from the last glacial maximum to the present. Forests 2017, 8, 232. [Google Scholar] [CrossRef]
- Peyre, G.; Balslev, H.; Font, X. Phytoregionalisation of the Andean paramo. PeerJ 2018, 6, e4786. [Google Scholar] [CrossRef]
- Escudero, A.; Giménez-Benavides, L.; Iriondo, J.M.; Rubio, A. Patch Dynamics and Islands of Fertility in a High Mountain Mediterranean Community. Arct. Antarct. Alp. Res. 2004, 36, 518–527. [Google Scholar] [CrossRef]
- Scherrer, D.; Körner, C. Topogaphically controlled thermal-habitat differentiation buffers alpine plant diversity against climate warming. J. Biogeogr. 2011, 38, 406–416. [Google Scholar] [CrossRef]
- Körner, C. Alpine Plant Life: Functional Plant Ecology of High Mountain Ecosystems; Springer: Berlin/Heidelberg, Germany, 2003. [Google Scholar]
- Rumpf, S.B.; Hulber, K.; Klonner, G.; Moser, D.; Schutz, M.; Wessely, J.; Willner, W.; Zimmermann, N.E.; Dullinger, S. Range dynamics of mountain plants decrease with elevation. Proc. Natl. Acad. Sci. USA 2018, 115, 1848–1853. [Google Scholar] [CrossRef] [PubMed]
- Enquist, B.; Condit, R.; Peet, R.; Schildhauer, M.; Thiers, B. Cyberinfrastructure for an integrated botanical information network to investigate the ecological impacts of global climate change on plant biodiversity. PeerJ Prepr. 2016, 4, e2615v2. [Google Scholar] [CrossRef]
- Wisz, M.S.; Hijmans, R.J.; Li, J.; Peterson, A.T.; Graham, C.H.; Guisan, A. Effects of sample size on the performance of species distribution models. Divers. Distrib. 2008, 14, 763–773. [Google Scholar] [CrossRef]
- Fois, M.; Cuena-Lombraña, A.; Fenu, G.; Bacchetta, G. Using species distribution models at local scale to guide the search of poorly known species: Review, methodological issues and future directions. Ecol. Model. 2018, 385, 124–132. [Google Scholar] [CrossRef]
- van Proosdij, A.S.J.; Sosef, M.S.M.; Wieringa, J.J.; Raes, N. Minimum required number of specimen records to develop accurate species distribution models. Ecography 2016, 39, 542–552. [Google Scholar] [CrossRef]
- Phillips, S.; Anderson, R.; Schapire, R. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Müller, W.A.; Jungclaus, J.H.; Mauritsen, T.; Baehr, J.; Bittner, M.; Budich, R.; Bunzel, F.; Esch, M.; Ghosh, R.; Haak, H.; et al. A Higher-resolution Version of the Max Planck Institute Earth System Model (MPI-ESM1.2-HR). J. Adv. Model. Earth Syst. 2018, 10, 1383–1413. [Google Scholar] [CrossRef]
- Xin, X. Performance of BCC-CSM2-MR in simulating summer climate changes in East Asia. Geophys. Res. Abstr. 2019, 21, 4711. [Google Scholar]
- Arias, P.A.; Ortega, G.; Villegas, L.D.; Martínez, J.A. Colombian climatology in CMIP5/CMIP6 models: Persistent biases and improvements. Rev. Fac. Ing. Univ. Antioq. 2021, 100, 75–96. [Google Scholar] [CrossRef]
- Crandall, K.A.; Tovar, C.; Arnillas, C.A.; Cuesta, F.; Buytaert, W. Diverging Responses of Tropical Andean Biomes under Future Climate Conditions. PLoS ONE 2013, 8, e63634. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Low, B.W.; Yeo, D.C.J. Novel methods to select environmental variables in MaxEnt: A case study using invasive crayfish. Ecol. Model. 2016, 341, 5–13. [Google Scholar] [CrossRef]
- Feng, X.; Park, D.S.; Liang, Y.; Pandey, R.; Papes, M. Collinearity in ecological niche modeling: Confusions and challenges. Ecol. Evol. 2019, 9, 10365–10376. [Google Scholar] [CrossRef] [PubMed]
- Syfert, M.M.; Smith, M.J.; Coomes, D.A. The effects of sampling bias and model complexity on the predictive performance of MaxEnt species distribution models. PLoS ONE 2013, 8, e55158. [Google Scholar] [CrossRef]
- Cobos, M.E.; Peterson, A.T.; Osorio-Olvera, L.; Jiménez-García, D. An exhaustive analysis of heuristic methods for variable selection in ecological niche modeling and species distribution modeling. Ecol. Inform. 2019, 53, 100983. [Google Scholar] [CrossRef]
- Dormann, C.F.; Elith, J.; Bacher, S.; Buchmann, C.; Carl, G.; Carré, G.; Marquéz, J.R.G.; Gruber, B.; Lafourcade, B.; Leitão, P.J.; et al. Collinearity: A review of methods to deal with it and a simulation study evaluating their performance. Ecography 2013, 36, 27–46. [Google Scholar] [CrossRef]
- Chatterjee, S.; Hadi, A.S. Regression Analysis by Example; Wiley: Hoboken, NJ, USA, 2006. [Google Scholar]
- Naimi, B.; Hamm, N.A.S.; Groen, T.A.; Skidmore, A.K.; Toxopeus, A.G. Where is positional uncertainty a problem for species distribution modelling? Ecography 2014, 37, 191–203. [Google Scholar] [CrossRef]
- Phillips, S.J.; Anderson, R.P.; Dudík, M.; Schapire, R.E.; Blair, M.E. Opening the black box: An open-source release of Maxent. Ecography 2017, 40, 887–893. [Google Scholar] [CrossRef]
- Phillips, S.; Dudík, M.; Schapire, R. A maximum entropy approach to species distribution modeling. In Proceedings of the Twenty-First International Conference on Machine Learning, Banff, AB, Canada, 4–8 July 2004; pp. 655–662. [Google Scholar]
- Kass, J.M.; Muscarella, R.; Galante, P.J.; Bohl, C.L.; Pinilla-Buitrago, G.E.; Boria, R.A.; Soley-Guardia, M.; Anderson, R.P. ENMeval 2.0: Redesigned for customizable and reproducible modeling of species’ niches and distributions. Methods Ecol. Evol. 2021, 12, 1602–1608. [Google Scholar] [CrossRef]
- Buffum, B.; McGreevy, T.J., Jr.; Gottfried, A.E.; Sullivan, M.E.; Husband, T.P. An Analysis of Overstory Tree Canopy Cover in Sites Occupied by Native and Introduced Cottontails in the Northeastern United States with Recommendations for Habitat Management for New England Cottontail. PLoS ONE 2015, 10, e0135067. [Google Scholar] [CrossRef] [PubMed]
- Lobo, J.M.; Jiménez-Valverde, A.; Real, R. AUC: A misleading measure of the performance of predictive distribution models. Glob. Ecol. Biogeogr. 2008, 17, 145–151. [Google Scholar] [CrossRef]
- West, A.M.; Kumar, S.; Brown, C.S.; Stohlgren, T.J.; Bromberg, J. Field validation of an invasive species Maxent model. Ecol. Inform. 2016, 36, 126–134. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy Function Approximation: A Gradient Boosting Machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Cox, D.R. The regression analysis of binary sequences. J. R. Stat. Soc. Ser. B 1958, 20, 215–232. [Google Scholar] [CrossRef]
- Webb, G.I. Naïve Bayes. In Encyclopedia of Machine Learning; Sammut, C., Webb, G.I., Eds.; Springer: Boston, MA, USA, 2011. [Google Scholar]
- Liu, Z. Linear Discriminant Analysis. In Encyclopedia of Systems Biology; Dubitzky, W., Wolkenhauer, O., Cho, K., Yokota, H., Eds.; Springer: New York, NY, USA, 2013. [Google Scholar]
- Fix, E.; Hodges, J.L. Discriminatory Analysis, Nonparametric Discrimination: Consistency Properties. Technol. Rep. 1951, 4, 238–247. [Google Scholar] [CrossRef]
- Cover, T.M.; Hart, P.E. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Kuhn, M. Building predictive models in R using the caret package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Spiers, J.A.; Oatham, M.P.; Rostant, L.V.; Farrell, A.D. Applying species distribution modelling to improving conservation based decisions: A gap analysis of Trinidad and Tobago’s endemic vascular plants. Biodivers. Conserv. 2018, 27, 2931–2949. [Google Scholar] [CrossRef]
- Liu, C.; Berry, P.M.; Dawson, T.P.; Pearso, R.G. Selecting thresholds of occurrence in the prediction of species distributions. Ecography 2005, 28, 385–393. [Google Scholar] [CrossRef]
- Hijmans, R.; Van Etten, J.; Cheng, J.; Mattiuzzi, M.; Sumner, M.; Greenberg, J.; Hijmans, M. Package ‘raster’. R Package 2015, 734, 473. [Google Scholar]
- Wickham, H.; Chang, W.; Wickham, M.H. Package ‘ggplot2’—Create elegant data visualisations using the grammar of graphics. R Package 2016, 2, 1–189. [Google Scholar]
- Duchicela, S.A.; Cuesta, F.; Pinto, E.; Gosling, W.D.; Young, K.R. Indicators for assessing tropical alpine rehabilitation practices. Ecosphere 2019, 10, e02595. [Google Scholar] [CrossRef]
- Leon-Garcia, I.V.; Lasso, E. High heat tolerance in plants from the Andean highlands: Implications for paramos in a warmer world. PLoS ONE 2019, 14, e0224218. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.C.R.; Sun, G.; Zhu-Barker, X.; Liang, Q.; Wu, N.; Horwath, W.R. Tree growth acceleration and expansion of alpine forests: The synergistic effect of atmospheric and edaphic change. Sci. Adv. 2016, 2, e1501302. [Google Scholar] [CrossRef] [PubMed]
- Lasso, E.; Matheus-Arbeláez, P.; Gallery, R.E.; Garzón-López, C.; Cruz, M.; Leon-Garcia, I.V.; Aragón, L.; Ayarza-Páez, A.; Curiel Yuste, J. Homeostatic Response to Three Years of Experimental Warming Suggests High Intrinsic Natural Resistance in the Páramos to Warming in the Short Term. Front. Ecol. Evol. 2021, 9, 615006. [Google Scholar] [CrossRef]
- Zhu, Z.; Piao, S.; Myneni, R.B.; Huang, M.; Zeng, Z.; Canadell, J.G.; Ciais, P.; Sitch, S.; Friedlingstein, P.; Arneth, A.; et al. Greening of the Earth and its drivers. Nat. Clim. Chang. 2016, 6, 791–795. [Google Scholar] [CrossRef]
- Correa-Díaz, A.; Romero-Sánchez, M.E.; Villanueva-Díaz, J. The greening effect characterized by the Normalized Difference Vegetation Index was not coupled with phenological trends and tree growth rates in eight protected mountains of central Mexico. For. Ecol. Manag. 2021, 496, 119402. [Google Scholar] [CrossRef]
- Scherrer, D.; Schmid, S.; Körner, C. Elevational species shifts in a warmer climate are overestimated when based on weather station data. Int. J. Biometeorol. 2010, 55, 645–654. [Google Scholar] [CrossRef]
- Körner, C.; Hiltbrunner, E. Why Is the Alpine Flora Comparatively Robust against Climatic Warming? Diversity 2021, 13, 383. [Google Scholar] [CrossRef]
- Chacón-Moreno, E.; Rodríguez-Morales, M.; Paredes, D.; Suárez del Moral, P.; Albarrán, A. Impacts of Global Change on the Spatial Dynamics of Treeline in Venezuelan Andes. Front. Ecol. Evol. 2021, 9, 615223. [Google Scholar] [CrossRef]
- Gottfried, M.; Pauli, H.; Futschik, A.; Akhalkatsi, M.; Barančok, P.; Benito Alonso, J.L.; Coldea, G.; Dick, J.; Erschbamer, B.; Fernández Calzado, M.a.R.; et al. Continent-wide response of mountain vegetation to climate change. Nat. Clim. Chang. 2012, 2, 111–115. [Google Scholar] [CrossRef]
- Weigend, M. Observations on the Biogeography of the Amotape-Huancabamba Zone in Northern Peru. Bot. Rev. 2002, 68, 38–54. [Google Scholar] [CrossRef]
- Cochrane, A.; Nicotra, A.; Ooi, M. Climate change: Alters plant recruitment from seed. Austral Ecol. 2019, 44, 931–934. [Google Scholar] [CrossRef]
- Harsch, M.A.; Bader, M.Y. Treeline form—A potential key to understanding treeline dynamics. Glob. Ecol. Biogeogr. 2011, 20, 582–596. [Google Scholar] [CrossRef]
- Rieseberg, L.H. Major Ecological Transitions in Wild Sunflowers Facilitated by Hybridization. Science 2003, 301, 1211–1216. [Google Scholar] [CrossRef]
- Little, C.J.; Wheeler, J.A.; Sedlacek, J.; Cortés, A.J.; Rixen, C. Small-scale drivers: The importance of nutrient availability and snowmelt timing on performance of the alpine shrub Salix herbacea. Oecologia 2016, 180, 1015–1024. [Google Scholar] [CrossRef]
- Sedlacek, J.; Bossdorf, O.; Cortés, A.J.; Wheeler, J.A.; van-Kleunen, M. What role do plant-soil interactions play in the habitat suitability and potential range expansion of the alpine dwarf shrub Salix herbacea? Basic. Appl. Ecol. 2014, 15, 305–315. [Google Scholar] [CrossRef]
- Cuesta, F.; Muriel, P.; Llambí, L.D.; Halloy, S.; Aguirre, N.; Beck, S.; Carilla, J.; Meneses, R.I.; Cuello, S.; Grau, A.; et al. Latitudinal and altitudinal patterns of plant community diversity on mountain summits across the tropical Andes. Ecography 2017, 40, 1381–1394. [Google Scholar] [CrossRef]
- Arnold, P.A.; Kruuk, L.E.B.; Nicotra, A.B. How to analyse plant phenotypic plasticity in response to a changing climate. New Phytol. 2019, 222, 1235–1241. [Google Scholar] [CrossRef] [PubMed]
- Velazco, S.J.E.; Galvão, F.; Villalobos, F.; De Marco Júnior, P. Using worldwide edaphic data to model plant species niches: An assessment at a continental extent. PLoS ONE 2017, 12, e0186025. [Google Scholar] [CrossRef] [PubMed]
- Llambí, L.D.; Rada, F. Ecological research in the tropical alpine ecosystems of the Venezuelan páramo: Past, present and future. Plant Ecol. Divers. 2019, 12, 519–538. [Google Scholar] [CrossRef]
- Cáceres, Y.; Llambí, L.D.; Rada, F. Shrubs as foundation species in a high tropical alpine ecosystem: A multi-scale analysis of plant spatial interactions. Plant Ecol. Divers. 2014, 8, 147–161. [Google Scholar] [CrossRef]
- Cuesta, F.; Llambí, L.D.; Huggel, C.; Drenkhan, F.; Gosling, W.D.; Muriel, P.; Jaramillo, R.; Tovar, C. New land in the Neotropics: A review of biotic community, ecosystem, and landscape transformations in the face of climate and glacier change. Reg. Environ. Chang. 2019, 19, 1623–1642. [Google Scholar] [CrossRef]
- Wheeler, J.A.; Schnider, F.; Sedlacek, J.; Cortés, A.J.; Wipf, S.; Hoch, G.; Rixen, C. With a little help from my friends: Community facilitation increases performance in the dwarf shrub Salix herbacea. Basic. Appl. Ecol. 2015, 16, 202–209. [Google Scholar] [CrossRef]
- Llambí, L.D.; Hupp, N.; Saez, A.; Callaway, R. Reciprocal interactions between a facilitator, natives, and exotics in tropical alpine plant communities. Perspect. Plant Ecol. Evol. Syst. 2018, 30, 82–88. [Google Scholar] [CrossRef]
- Seehausen, O. Hybridization and adaptive radiation. Trends Ecol. Evol. 2004, 19, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Payseur, B.A.; Rieseberg, L.H. A genomic perspective on hybridization and speciation. Mol. Ecol. 2016, 25, 2337–2360. [Google Scholar] [CrossRef]
- Pineda, Y.M.; Cortés, A.J.; Madriñán, S.; Jiménez, I. The Nature of Espeletia Species. bioRxiv 2020, 9, 318865. [Google Scholar] [CrossRef]
- Pérez-Escobar, O.A.; Cámara-Leret, R.; Antonelli, A.; Bateman, R.; Bellot, S.; Chomicki, G.; Cleef, A.; Diazgranados, M.; Dodsworth, S.; Jaramillo, C.; et al. Mining threatens Colombian ecosystems. Science 2018, 359, 1475. [Google Scholar] [CrossRef]
- Vásquez, D.L.A.; Balslev, H.; Sklenář, P. Human impact on tropical-alpine plant diversity in the northern Andes. Biodivers. Conserv. 2015, 24, 2673–2683. [Google Scholar] [CrossRef]
- Baptiste, B.; Pinedo-Vasquez, M.; Gutierrez-Velez, V.H.; Andrade, G.I.; Vieira, P.; Estupiñán-Suárez, L.M.; Londoño, M.C.; Laurance, W.; Lee, T.M. Greening peace in Colombia. Nat. Ecol. Evol. 2017, 1, 102. [Google Scholar] [CrossRef] [PubMed]
- Cortés, A.J.; Wheeler, J.A. The Environmental Heterogeneity of Mountains at a Fine Scale in a Changing World. In Mountains, Climate, and Biodiversity; Hoorn, C., Perrigo, A., Antonelli, A., Eds.; Wiley: Hoboken, NJ, USA, 2018. [Google Scholar]
- Toivonen, J.M.; Horna, V.; Kessler, M.; Ruokolainen, K.; Hertel, D. Interspecific variation in functional traits in relation to species climatic niche optima in Andean Polylepis (Rosaceae) tree species: Evidence for climatic adaptations. Funct. Plant Biol. 2014, 41, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Barghi, N.; Hermisson, J.; Schlötterer, C. Polygenic adaptation: A unifying framework to understand positive selection. Nat. Rev. Genet. 2020, 21, 769–781. [Google Scholar] [CrossRef]
- Cortés, A.J.; López-Hernández, F.; Blair, M.W. Genome–Environment Associations, an Innovative Tool for Studying Heritable Evolutionary Adaptation in Orphan Crops and Wild Relatives. Front. Genet. 2022, 13, 910386. [Google Scholar] [CrossRef] [PubMed]
- Aitken, S.N.; Bemmels, J.B. Time to get moving: Assisted gene flow of forest trees. Evol. Appl. 2016, 9, 271–290. [Google Scholar] [CrossRef]
- Aitken, S.N.; Whitlock, M.C. Assisted Gene Flow to Facilitate Local Adaptation to Climate Change. Annu. Rev. Ecol. Evol. Syst. 2013, 44, 367–388. [Google Scholar] [CrossRef]
- Breed, M.F.; Harrison, P.A.; Blyth, C.; Byrne, M.; Gaget, V.; Gellie, N.J.C.; Groom, S.V.C.; Hodgson, R.; Mills, J.G.; Prowse, T.A.A.; et al. The potential of genomics for restoring ecosystems and biodiversity. Nat. Rev. Genet. 2019, 20, 615–628. [Google Scholar] [CrossRef]
- Arenas, S.; Cortés, A.J.; Mastretta-Yanes, A.; Jaramillo-Correa, J.P. Evaluating the accuracy of genomic prediction for the management and conservation of relictual natural tree populations. Tree Genet. Genomes 2021, 17, 12. [Google Scholar] [CrossRef]
- Cortés, A.J.; López-Hernández, F.; Osorio-Rodriguez, D. Predicting thermal adaptation by looking into populations’ genomic past. Front. Genet. 2020, 11, 564515. [Google Scholar] [CrossRef] [PubMed]
- Walters, S.J.; Robinson, T.P.; Byrne, M.; Nevill, P. Seed sourcing in the genomics era: Multispecies provenance delineation for current and future climates. Restor. Ecol. 2022, 30, e13718. [Google Scholar] [CrossRef]
- Parker, K.A.; Seabrook-Davison, M.; Ewen, J.G. Opportunities for Nonnative Ecological Replacements in Ecosystem Restoration. Restor. Ecol. 2008, 18, 269–273. [Google Scholar] [CrossRef]
- Ramirez-Villegas, J.; Cuesta, F.; Devenish, C.; Peralvo, M.; Jarvis, A.; Arnillas, C.A. Using species distributions models for designing conservation strategies of Tropical Andean biodiversity under climate change. J. Nat. Conserv. 2014, 22, 391–404. [Google Scholar] [CrossRef]
- Leroy, T.; Louvet, J.M.; Lalanne, C.; Le Provost, G.; Labadie, K.; Aury, J.M.; Delzon, S.; Plomion, C.; Kremer, A. Adaptive introgression as a driver of local adaptation to climate in European white oaks. New Phytol. 2020, 226, 1171–1182. [Google Scholar] [CrossRef] [PubMed]
- Zellweger, F.; De Frenne, P.; Lenoir, J.; Vangansbeke, P.; Verheyen, K.; Bernhardt-Römermann, M.; Baeten, L.; Hédl, R.; Berki, I.; Brunet, J.; et al. Forest microclimate dynamics drive plant responses to warming. Science 2020, 368, 772–775. [Google Scholar] [CrossRef] [PubMed]
- Peyre, G.; Lopez, C.; Diaz, M.D.; Lenoir, J. Climatic refugia in the coldest neotropical hotspot, the Andean páramo. bioRxiv 2022, 11, 517325. [Google Scholar] [CrossRef]
- Gentili, R.; Baroni, C.; Panigada, C.; Rossini, M.; Tagliabue, G.; Armiraglio, S.; Citterio, S.; Carton, A.; Salvatore, M.C. Glacier shrinkage and slope processes create habitat at high elevation and microrefugia across treeline for alpine plants during warm stages. Catena 2020, 193, 104626. [Google Scholar] [CrossRef]
- Petrík, P.; Grote, R.; Gömöry, D.; Kurjak, D.; Petek-Petrik, A.; Lamarque, L.J.; Sliacka Konôpková, A.; Mukarram, M.; Debta, H.; Fleischer, P. The Role of Provenance for the Projected Growth of Juvenile European Beech under Climate Change. Forests 2022, 14, 26. [Google Scholar] [CrossRef]
- Sinclair, S.J.; White, M.D.; Newell, G.R. How useful are species distribution models for managing biodiversity under future climates? Ecol. Soc. 2010, 15, 8. [Google Scholar] [CrossRef]
- Tovar, C.; Melcher, I.; Kusumoto, B.; Cuesta, F.; Cleef, A.; Meneses, R.I.; Halloy, S.; Llambí, L.D.; Beck, S.; Muriel, P.; et al. Plant dispersal strategies of high tropical alpine communities across the Andes. J. Ecol. 2020, 108, 1910–1922. [Google Scholar] [CrossRef]
- Peyre, G. What Does the Future Hold for Páramo Plants? A Modelling Approach. Front. Ecol. Evol. 2022, 10, 896387. [Google Scholar] [CrossRef]
- Warren, D.L.; Seifert, S.N. Ecological niche modeling in Maxent: The importance of model complexity and the performance of model selection criteria. Ecol. Appl. 2011, 21, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.M.; Chalmandrier, L.; Lenoir, J.; Burgess, T.I.; Essl, F.; Haider, S.; Kueffer, C.; McDougall, K.; Milbau, A.; Nuñez, M.A.; et al. Lags in the response of mountain plant communities to climate change. Glob. Chang. Biol. 2017, 24, 563–579. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, A.; Meneses, R.I.; Rabatel, A.; Soruco, A.; Dangles, O.; Anthelme, F. Time lag between glacial retreat and upward migration alters tropical alpine communities. Perspect. Plant Ecol. Evol. Syst. 2018, 30, 89–102. [Google Scholar] [CrossRef]
- Yackulc, C.B.; Nichols, J.D.; Reid, J.; Der, R. To predict the niche, model colonization and extinction. Ecology 2015, 96, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Buisson, L.; Thuiller, W.; Casajus, N.; Lek, S.; Grenouillet, G. Uncertainty in ensemble forecasting of species distribution. Glob. Chang. Biol. 2010, 16, 1145–1157. [Google Scholar] [CrossRef]
- Thornthwaite, C.W.; Mather, J.R. The Water Balance. Climatology 1955, 8, 1–104. [Google Scholar]
- Thornthwaite, C.W.; Mather, J.R. Instructions and Tables for Computing Potential Evapotranspiration and the Water Balance. Climatology 1957, 10, 185–311. [Google Scholar]
- Zellweger, F.; De Frenne, P.; Lenoir, J.; Rocchini, D.; Coomes, D. Advances in Microclimate Ecology Arising from Remote Sensing. Trends Ecol. Evol. 2019, 34, 327–341. [Google Scholar] [CrossRef]
- Guevara-Escudero, M.; Osorio, A.N.; Cortés, A.J. Integrative Pre-Breeding for Biotic Resistance in Forest Trees. Plants 2021, 10, 2022. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Xiao, P.; Jiang, Y.; Dong, M.; Chen, Z.; Li, H.; Hu, Z.; Lei, A.; Wang, J. Transgenerational Epigenetic Inheritance under Environmental Stress by Genome-Wide DNA Methylation Profiling in Cyanobacterium. Front. Microbiol. 2018, 9, 1479. [Google Scholar] [CrossRef] [PubMed]
- Boskovic, A.; Rando, O.J. Transgenerational Epigenetic Inheritance. Annu. Rev. Genet. 2018, 52, 21–41. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bedoya-Canas, L.E.; López-Hernández, F.; Cortés, A.J. Climate Change Responses of High-Elevation Polylepis Forests. Forests 2024, 15, 811. https://doi.org/10.3390/f15050811
Bedoya-Canas LE, López-Hernández F, Cortés AJ. Climate Change Responses of High-Elevation Polylepis Forests. Forests. 2024; 15(5):811. https://doi.org/10.3390/f15050811
Chicago/Turabian StyleBedoya-Canas, Larry E., Felipe López-Hernández, and Andrés J. Cortés. 2024. "Climate Change Responses of High-Elevation Polylepis Forests" Forests 15, no. 5: 811. https://doi.org/10.3390/f15050811
APA StyleBedoya-Canas, L. E., López-Hernández, F., & Cortés, A. J. (2024). Climate Change Responses of High-Elevation Polylepis Forests. Forests, 15(5), 811. https://doi.org/10.3390/f15050811








